Abstract
The Kemp elimination is prototypical reaction used to study proton abstraction from carbon. Several hydrophobic systems are known to accelerate this reaction, including two classes of computationally-designed enzymes. However, it is unclear whether these computationally-designed enzymes establish specific interactions with their substrates, as natural enzymes do, or if most of the rate acceleration is due to the hydrophobicity of the substrate. We used a simple system composed of cationic micelles and a long chain base (such as lauryl phosphate or lauric acid) to measure the rate acceleration for the Kemp elimination. Remarkably, we found that this simple system can accelerate the reaction by 4 orders of magnitude, approaching the rates of more complex designed systems. Use of different substrates suggests that the reaction takes place at the interface between the micellar head and water (the Stern layer) with the long-chain base embedded in the micelle and the substrate in the aqueous solution. Thus, we suggest that significant rate accelerations can be achieved regardless of the precise positioning of substrates. Because natural enzymes use specific interactions to position their substrates, we propose that acceleration of the Kemp elimination is not a suitable benchmark for the success of the design process, and we suggest that more complex reactions should be used.
Keywords: Kemp elimination, micelles, enzyme design, catalysis, hydrophobic interactions
INTRODUCTION
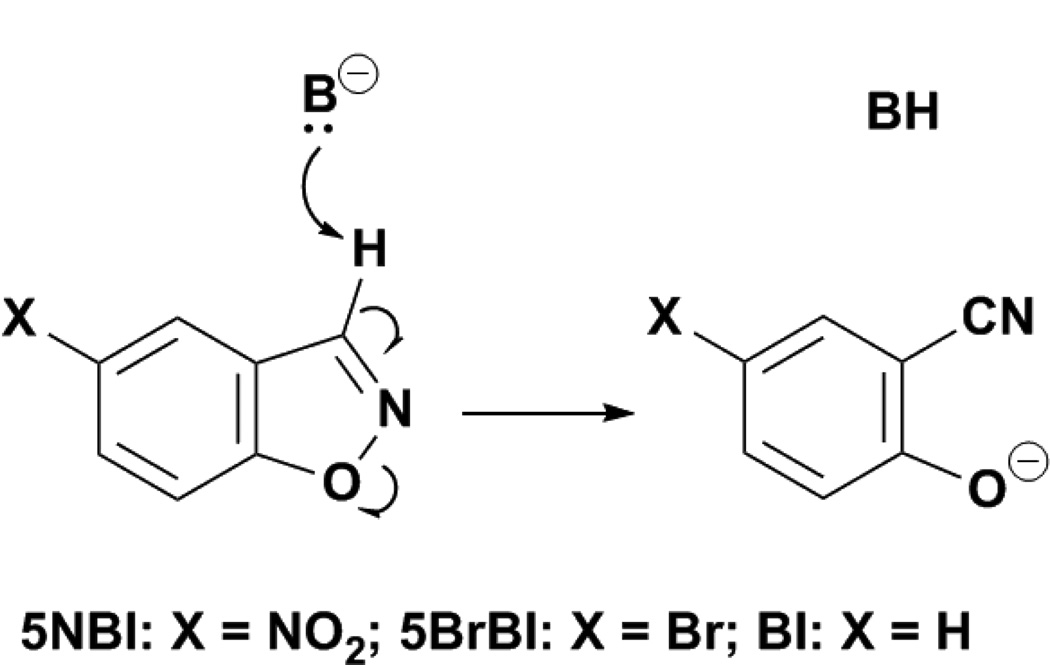
The Kemp elimination is a well-studied reaction in which a catalytic base abstracts a proton from a benzisoxazole ring. This abstraction results in ring-opening, forming the cyanophenol product (FIGURE 1).[1–3] In contrast to many other proton transfer reactions, the negative charge that forms at the transition state is delocalized throughout the aromatic system, making the Kemp elimination a relatively easy decarboxylation to catalyse. Indeed, many systems have been used to accelerate the Kemp elimination: carboxylic acids in aprotic solvents,[2,4] vesicles and micelles,[5] cyclodextrines,[6] synzymes,[2,7] cavitands,[8] bovine serum albumin,[9,10] catalytic antibodies,[11] minimally-designed peptides,[12] and computationally-designed enzymes[13,14] have been shown to possess significant Kemp eliminase activity.
Fig. 1.
Schematic representation of the Kemp elimination.
Although this reaction is catalysed by a wide array of systems, direct comparisons between the catalytic parameters derived from different systems are not always meaningful, because different bases accelerate this reaction with significantly different rate constants. For example, the aqueous reaction of 5-nitrobenzisoxazole (5NBI) proceeds with second-order rate constants (k2) of 6 × 10−5 M−1s−1, 1 × 10−1 M−1s−1, and 15 M−1s−1 with respect to acetate,[15] primary amines,[2] and hydroxide.[3] Organic solvents such as acetonitrile and DMSO can accelerate this reaction by several orders of magnitude when the catalytic base is a (negatively) charged species such as acetate.[2,4]
The micelle-assisted Kemp elimination was studied in the past with respect to hydroxide.[5] Unsurprisingly, the effect of micelles was found to be minimal (~5-fold relative to the aqueous reaction), as the highly hydrophilic hydroxyl ion is not predicted to significantly interact with micelles. In this work, we wanted to test whether more hydrophobic catalysts, such as the ones with a long-chain carbon tail that might intercalate within the micelle, can provide substantial rate acceleration to the micelle-assisted Kemp elimination.
EXPERIMENTAL
Materials
5-Nitrobenzisoxazole and 5-bromobenzisoxazole were purchased from Ark Pharm. 5-bromobenzisoxazole was recrystallized from methanol. Benzisoxazole and hydroxylamine hydrochloride were purchased from Alfa Aesar. 2-Hydroxy-1-benzaldehyde, triphenylphosphine, and DDQ were from Sigma-Aldrich. CTAC, sodium laurate, and dodecyl phosphate were purchased from TCI. Dodecyl phosphate was recrystallized twice from isoctanol. Naphthisoxazole was prepared in two steps from 2-hydroxy-1-benzaldehyde as described below, using slightly modified literature procedures.[16,17]
Synthesis of 2-hydroxynaphthalene-1-carbaldehyde oxime (1)
2-Hydroxy-1-benzaldehyde (2.5g, 14.5 mmol) was dissolved in 7 ml of ethanol and mixed with 7 ml of ethanolic NH2OH·HCl (2.9 g, 33 mmol) and 7 ml of aqueous sodium acetate (4.1 g, 50 mmol). The solution was stirred at room temperature and monitored by TLC (70:30 hexanes/ethyl acetate). After 3 hours the solution was put in fridge and left overnight. The green precipitate was washed with cold ethanol and hot water, then dried under vacuum and used without further purification. Yield in 1: 1.3 g (7.0 mmol, 48%). The 1H NMR spectra in CDCl3was consistent with the one reported in the literature,[16] with peaks at 10.80 ppm (s, 1H), 9.15 ppm (s, 1H), 7.97 ppm (d, 1H), 7.79 ppm (d, 1H), 7.56 ppm (t, 1H), 7.37 ppm (t, 1H), 7.26 ppm (s, 1H), and 7.21 ppm (d, 1H).
Synthesis of naphtho[1,2-d]isoxazole
To a solution of 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ, 341 mg, 1.5 mmol) and triphenylphosphine (Ph3P, 394 mg, 1.5 mmol) in CH2Cl2 (5 mL) was added 1 (187 mg, 1.0 mmol) at room temperature. The resulting solution was stirred for an additional 10 minutes. Solvent was removed via rotary evaporation, and the crude reaction mixture was purified using SiO2 column chromatography with 50% hexane / 50% CH2Cl2 as the eluent. Solvents were again removed via rotary evaporation, and the product was isolated as a white solid in 59% yield. 1H NMR δ (CDCl3): 9.12 ppm (s, 1H), 8.17 ppm (d, 1H), 8.00 ppm (d, 1H), 7.97 ppm (d, 1H), 7.74 ppm (d, 1H), 7.70 ppm (t, 1H), 7.58 ppm (t, 1H).
General kinetic methods
Reactions were run in 25 mM buffer (MES, MOPS, HEPES, or phosphate) at 20 °C. We used plastic disposable UV microcuvettes (from Brand) for all trials. Typically, 25 µL of an aqueous buffer solution were mixed with 12.0 µL of a 47 mM aqueous solution of CTAC, a variable amount of catalyst (0–6 µL of a stock solution in methanol), and water to reach 500 µL. Reactions were started by addition of the substrate (1–6 µL of stock solutions in methanol) and monitored using a Cary 50 UV spectrophotometer at the following wavelengths: 380 nM for 5-nitrobenzisoxazole (ε = 18,400 M−1cm−1); 349 nm for 5-bromobenzisoxazole (ε = 8,500 M−1cm−1); 323 nm for benzisoxazole (ε = 5,080 M−1cm−1); 370 nm for naphthisoxazole (ε = 10,000 M−1cm−1). Addition of methanol without catalyst did not accelerate the reactions.
RESULTS AND DISCUSSION
Because the catalytic base should bear at least a partial negative charge to be active, we decided to use the positively-charged cetyltrimethylammonium chloride (CTAC) as the micellar agent to ensure an attractive interaction with the hydrophilic heads of the long-chain base. The literature value for the critical micelle concentration (c.m.c.) of CTAC is 1.35 mM,[18] but we found a maximum activity at [CTAC] = 1.1 mM. It is possible that small experimental errors in our stock solution account for this discrepancy; however, micelles were clearly formed because of the significant rate acceleration provided by our system (see below).
In the absence of added base, Kemp elimination of 5NBI proceeded with a second-order rate constant (k2) in hydroxide of 290 M−1s−1. This value is slightly larger than the reported value of 80 M−1s−1,[5] possibly because of the differences in the experimental conditions (previous results were obtained around pH 11, while our results were obtained over a wider range of pH values). The measured k2 for the Kemp elimination in the absence of micelles was found to be 15 M−1s−1, in very good agreement with the previously reported value of 14.7 M−1s−1.[5] Thus, the micellar environment provides 5–20 fold rate acceleration to the Kemp elimination when hydroxide is used as the base. Because polar aprotic solvents significantly accelerate the Kemp elimination, this small effect is consistent with the reaction occurring in a slightly more hydrophobic environment than water, such as the one present at the micelle/water interface.[5]
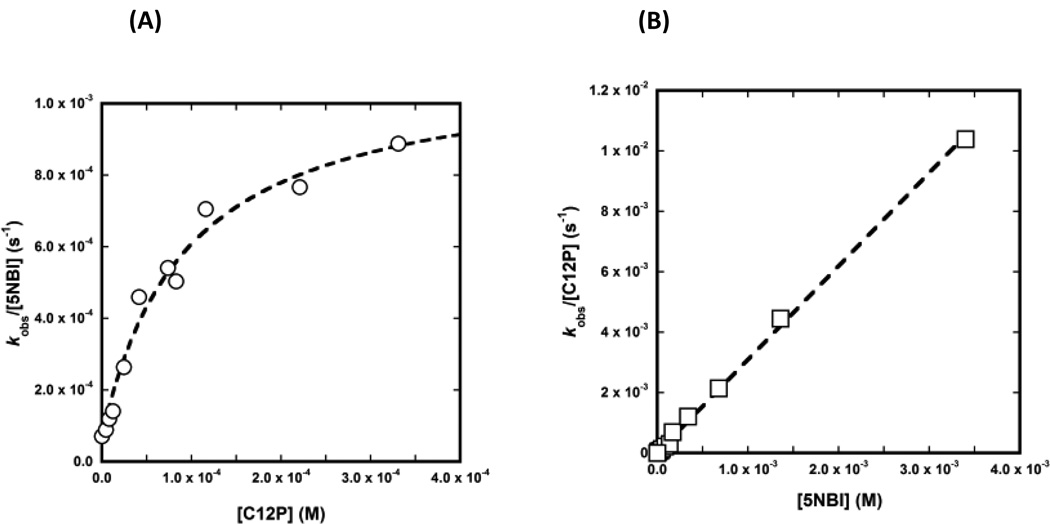
When we added dodecyl phosphate (C12P) to the reaction medium (at pH 6.6), we measured an increase in the velocity of the Kemp elimination of 5NBI with an apparent dissociation constant of about 93 µM and a maximum first-order rate constant of 1.0 × 10−3 s−1 (Figure 2A). Because C12P is significantly hydrophobic (the calculated octanol/water partition coefficient for the neutral species of C12P is ~ 104),[19] we expected this compound to be present embedded within the micelle rather than in bulk water. Considering an aggregation number of 88,[18] the concentration of CTAC micelles when [CTAC] = 1.1 mM is estimated to be 12.5 µM. Thus, our results suggest that about 7 molecules of C12P associate with a CTAC micelle to achieve half of the maximum velocity. In contrast, we did not observe any saturation in 5NBI up to a concentration of 3.0 mM (Figure 2B). Thus, it is possible that the linear long-chain of CTAC allows efficient packing and intercalation within the micelle, while the bulkier and less hydrophobic phenyl ring of 5NBI prevents packing and constraints the substrate to the micelle-water interface. From the results above, we calculated the value of k2 for the micellar-assisted encounter of C12P and 5NBI at pH 6.6 to be 5.8 M−1s−1.
Fig. 2.
Effect of changing the concentration of (a) C1P and (b) 5NBI on the rate of the Kemp elimination in the presence of 1.1. mM CTAC.
Importantly, an increase in butyl phosphate concentration over a much larger range did not result in significant differences between the observed velocities in the presence and absence of micelles (data not shown). This result suggests that the hydrophobic interactions between the micelle and the catalyst are important for the rate acceleration, supporting the hypothesis that C12P intercalates between CTAC molecules, as observed with other detergents.[20]
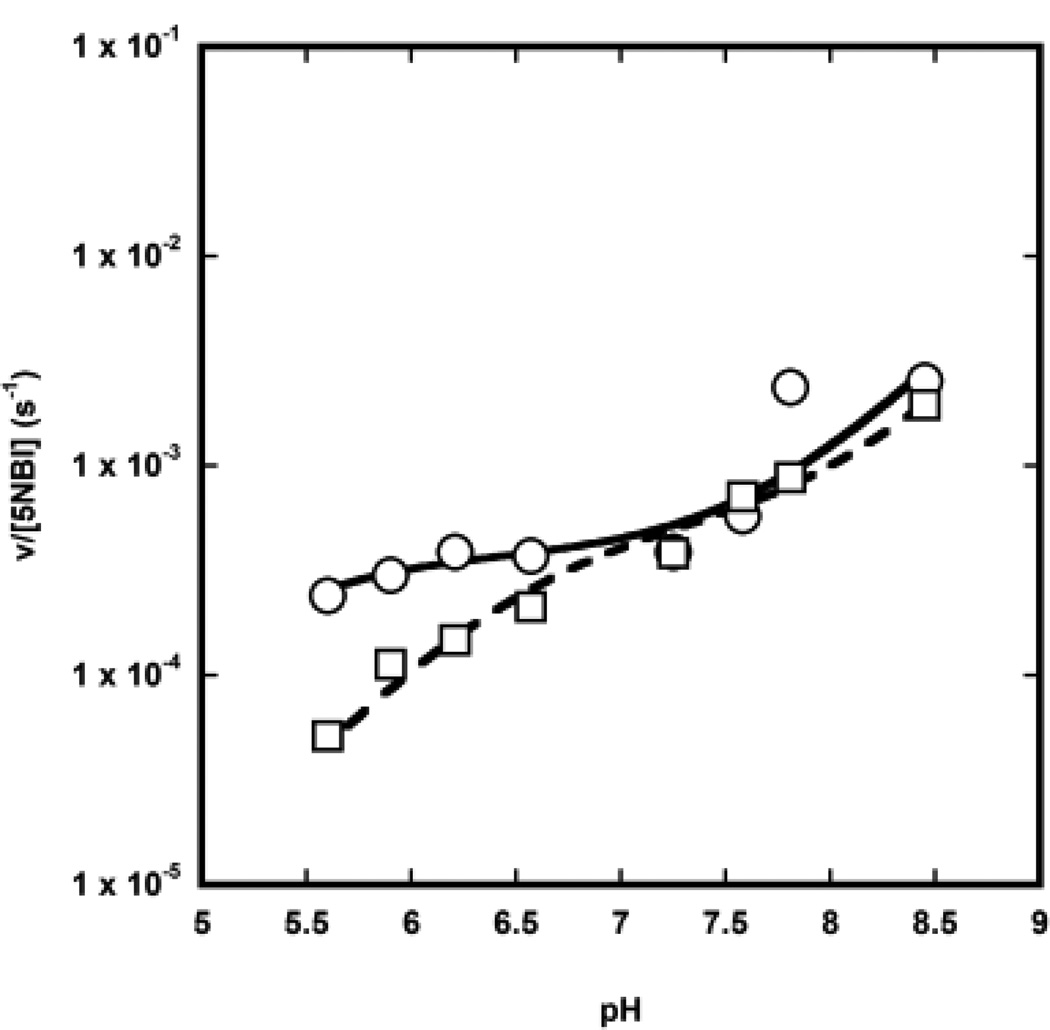
In order to determine whether the monoanionic or the dianionic form of C12P were the reactive species, we varied the pH keeping the base concentration fixed at 42 µM. Because the pKa of dodecyl phosphate is 7.0,[21] in case the dianion was the reactive species we expected the observed velocity to be constant above pH 7.0, as most of the phosphate would be in the productive dianionic state above such pH. On the contrary, if the monoanion were the reactive species we expected the observed velocity to drop above pH 7.0 because the concentration of the monoanion would significantly decrease above the pKa value.
The flat pH-rate profile observed between pH ~ 6.0 and 7.7 (Figure 3) strongly suggests that the monoester dianion is the reactive species. However, the pH-rate profile shown in Figure 3 presents two additional interesting features. First, the apparent pKa value of C12P is below 5.3, significantly different from the reported pKa value for C12P of 7.0 in the absence of micelles. This reduced pKa value suggests that C12P is indeed associated with the micelle, as expected by the hydrophobicity of its tail, so that the phosphoryl group of C12P is in proximity of the positively-charged trimethylammonium group of CTAC. This proximity with the positive charge will make it more likely for the phosphate to lose its protons, thereby reducing the pKa. Second, the observed velocity increases above pH 7.7. This is due to the hydroxide-catalysed reaction that starts to be prevalent above this pH. The pH-rate profile at 4.2 µM C12P can be fit using a kinetic pKa of 6.6, which is closer, but not identical, to the pKa of C12P in solution. This last result suggests even at 4.2 µM the base is partly associated with the micelle.
Fig. 3.
pH-rate profile for the Kemp elimination of 5NBI in the presence of 1.1. mM CTAC and 42 µM (open circles) or 4.2 µM (open squares) C12P.
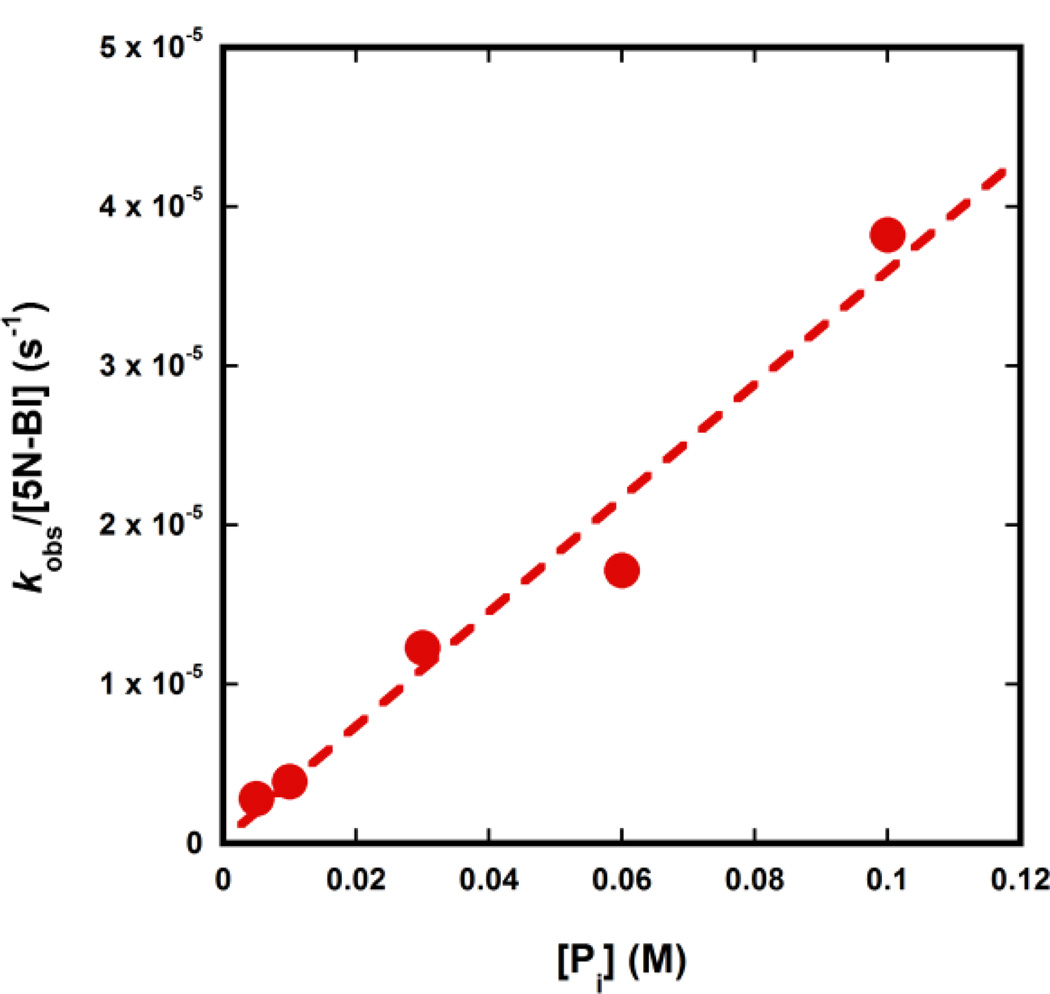
In order to determine the rate acceleration provided by the micellar system, we needed to compare the values of k2 for the C12P-catalysed Kemp elimination in the presence and in the absence of micelles. However, this comparison is problematic for two reasons: first, C12P is poorly soluble in water, and it is not possible to achieve high concentrations of this base. Second, although the c.m.c. of C12P is higher than the concentrations used herein, detergents can form premicelles, which can also accelerate chemical reactions.[22–24] Because of these two potential problems, we decided to compare the measured value of k2 to that of the inorganic phosphate-catalysed Kemp elimination. Reactions of inorganic phosphate did not show pH-dependence above pH 6.8 (data not shown), suggesting that also in this case the dianion was the reactive species. At pH 7.0, which is in the plateau region, we measured a value of k2 for the phosphate-catalysed Kemp elimination in aqueous solution of 3.6 × 10−4 M−1s−1 (Figure 4). Because the value of k2 for the micellar-assisted encounter of C12P and 5NBI at pH 6.6 is 5.8 M−1s−1 (see above), this result implies that the rate acceleration brought about by the cationic micellar system is about 16,000-fold.
Fig. 4.
Effect of inorganic phosphate on the Kemp elimination of 5NBI in the absence of micelles at pH 7.0.
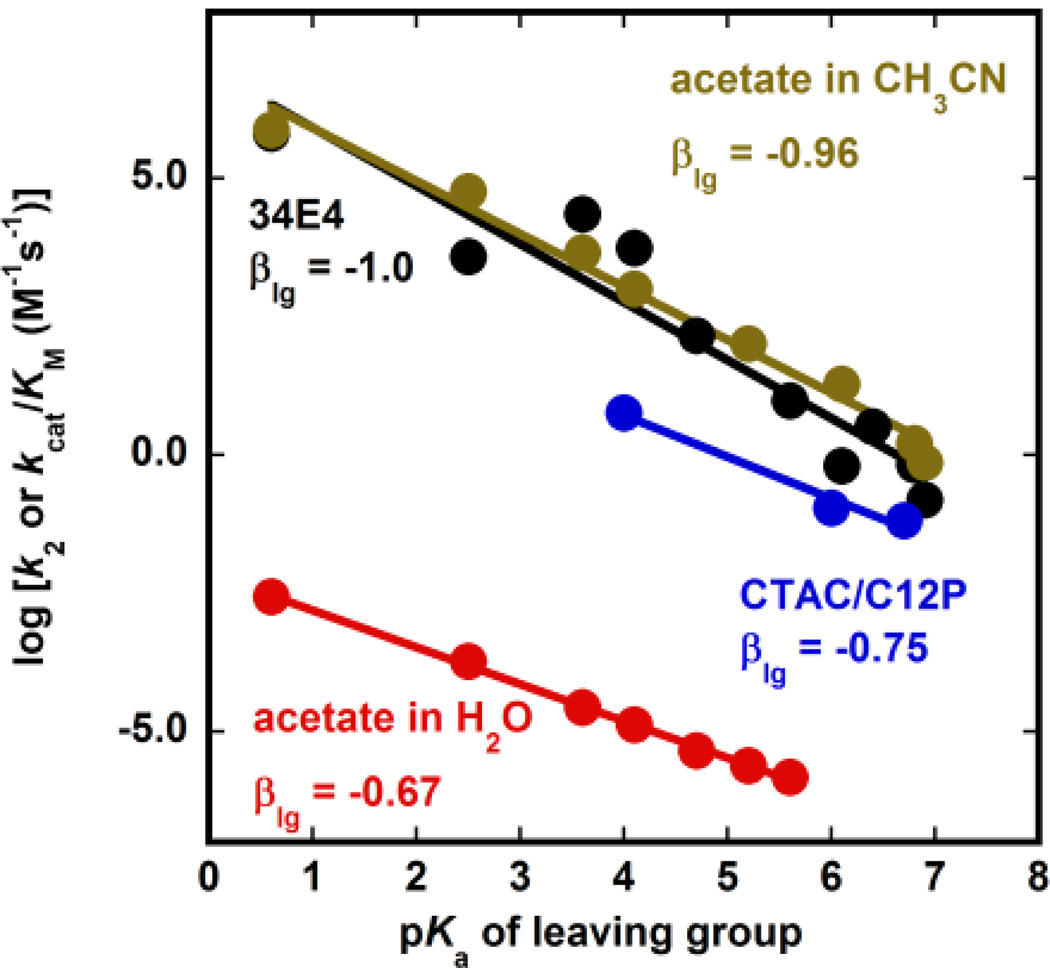
Reactions in micelles usually occur at the interface between water and the hydrophilic head, a region also called the Stern layer. This region has characteristics more similar to water than to the highly hydrophobic interior of the micelle, and the dielectric constant of the Stern layer has been often compared to that of ethanol.[25] To determine whether the Kemp elimination takes place at the Stern layer, we measured the values of k2 for two other substrates, 5-bromobenisoxazole (5BrBI) and benzisoxazole (BI). These two substrates have reduced ability to delocalize the negative charge present in the product-like transition state and thus are expected to react slower than 5NBI, with pKa values of 5.9 and 7.0 for the products of the reactions of 5BrBI and BI, respectively. Brønsted plots that relate the pKa of the cyanophenol product to the observed rate constant of the parent substrate have been determined in several environments, including aqueous solution,[2] acetonitrile,[2] catalytic antibodies,[26] and the computationally-designed Kemp eliminase HG3.17.[15] The slopes of these plots, commonly referred to as βLG, were found to be −0.67, −0.96, −1.0, and −1.4, respectively. When we plotted the observed values of k2 as a function of the pKa of the product in the micelle-assisted Kemp elimination, we found a slope of −0.75, which is very similar to that observed in water (Figure 5). This result suggests that the reaction takes place in an environment with water-like characteristics, such as the Stern layer, and not in the interior of the micelle.
Fig. 5.
Brønsted plot for the Kemp elimination of 5NBI catalysed by acetate in water (red, ref. 2), C12P in CTAC micelles (blue, this work), the catalytic antibody 34E4 (black, ref. 26), and acetate in acetonitrile (gold, ref. 2).
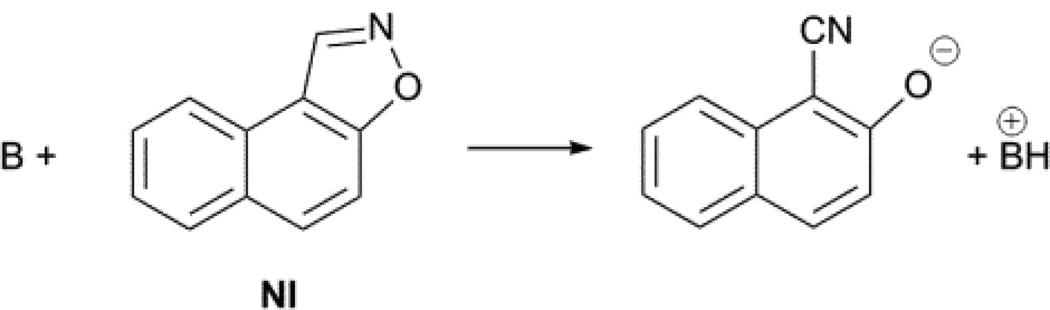
To understand whether the benzisoxazole substrate is also associated with the micelle, we compared the C12P-catalysed Kemp elimination of BI (which contains one phenyl ring) to that of naphthisoxazole (NI, which contains two phenyl rings, Figure 6). The calculated octanol/water partition coefficients are 101.6 and 102.8 for BI and NI, respectively.[19] If the substrate were embedded in the micelle, we would expect a significant increase in the value of k2 for NI, which is significantly more hydrophobic than BI. In contrast to this prediction, we found that the two substrates reacted with very similar k2 values (0.071 and 0.068 M−1s−1 for BI and NI, respectively). This result is in agreement with the lack of saturation in 5NBI reported above, and suggests that the benzisoxazole substrate is not significantly associated with the micelle. Thus, our results are consistent with a model in which the anionic base intercalates within the micelle with the long hydrophobic chain and leaves the negatively-charged head to interact with the positively-charged head of the micelle, with the substrate present in the Stern layer rather than embedded in the micelle.
Fig. 6.
Kemp elimination of naphthisoxazole (NI).
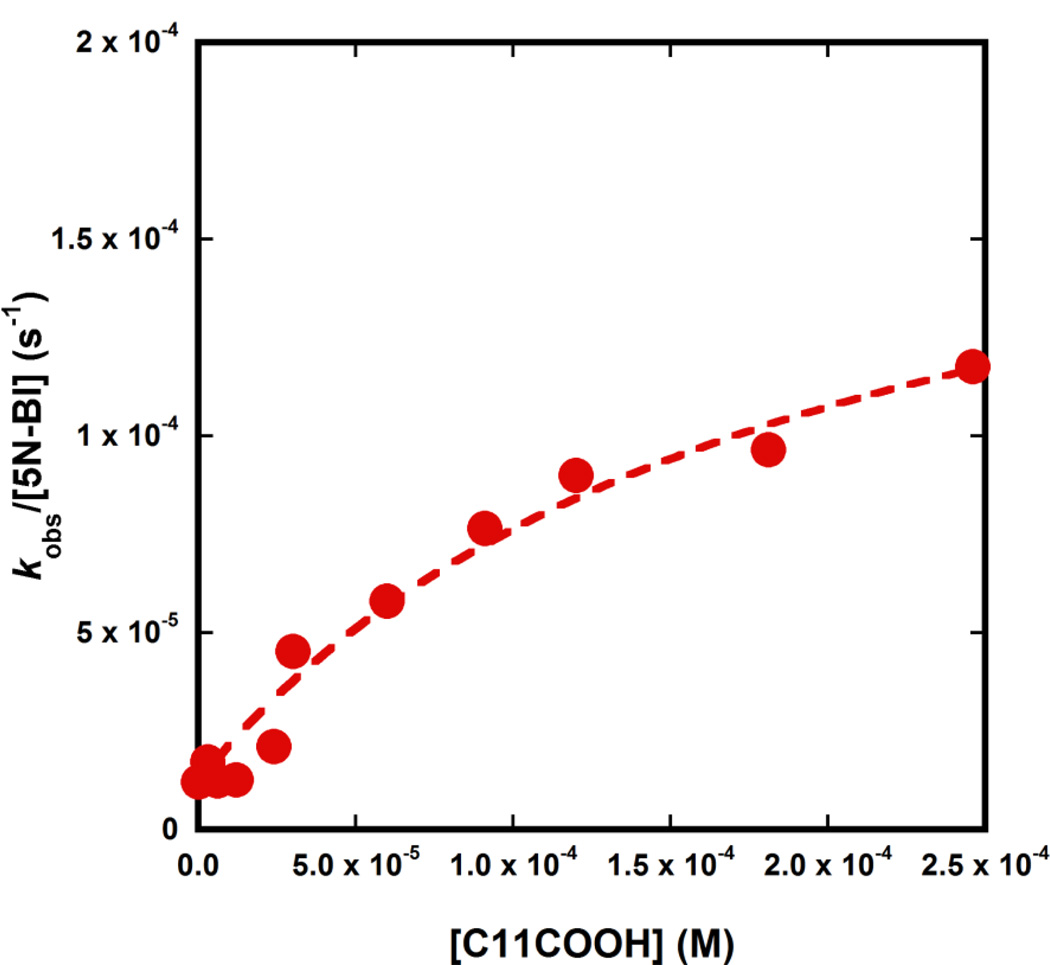
Other compounds such as dodecylamine and dodecanol did not promote the Kemp elimination above background levels (data not shown), but dodecanoic acid (lauric acid, C11COOH) provided significant rate-acceleration. We found that at pH 6.6 and in the presence of CTAC micelles, C11COOH catalyses the reaction with a k2 value of 0.98 M−1s−1 (Fig. 7). Because C11COOH is even less soluble than dodecyl phosphate in aqueous solutions, we compared the k2 value reported above to that of acetate in aqueous solution (5.8 × 10−5 M−1s−1).[15] Thus, the rate acceleration was about 17,000-fold in case of C11COOH, which virtually identical to the value of 16,000 measured when C12P was used as the catalyst. On the contrary, we found no effect upon addition of 2-naphthaleneacetic acid. This result is consistent with the hypothesis that a long hydrophobic tail is needed to associate the base with the CTAC micelle.
Fig. 7.
Effect of lauric acid (C11COOH) on the Kemp elimination of 5NBI at pH 6.6 in the presence of 1.1. mM CTAC.
The 104-rate acceleration measured in the micellar system is only 2–3 orders of magnitude smaller than the rate acceleration provided by the most successful computationally-designed Kemp eliminases (106 for KE59 and 107 for HG3).[15] The measured second-rate constant for a bimolecular reaction in micelles can simply reflect of the increased concentration of reactants in the hydrophobic surface of the micelle.[27] However, the comparison of k2 values for reactions of BI and NI suggests that these substrates are not significantly associated with the micelle, and that the rate acceleration measured herein is due to other factors. In particular, the orientation of the catalytic base is fixed by the micellar environment, and this reduction in entropy might significantly contribute to the rate acceleration. Because the micelle provides no specific interactions to orient the benzisoxazole substrate relative to the catalytic base, our results suggest that precise positioning of the substrate might not be essential to produce significant rate accelerations in the Kemp elimination.
CONCLUSIONS
In this paper, we have shown that the Kemp elimination can be significantly accelerated by long-chain bases associated with micelles. As observed in other systems, the long-chain base seems to be oriented with its tail inside the micelle,[28] while the aromatic substrate seems to be associated with the Stern layer. Thus, our results suggests that relatively loose and non-specific interactions with the substrate can provide at least 4 orders of magnitude rate acceleration even in a non-hydrophobic environment when the catalytic base is easily accessible. This rate acceleration is larger than the ~ 20-fold rate accelerations measured for hydrolyses of benzoyl chlorides and benzenesulfonyl chlorides in the presence of CTAC,[29] and of the 30- and 100-fold measured for hydrolyses of aromatic phosphate triesters[30] and diesters[31] in CTAC; in fact, it is more similar to the 105-fold rate accelerations achieved by metallomicelles.[32,33]
Our results parallel and expand the findings of Korendovych et al. that placed a single Glu/Asp in the very hydrophobic binding site of calmodulin to achieve significant rate acceleration of 3 × 105,[12] which is very similar to what we measured with an even simpler system such as the micelle/long chain base system. The simple micellar system described herein, with its value of k2 = 0.98 M−1s−1 for lauric acid, is clearly less efficient than more complex systems that use a carboxylate base, such as the above-mentioned rationally-redesigned calmodulin (kcat/KM = 5.8 M−1s−1)[12] and the computationally-designed KE-59 (kcat/KM = 160 M−1s−1)[14] and HG3 (kcat/KM = 1,300 M−1s−1),[13,15] but can nonetheless provide significant rate acceleration in spite of the simplicity of the system. These more complex, proteinaceous systems present the obvious advantage that they can be evolved to significantly increase their catalytic efficiencies.[14,15,34] Nevertheless, these increases have been somehow modest (100-fold) and probably due to increase in hydrophobicity of the active site, rather than to the establishment of specific interactions.[35,36] Further, despite of this increase, the rates of the evolved Kemp eliminases remain well-below the diffusion limit, as elegantly pointed out by Korendovych and DeGrado.[36]
Our results reinforce the idea that the Kemp elimination is a simple reaction that does not necessitate specific, enzyme-like interactions to be greatly accelerated. In addition, they suggest that precise positioning of active site residues might not be essential for the observed catalysis of Kemp eliminases. Therefore, we suggest that the Kemp elimination should not be used as a benchmark for the success of the design process, and that more complex reactions, that are not especially sensitive to the presence of a hydrophobic environment, should be used in future design attempts.
Acknowledgments
This work was supported by a grant to the College of Charleston from the Howard Hughes Medical Institute through the Pre-college & Undergraduate Science Education Program to the College of Charleston, by an IDeA grant from NIGMS/NIH (P20GM103499) to the College of Charleston, by URCA grants (College of Charleston) to E.S., J.S., and M.F., by an MRI grant (1429308) from the National Science Foundation to the College of Charleston, and by a Cottrell College Science Award (22490) from Research Corporation for Science Advancement to M.F.
REFERENCES
- 1.Casey ML, Kemp DS, Paul KG, Cox DD. J Org. Chem. 1973;38:2294. [Google Scholar]
- 2.Hollfelder F, Kirby AJ, Tawfik DS. J Org. Chem. 2001;66:5866. doi: 10.1021/jo015723v. [DOI] [PubMed] [Google Scholar]
- 3.Kemp DS, Casey ML. J Am. Chem. Soc. 1973;95:6670. [Google Scholar]
- 4.Kemp DS, Cox DD, Paul KG. J Am. Chem. Soc. 1975;97:7312. [Google Scholar]
- 5.Perez-Juste J, Hollfelder F, Kirby AJ, Engberts JBFN. Org. Lett. 2000;2:127. doi: 10.1021/ol991215k. [DOI] [PubMed] [Google Scholar]
- 6.McCracken PG, Ferguson CG, Vizitiu D, Walkinshaw CS, Wang Y, Thatcher GRJ. J Chem. Soc. Perkin Trans. 2. 1999:911. [Google Scholar]
- 7.Hollfelder F, Kirby AJ, Tawfik DS. J Am. Chem. Soc. 1997;119:9578. [Google Scholar]
- 8.Merski M, Shoichet BK. P Natl. Acad. Sci. U.S.A. 2012;109:16179. doi: 10.1073/pnas.1208076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollfelder F, Kirby AJ, Tawfik DS. Nature. 1996;383:60. doi: 10.1038/383060a0. [DOI] [PubMed] [Google Scholar]
- 10.Hollfelder F, Kirby AJ, Tawfik DS, Kikuchi K, Hilvert D. J Am. Chem. Soc. 2000;122:1022. [Google Scholar]
- 11.Thorn SN, Daniels RG, Auditor MTM, Hilvert D. Nature. 1995;373:228. doi: 10.1038/373228a0. [DOI] [PubMed] [Google Scholar]
- 12.Korendovych IV, Kulp DW, Wu YB, Cheng H, Roder H, DeGrado WF. P Natl. Acad. Sci. U.S.A. 2011;108:6823. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, Thomas LM, Hilvert D, Houk KN, Mayo SL. P Natl. Acad. Sci. U.S.A. 2012;109:3790. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 15.Blomberg R, Kries H, Pinkas DM, Mittl PRE, Grutter MG, Privett HK, Mayo SL, Hilvert D. Nature. 2013;503:418. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 16.Dale TJ, Sather AC, Rebek J. Tetrahedron Lett. 2009;50:6173. [Google Scholar]
- 17.Iranpoor N, Firouzabadi H, Nowrouzi N. Tetrahedron Lett. 2006;47:8247. [Google Scholar]
- 18.Mata J, Varade D, Bahadur P. Thermochim. Acta. 2005;428:147. [Google Scholar]
- 19.VCCLAB, Virtual Computational Chemistry Laboratory. 2005 doi: 10.1007/s10822-005-8694-y. http://www.vcclab.org. [DOI] [PubMed] [Google Scholar]
- 20.Lee BS, Nome F. Langmuir. 2000;16:10131–10136. [Google Scholar]
- 21.Carnali JO, Pethica BA. J Phys. Chem. B. 2006;110:24936. doi: 10.1021/jp062731c. [DOI] [PubMed] [Google Scholar]
- 22.Buckingham SA, Garvey CJ, Warr GG. J Phys. Chem. 1993;97:10236. [Google Scholar]
- 23.Brinchi L, Di Profio P, Germani R, Giacomini V, Savelli G, Bunton CA. Langmuir. 2000;16:222. [Google Scholar]
- 24.Brinchi L, Di Profio P, Germani R, Goracci L, Savelli G, Gillitt ND, Bunton CA. Langmuir. 2007;23:436. doi: 10.1021/la061807t. [DOI] [PubMed] [Google Scholar]
- 25.Cordes EH. Pure Appl. Chem. 1978:50. [Google Scholar]
- 26.Hu YF, Houk KN, Kikuchi K, Hotta K, Hilvert D. J Am. Chem. Soc. 2004;126:8197. doi: 10.1021/ja0490727. [DOI] [PubMed] [Google Scholar]
- 27.Scrimin P, Tecilla P, Tonellato U, Bunton CA. Colloids Surf. A. 1998;144:71. [Google Scholar]
- 28.Alawadi N, Williams A. J Org. Chem. 1990;55:2001. [Google Scholar]
- 29.Bunton CA. Adv. Colloid Interface Sci. 2006;123–126:333–343. doi: 10.1016/j.cis.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Bunton CA, Robinson LB. J Org. Chem. 1969;34:773–780. [Google Scholar]
- 31.Bunton CA, Diaz S, Hellyer JM, Ihara Y, Ionescu LG. J Org. Chem. 1975;40:2313–2317. [Google Scholar]
- 32.Menger FM, Gan LH, Johnson E, Durst DH. J Am. Chem. Soc. 1987;109:2800–2803. [Google Scholar]
- 33.Gruber B, Kataev E, Aschenbrenner J, Stadlbauer S, König B. J Am. Chem. Soc. 2011;133:20704–20707. doi: 10.1021/ja209247w. [DOI] [PubMed] [Google Scholar]
- 34.Khersonsky O, Kiss G, Rothlisberger D, Dym O, Albeck S, Houk KN, Baker D, Tawfik DS. P Natl. Acad. Sci. U.S.A. 2012;109:10358. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frushicheva MP, Mills MJL, Schopf P, Singh MK, Prasad RB, Warshel A. Curr. Opin. Chem. Biol. 2014;21:56–62. doi: 10.1016/j.cbpa.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korendovych IV, DeGrado WF. Curr. Opin. Struc. Biol. 2014;27:113–121. doi: 10.1016/j.sbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]