Abstract
BMI1 oncogene is a catalytic member of epigenetic repressor polycomb group proteins. It plays a critical role in the regulation of gene expression pattern and consequently several cellular processes during development, including cell cycle progression, senescence, aging, apoptosis, angiogenesis, and importantly self-renewal of adult stem cells of several lineages. Preponderance of evidences indicates that deregulated expression of PcG protein BMI1 is associated with several human malignancies, cancer stem cell maintenance, and propagation. Importantly, overexpression of BMI1 correlates with therapy failure in cancer patients and tumor relapse. This review discusses the diverse mode of BMI1 regulation at transcriptional, posttranscriptional, and posttranslational levels as well as at various critical signaling pathways regulated by BMI1 activity. Furthermore, this review highlights the role of BMI1 as a biomarker and therapeutic target for several subtypes of hematologic malignancies and the importance to target this biomarker for therapeutic applications.
Keywords: BMI1, hematologic malignancy, biomarker
Introduction
Embryonic development requires cell fate decision governed by regulated gene expression pattern.1 These gene expression patterns are largely regulated by reprogrammable epigenetic regulatory mechanisms that control the local chromatin conformation.2 The repeated specific pattern of gene expression established by epigenetic machinery during multiple rounds of cell division establishes cellular identity. Epigenetic regulation is significantly achieved by modulating nucleosome dynamics through histone tail modifications. The major molecular machines that dictate gene expression pattern through histone tail modification and chromatin modulations are global epigenetic modifiers, such as trithorax group (TrxG) and polycomb group (PcG) proteins, which trigger transcriptional activation and repression of target gene, respectively.3 The counteracting activities of TrxG and PcG proteins regulate chromatin dynamics and gene expression cascade to retain cellular memory throughout the life of the organism. An imbalance in this dynamics has been associated with compromised gene expression pattern as well as with an increased risk of cancer.3–5 In this regard, several studies corroborate the dysregulation of PcG proteins as a diagnostic and prognostic marker of several types of cancer, including hematologic malignancies.6 PcG proteins form primarily two large multisubunit polycomb-repressive complexes (PRCs), namely, PRC1 and PRC2. These complexes in association with other epigenetic markers establish target gene repression through histone tail posttranslational modifications.7–9 For each complex, there are defined core subunits responsible for the bonafide activity of the complex and some variable subunits that plausibly dictate the identity of the target gene in a lineage- and context-dependent manner.10–12 Generally, PRC2 initiates histone tail modification by mono-, di-, and trimethylation of histone 3 at lysine 27 (H3K27me) residue with the help of its catalytic subunit enhancer of zeste homolog 2 (EZH2) methyl transferase enzyme, and this modification promotes the recruitment of PRC1 through its chromodomain-binding proteins, which then recognizes the H3K27me marker established by PRC2.7,13 PRC1 is composed of RING1A/B, PH1, PH2, CBX2, 4, 6, 7, and 8, and many other variable subunits. This composition varies depending on the lineage of cells, and how the diverse composition impacts target gene identification for a specific cell types is a pending conundrum that needs further research.14,15 PRC1 catalyzes monoubiquitination of histone 2A at lysine 119 residue (H2AK119ub) to maintain target gene repression with the help of its catalytic subunit RING1A/B, which is an E3 ligase. RING1A/B protein activity is significantly enhanced by its association with another PRC1 component BMI1 (B-cell specific moloney murine leukemia virus integration region 1). BMI1 was essentially identified as a cMyc proto-oncogene, which cooperates with cMyc in oncogenesis of murine B-cell lymphoma.16 BMI1-directed PRC1 activity is counterbalanced by selective H2AK119 deubiquitination mediated by ubiquitin-specific protease (USP16).17 Emerging studies establish that BMI1 has important function as a diagnostic and prognostic biomarker of several lineages of cancer, including hematologic malignancies.18–22 This review summarizes the role of BMI1 as a biomarker of hematologic malignancies, the regulatory mechanism that controls BMI1 expression and cellular pathways that are perturbed by aberrant expression and activity of BMI1 leading to oncogenic transformations.
BMI1 Gene and Protein Structure
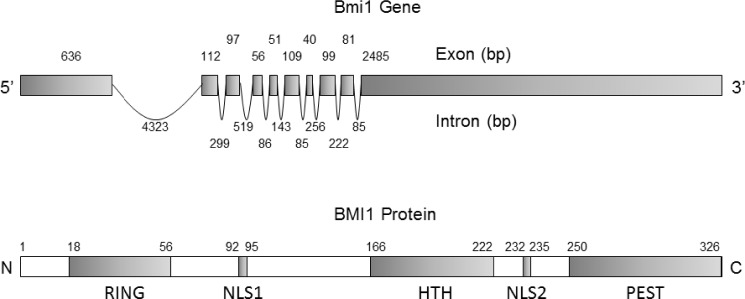
The human Bmi1 gene, composed of 10 exons and 9 introns, is localized on the short arm of chromosome 10 (10p11.23) and encodes 37 kDa protein composed of 326 amino acids.23 BMI1 protein structure is highly evolutionarily conserved and possesses significant homology with another PcG protein Mel 18, a transcriptional repressor of Bmi1 gene.24,25 BMI1 and Mel18 share 65% amino acid identify. Both proteins possess similar arrangement of protein domains, including N-terminal RING finger domain and C-terminal helix-turn-helix and prolinerich domains. Furthermore, proteomic analysis of interactome composition of these two proteins suggests that they interact with a similar repertoire of proteins.25 However, in PRC1, they exhibit strikingly opposite function, including negative regulation of BMI1 by Mel18 and consequently promoting senescent phenotype.26,27 BMI1 protein has several defined domains and motifs, including N-terminal RING finger domain (RF), a central helix-turn-helix (HTH) domain, a carboxyl terminal PEST-like domain, and two nuclear localization signals KRRR and KRMK.28 Figure 1 elucidates Bmi1 gene and BMI1 protein structure. The RF domain of BMI1 is required for its association with RING1B E3 ligase, a catalytic component of PRC1, to activate PRC1 activity.29 The RF and HTH domains are important for BMI1 localization at DNA strand break and crucial for the recruitment of DNA damage repair machinery, prevention of cellular senescence, and cancer cell survival.30–32 The C-terminal PEST domain is rich in proline (P), glutamic acid (E), serine (S), and threonine (T) and is critical for BMI1 protein turnover. We have previously demonstrated that deletion of PEST domain prevents BMI1 degradation and promotes its oncogenic potential, including epithelial-to-mesenchymal transition (EMT).28
Figure 1.
Gene and protein structure of BMI1. (A) Bmi1 gene structure composed of 10 exon and 9 introns. (B) Schematic elucidation of different domains of BMI1 protein critical for its function starting from N terminus RING domain required for its interaction with RING1A/B protein, Nuclear localization signal 1 (NLS1), Helix-turn-helix (HTH) domain required for interaction with E4F1, Nuclear localization signal 2 (NLS2), and finally, Proline (P), Glutamic acid (E)-, Serine (S)- and Threonine (T)-rich domain (PEST), which is required for its regulated protein turnover.
BMI1 Expression and Regulation
BMI1 is expressed ubiquitously in almost all types of tissues. However, its expression levels are high in brain, lungs, thymus, kidney, gonads, salivary glands, placenta, blood, bone marrow, and stem cells of several lineages.33 It is overexpressed in several cancer subtypes and serves as a biomarker for these cancer types. BMI1 plays a critical role in cellular physiology and hence the transcript and protein levels of BMI1 are tightly regulated in diverse cell types. For the past several years, many investigators are trying to understand the regulation of BMI1 at transcriptional, posttranscriptional, and posttranslational levels. However, the comprehensive knowledge on BMI1 regulation by different mechanisms is still scarce.
Transcriptional regulation
Bmi1 gene expression is regulated by the number of transcription factors in a context-and lineage-dependent manner. BMI1 expression is positively regulated by transcription factor Sp1, Twist1, FoxM1, ZEB1, E2F1, SALL4, Myc-N, c-Myc, and HDACs, whereas its expression is negatively regulated by Mel18, Nanog, and KLF4.26,27,34–43 E2F-1, Myc-N, and c-Myc trans-activates BMI1 by binding to its promoter and thus upregulating its expression in neuroblastoma cells.34,38 Similarly, SALL4 and Sp1 transcription factors bind to a specific region of the BMI1 promoter and upregulate its expression in normal and malignant cells of leukemic stem and nasopharyngeal origin, respectively.39,40 We have shown that HDAC inhibitor downregulates BMI1 expression in breast epithelial and breast cancer cells, suggesting that HDACs positively regulate BMI1 expression.37 Furthermore, forkhead box transcription factor, FOXM1, positively regulates BMI1 expression in neuroblastoma cells.36 Twist1 and Zeb1 are EMT markers and positively regulate BMI1 expression to maintain stemness of the cell and EMT in head and neck cancer. Twist 1 directly regulates BMI1 expression by regulating its transcription, whereas ZEB1 indirectly regulates BMI1 expression by repressing microRNA (miRNA) 200, an inhibitor of BMI1.21,35,42,44 Furthermore, BMI1 is also repressed by Nanog in embryonic stem cells.43
Posttranscriptional regulation of mRNA
Gene expression is regulated at different levels, including regulation of mRNA activity after synthesis. This regulation may operate at the level of RNA processing for maturation, its transport to correct subcellular localization, its stability and finally translation of its coded information in to protein. These regulations occur through interaction of cellular factors with mRNA sequence elements located within 5′- and 3′-untranslated regions of unprocessed mRNA, including secondary structures, internal ribosome entry sites, and poly-A tail. The posttranscriptional regulations are also achieved through the activity of target-specific microRNA expression. The miRNAs are short 21–23 nucleotide RNA sequences that control the level of expression of target gene through controlling transcription as well as mRNA stability. Several miRNAs are implicated in the regulation of BMI1 expression. Deregulation of BMI1 associated miRNA can be exploited as a biomarker of cancer progression (Table 1).
Table 1.
The microRNAs (miRNAs) that regulate BMI1 expression and may serve as biomarker for associated cancer.
| S. NO. | CANCER SUBTYPE | miRNA INVOLVED AND ITS MODE OF BMI1 REGULATION | REF. | |
|---|---|---|---|---|
| 1 | Non-Small Cell Lung Cancer (NSCLC) | miR-452 | Inhibit metastasis by down regulating BMI1 | 110 |
| miR-203 | Downregulate BMI1 expression and inhibit NSCLC cell proliferation | 111 | ||
| miR-487b | Suppress BMI1 and other important cellular target to prevent primary lung cancer | 112 | ||
| 2 | Brain cancer | miR-130b | Enhances BMI1 expression and stem cell-like properties in glioblastoma | 113 |
| miR-128a | Brain-specific miRNA that suppress BMI1 expression to prevent glioblastoma progression | 114–117 | ||
| miR-218 | Inhibits glioma progression and glioma stem cell self-renewal by suppressing BMI1 | 118 | ||
| miR-128 | Overexpression of miR-128 could suppress pituitary GH3 tumor growth via downregulating BMI1 expression | 119 | ||
| miR-128a | Inhibit medulloblastoma cancer growth by inhibiting BMI1 expression | 120 | ||
| 3 | Head and Neck Squamous Cell Carcinoma (HNSCC) | miR-494 | Inhibit BMI1 expression and reduce cancer stemness | 121 |
| miR-200c | Downregulate BMI1 to inhibit HNSCC progression | 122 | ||
| 4 | Breast cancer | miR-495 | Inhibit BMI1 expression and suppress breast cancer cell proliferation | 123 |
| miR-31 | BMI1 and miR-31 represses each other in a cross negative feedback loop | 124 | ||
| miR-200c/141 cluster | Downregulate BMI1 to trigger senescence | 125 | ||
| miR-22 | It suppresses miR-200 precursor, thereby enhances BMI1 expression | 126 | ||
| miRNA-200c and 203 | Repress BMI1 expression to prevent cancer progression and cancer stem cell stem cell self-renewal | 127–129 | ||
| miR-34a | Downregulate Bcl-2 and BMI1 to prevent breast cancer | 130 | ||
| miR-128-2 | Repress BMI1 to inhibit mammary epithelial cell transformation | 131,132 | ||
| miR-200 family, miR-15/16, miR-103, miR-107, miR-145, miR-335 and miR-128b | Downregulation of BMI1 to inhibit breast cancer progression | 133 | ||
| 5 | Colorectal cancer | miR-215 | It represses BMI1 expression and reported to be down regulated in colorectal cancer | 134 |
| miR-218 | Inhibit colon cancer by downregulating BMI1 | 135 | ||
| 6 | Gallbladder cancer | miR-218 | Long non-coding RNA CCAT1 promotes gallbladder cancer via negative regulation of miR-218-5p. miR-218-5p downregulate BMI1 expression | 136 |
| miR-200 | Repress EMT by downregulating BMI1 expression in bladder cancer | 137 | ||
| 7 | Melanoma | miR-203 | Inhibit melanoma metastasis by downregulating BMI1 | 138 |
| miR-218 | Downregulate BMI1 to inhibit melanoma progression | 139 | ||
| miR-200c | Inhibit melanoma progression through BMI1 downregulation | 140 | ||
| 8 | Hepatocellular carcinoma | miR-218 | Downregulate BMI1 to inhibit liver cancer progression | 141,142 |
| 9 | Nasopharyngeal carcinoma | miR-320a | Downregulate BMI1 and thus play tumor suppressor role in NPC | 143 |
| 10 | Pancreatic ductual adenocarcinoma | miR-183 | Downregulate BMI1 to inhibit PDAC progression | 144 |
| miR-135a | Downregulate BMI1 to inhibit PDAC progression | 145 | ||
| miR-15a | Inhibit progression and metastasis of PDAC by down-regulating BMI1 expression | 146 | ||
| 11 | Prostate cancer | miR-128 | Suppresses prostate cancer by inhibiting BMI1 mediated cancer stem cell properties | 147 |
| miR-200b | Suppresses prostate cancer by regulating BMI1 | 148 | ||
| 12 | Thymic lymphoma | miR-200c | Downregulation of BMI1 by miR-200c prevent radiation induced thymic lymphoma | 42,149 |
| 13 | Gastric cancer | miR-30e | Loss of miR30e* trigger increased BMI1 expression and promote gastric cancer | 150 |
| 14 | Esophageal cancer | miR-203 | Inhibit progression and metastasis of esophageal cancer stem cell by suppressing BMI1 | 151 |
| 15 | Lymphoma | miR-16 | Negatively regulate BMI1 and downregulated in MCL side population | 99 |
| 16 | Ovarian cancer | miR-15a and miR-16 | Downregulate BMI1 expression to inhibit ovarian cancer progression | 152 |
| 17 | Renal cancer | miR-708 | Suppresses BMI1 expression to prevent Renal cancer progression | 153 |
| 18 | Endometrial cancer | miR-194 | Downregulates BMI1 expression to inhibit endometrial cancer | 62 |
| 19 | Tongue cancer | miR-200b and miR-15b | Target BMI1 repression and their loss induce EMT and chemoresistance in tongue cancer cells | 154 |
Posttranslational regulation of BMI1 protein
Posttranslational regulations can be of two types: (1) reversible regulation and (2) irreversible regulations. The reversible regulation can be achieved through posttranslational modification, whereas irreversible regulation is achieved through protein degradation majorly operated by ubiquitin proteasome system (UPS). The reversible protein modifications such as acetylation, phosphorylation, sumoylation, or ubiquitination can either alter the protein function or often direct protein to irreversible type of regulation through UPS-mediated degradation of protein. Posttranslational regulation of BMI1 is not significantly characterized. BMI1 is reported to be modified by several factors in context- and lineage-dependent manner. CBX4 triggers BMI1 sumoylation at lysine 88 during DNA damage to trigger its recruitment at damaged site.45 BMI1 is phosphorylated by several kinases in a context-dependent manner for different physiological consequences. Phosphorylation of BMI1 by 3pk (MAPKAP kinase 3), an intermediate member of MAPK signaling pathway, regulates the dynamics of BMI1 dissociation with chromatin and thus controls BMI1-driven repression program for the regulation of differentiation and developmental processes.46 Furthermore, AKT kinase phosphorylates BMI1 and increases its oncogenic potential in prostate cancer in an Ink4a/Arf independent manner.47 On the contrary, BMI1 phosphorylation at S361 by AKT triggers its dissociation from chromatin, thus hindering its oncogenic activity.48 BMI1 is also regulated at posttranslational level by ubiquitin proteasome system-mediated protein degradation. Our studies have shown that BMI1 C-terminal PS domain is critical for BMI1 stability and within this region a consensus motif DSGSDKANS is recognized by an E3 ligase β-TrCP, which regulate BMI1 protein turnover and consequent oncogenic activity.49 Increased stability of BMI1 triggers EMT of mammary epithelial cells, leading to metastatic phenotype.28 Interestingly, C-terminal PS domain deletion mutant of BMI1 seems to be more stable and oncogenic than BMI1 mutant that cannot be recognized by β-TrCP, suggesting plausible role of yet other unidentified E3 ligase(s) in BMI1 degradation and consequent modulation of downstream signaling.
BMI1 Role in Cellular Physiology
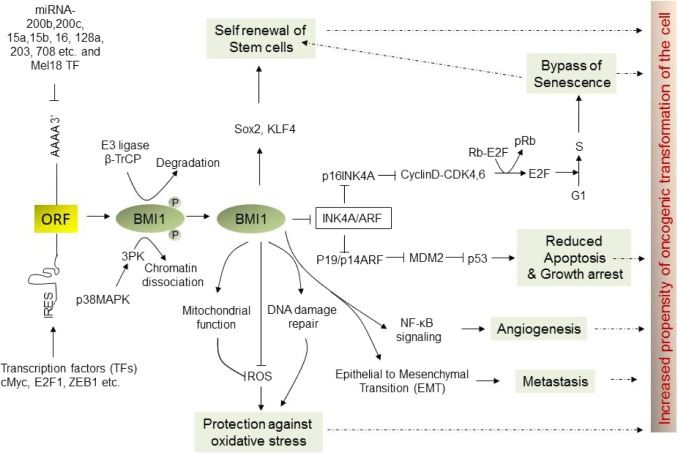
Through repression of target gene expression in a lineage-and context-dependent manner, BMI1 regulates a myriad of cellular processes critical for cell growth, cell fate decision, development, senescence, aging, DNA damage repair, apoptosis, and self-renewal of stem cells (Fig. 2).50 BMI1-knockout mice exhibited more than 50% of survival by third day after birth and those that survived showed skeletal/spinal growth defects, reduced density in cerebellum and neural layers, including other neurological abnormalities, and importantly hematopoietic abnormalities.51 The most studied and validated BMI1 target is Ink4a/Arf locus, which encodes three tumor suppressor: p16Ink4a, p14Arf, and p19Arf. Functional inactivation of BMI1 frequently promotes senescence (a cellular state in which cell ceases to proliferate and eventually die) with loss of G1 to S phase transition during cell cycle, majorly attributed to the derepression of the Ink4a/Arf locus. Hyperactivation of BMI1 bypasses cellular senescence and consequently promotes tumorigenesis by promoting cell cycle progression through repression of cell cycle checkpoint p16 and p21. Additionally, BMI1-mediate repression of p19, which otherwise induces p53 and impairs cell cycle arrest and apoptosis. Thus, collectively BMI1 regulates p53-pRb axis through repression of Ink4a/Arf locus, which has been described as the principle barrier to the initiation and maintenance of neoplastic transformation. Hyperactivation of BMI1, through various mechanisms, bypasses this barrier, promoting cellular transformation. Bmi1 also plays a critical role in preventing oxidative DNA damage and in preventing DNA damage response pathways. BMI1 and RING2 are recruited at the side of DNA double strand breaks (DSBs) and contribute to the ubiquitination of y-H2AX. BMI1 loss hinders recruitment of DNA repair machinery components, including 53BP1, BRCA1, and RAP89, at the site of DSBs and impairs homologous recombination causing accumulation of cells in G2M phase. Many cancer chemo and radiotherapies rely on DNA-damage-mediated cellular toxicity. Hence, increased expression or activity of BMI1 may also serve as a biomarker of resistance to DNA-damaging therapy.52–54 Apart from repression of Ink4a/Arf locus and DNA damage repair mechanisms, BMI1 also targets several other loci and controls myriad of cellular processes leading to oncogenesis of various lineages of cell.50,55,56
Figure 2.
Upstream and downstream signaling pathways of BMI1. BMI1 is critical for normal cellular physiology and its deregulation through several mechanisms at transcriptional and translational levels as delineated in this figure promotes oncogenic transformation of cell through aberrant cellular signaling pathways.
BMI1 Role as a Biomarker of Cancer
BMI1 plays a significant role in several critical cellular processes, which when go awry lead to cancer progression. BMI1 is dysregulated in several types of cancer and overexpressed in several lineage of cancer stem cells.50,57–69 Due to its role in several cellular processes critical for normal cellular physiology and its frequent dysregulation in various types of cancer, BMI1 expression and hyperactivation have been considered as a biomarker for several types of cancer. This review addresses the involvement of BMI1 in several types of hematologic malignancies and its prospects as a biomarker and therapeutic target.
BMI1 Dysregulations in Hematologic Malignancies
The bone marrow produces blood stem cells (immature cells) that mature into blood cells over time while undergoing several rounds of differentiation. A blood stem cell may mature either into myeloid progenitor cell or into lymphoid progenitor cell. The lymphoid progenitor cell may further differentiate into lymphoblast cell that eventually form B-lymphocyte, T-lymphocyte, or natural killer cells. A myeloid progenitor cell differentiates into red blood cell, platelets, or white blood cells. Clonal proliferation of these cells at the level of stem, progenitor, and their different maturation stages due to aberrant signaling cues are responsible for evolution and propagation of hematologic malignancies. Similar to several other forms of cancer, BMI1 is also implicated in the uncontrolled proliferation of hematopoietic cells, self-renewal of hematopoietic stem cell and cancer stem cell, leading to malignant transformation. In fact, BMI1 deficiency compromises hematopoietic stem cell function, and it is indispensable for normal and leukemic stem and progenitor cell self-renewal.70,71 However, the role of BMI1, exploitation of its dysregulation as biomarker and therapeutic prospect of targeting BMI1 in various types of hematologic malignancies, has not been extensively discussed in the literature. The expression and activity of BMI1 in hematologic malignancies are regulated by several different mechanisms and can be utilized both as a biomarker and a therapeutic target.
BMI1 and Myeloid Malignancies
Myelodysplastic syndrome
Myelodysplastic syndrome (MDS) is a group of bone marrow disease with ineffective hematopoiesis and increased apoptosis of early and mature hematopoietic cells. The disease is very heterogeneous, and the prognosis is highly variable. MDS is highly susceptible for the transformation into acute myeloid leukemia or AML.72 Identification of biomarker for MDS is critical for early diagnosis and prognosis. BMI1 is a stem cell self-renewal marker of multiple lineages of stem cells, and since MDS is a disease of stem, progenitor, and blast cells, BMI1-associated abnormalities were investigated in MDS patient samples. In a study of 51 MDS and MDS-AML patients, high expression of BMI1 was observed in blast cell population and their variants with potential for transformation as compared to control. Particularly, higher positivity of BMI1 was reported in refractory anemia with excess blasts (RAEB), RAEB in transformation, and MDS-acute myeloid leukemia (MDS-AML) as compared to refractory anemia (RA) and RA with ringed sideroblast (RARS) cells. BMI1 has also been reported to prevent apoptosis in MDS-l. According to several emerging independent studies, it seems possible to stratify MDS population based on BMI1 expression level, and BMI1 expression might serve as biomarker for MDS prognosis.73,74
Acute myeloid leukemia
AML is also called acute myelocytic leukemia, acute myelogenous leukemia, acute nonlymphocytic leukemia, or acute granulocytic leukemia. It is also a cancer of myeloid line of blood cells. In this disease, patients have too many immature white blood cells in bone marrow, which are not able to undergo maturation. Several studies confirm the high expression of BMI1 in AML patients, which may contribute to leukemogenesis and unfavorable prognosis.75,76 A study performed with 64 AML patients corroborates the prognostic value of BMI1 in this disease. According to the findings from the study, patients with lower BMI1 positivity (≤55%, n = 33) had significantly longer overall survival (OS), cancer-specific survival (CSS), relapse free survival (RFS) and remission duration (RD) as compared to the patients harboring higher BMI1 levels (≥55%, n = 31). Furthermore, among the patients with secondary AML (n = 18), low BMI1 expression (n = 8) correlated with better OS and among patients with de novo AML (n = 46), and lower expression of BMI1 correlated with significantly higher OS, CSS, RFS, and RD.77 Another independent study published in the 56th American Society of Hematology conference reported prognostic value of BMI1 expression in 511 newly diagnosed AML patients.78 This study suggests that BMI1 expression was higher in AML with unfavorable cytogenetics (n = 252) compared to intermediate (n = 225) or favorable cytogenetics (n = 34). Higher BMI1 levels were associated with shorter median overall survival (OS). Furthermore, BMI1 inhibition by pharmacologic approaches triggered cytotoxicity in AML and acute lymphoid leukemia (ALL) cells by inducing apoptosis, suggesting that targeting BMI1 activity might be of therapeutic advantage for AML. BMI1 repression in human AML CD34+ cells de-represses p16Ink4a and p14Arf tumor suppressor and impairs their self-renewal capacity in culture. All these independent studies corroborate that expression of BMI1 is an independent biomarker, prognostic factor, and therapeutic target for AML.
Chronic myeloid leukemia
Chronic myeloid leukemia (CML) also known as chronic myelogenous leukemia is a disease of mature white blood cells, which mostly affects adults and is rarely seen in children. Average onset occurs around 60 years of age. CML undergo several stages of development starting from chronic phase to accelerated phase (and further to blast crisis).79,80 The disease is characterized by a balanced genetic translocation, t(9;22)(q34;q11.2), leading to a fusion of the Abelson (ABL) oncogene with breakpoint cluster region (BCR) gene.81–83 This fusion, also known as the Philadelphia chromosome, results into a chimeric oncoprotein Bcr-Abl, a constitutively active tyrosine kinase responsible for growth promoting cellular signaling. Despite a consistent molecular abnormality of Bcr-Abl fusion, CML exhibits marked heterogeneity in disease prognosis. Bcr-Abl collaborates with BMI1 for the leukemic transformation of CD34+ B-cells.75,84,85 BMI1 has previously been reported to cooperate with other oncogenes to induce malignant transformations and its overexpression contributes to disease aggressiveness.73,86,87 Hence, from conceptual standpoint, BMI1 might serve as another biomarker for CML transformation. Several independent studies corroborate that BMI1 expression was significantly higher in AML and CML patients as compared to healthy donors.84,85 Kaplan–Meier analysis on these samples exhibits strong association between relative BMI1 overexpression and significantly shorter OS and DFS. Interestingly, myriad of mechanisms contributes to BMI1 overexpression and ensuing downstream signaling to promote CML progression. E2F1 transcription factor, a direct positive regulator of BMI1, was also reported to be overexpressed in CML patient samples. Furthermore, this study reported an inverse correlation between BMI1 low expression and PR3 (proteinase-3, an independent prognostic marker of CML) high expression, which was associated with OS of CML patients.84 BMI1 prevents autophagy of CML cells by directly silencing tumor suppressor CCNG2/cyclin G2 and Ink4a/Arf locus along with regulating several differentiation makers including B-lymphoid differentiation genes p16, Ebf1, Pax5, and Ikzf1, to promote CML disease progression.75,88,89 Among CML patients, BMI1 expression was relatively lower in chronic phase as compared to advanced disease stages, such as accelerated or blastic CML. These relative expression data suggest that BMI1 overexpression contributes to disease aggressiveness and can be utilized as a biomarker of prognostic value to monitor CML progression at advanced stages.84,85
BMI1 and Lymphoid Malignancies
Acute lymphoblastic leukemia
Acute lymphoblastic leukemia (ALL) is the most common cancer among children.90 In ALL, the malignant transformation of lymphoid progenitor cell result into either B-cell lineage of ALL (B-ALL) or T-cell lineage of ALL (T-ALL). B-ALL contribute major fraction of ALL population. Being epigenetic modulator and stem cell self-renewal marker, BMI1 can further reprogram and transform CML B-lymphoid progenitors into leukemia initiating cells, leading to B-lymphoid leukemia (B-ALL) in vivo. However, survival of such leukemic B-lymphoid progenitors significantly depends upon Bcr-Abl fusion, suggesting a functional synergism between these two molecular markers of CML for disease progression.89 Further, whole exome sequencing identified BMI1 mutation in early T-cell precursor-ALL and overexpressed in CALM-AF10+ T-ALL (clathrin-assembly protein-like lymphoid-Myeloid leukemia gene to AF10), respectively, suggesting its role in T-cell lineage of ALL as well.91,92 Thus, emerging evidences are tempting to speculate that BMI1 may serve as biomarker for diagnosis and evolution of ALL disease of B-cell and T-cell origin and suggest a need for detail analysis in this regard using large cohort of ALL clinical samples.
Chronic lymphocytic leukemia
Chronic lymphocytic leukemia (CLL) is a clonal lymphoproliferative disorder characterized by immunophenotype of CD19+ CD5+ CD23+ peripheral lymphocytes weakly expressing CD20 and CD79b, and surface immunoglobulin (sIg).93 It is a slow growing B-cell malignancy with highly variable clinical course and hence, there is a need for the identification of novel molecular biomarker of this disease. In CLL, BMI1 is recurrently targeted by chromosomal rearrangement involving a novel t(10;14)(p12;q32)/IgH-BMI1 and its IG variants. The rearrangement of IGH-BMI1 was validated by FISH using probes centromeric to IGH and telomeric to BMI1. The median survival of CLL patients with t(10;14)(p12;q32) was 8.5 months, suggesting an aggressive behavior of the translocation. These aberrations were consistently acquired during disease progression; high-grade transformation of leukemia (Richter syndrome) exhibited a gain-of-function, and thus, BMI1 translocation can be utilized as biomarker for CLL with aggressive phenotype.94
Mantle cell lymphoma
Mantle cell lymphoma (MCL) is an indolent CD5+ lymphoma of naïve pre-germinal center B-cell, which is mostly characterized by t(11;14)(q13;q32) translocation, leading to dysregulation of cyclin D 1.95–97 Additionally, several other genomic alterations were identified in MCL leading to dysregulation of BMI1. The BMI1 expression pattern in normal hematopoietic cells is tightly regulated along the progressive stages of cellular differentiation, and accordingly, B-cell lymphomas also in part maintain this pattern of expression profile, such as MCL and CLL. These neoplasms derived from naïve pre-germinal center cells exhibit high expression levels of BMI1 as compared with follicular germinal center-derived follicular lymphoma (FL) and LCL.98 FISH and SNP array analyses of BMI1 in MCL exhibited abnormal hybridization pattern and structural abnormalities involving wide range of genes. The abnormalities identified in 17 cases were heterogeneous including reciprocal and nonreciprocal translocations, deletions, and insertions, constituting complex karyotypes. However, unlike CLL, these aberrations did not involve Ig loci.94 In another study, it was observed that BMI1 expression was significantly higher in MCL clinical specimen and further increased in MCL side population (SP) in patient samples and MCL cell lines as compared with CD5+ B-cell. MCL SP exhibits clonogenicity as well as self-renewal capacity and downregulates proapoptotic genes, BCL2L11/Bim and PMAIP1/Noxa, via increased BMI1 expression and activity. The increased BMI1 expression in MCL SP was more dramatic at the time of recurrence than at initial diagnosis.99 The high and aberrant expression of BMI1 in MCL and its SP is associated with poor prognosis and may contribute to pathogenesis of the disease. Hence, BMI1 expression may serve as biomarker for diagnosis, prognosis, and potential therapeutic target for treatment of aggressive form of MCL.
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is the most common form of non-Hodgkin lymphoma (NHL) in the adult population, representing approximately 30%–40% of all NHLs, and accounting for more than 80% cases of aggressive lymphomas in the world. DLBCL is a heterogeneous group of B-cell lymphoma with different genetic backgrounds, morphologic features, and responses to therapy. Gene expression profiling and immunohistochemical studies can allow further subgrouping of DLBCL into different subtypes, including germinal center B-cell-like (GCB) DLBCL, activated B-cell-like (ABC) DLBCL, and primary mediastinal large B-cell lymphoma (PML-BCL). Despite recent improvements, DLBCL still represents a major clinical challenge due to the heterogeneous nature of the disease and a substantial proportion of patient’s still experience poor outcome.100 Hence, better understanding of germinal center-derived lymphoma biology and identification of novel molecular biomarker and targets are crucial for the improvement of the current therapeutic options. Targeted expression profiling in B-cell lymphoma suggests that BMI1 is preferentially expressed in B-cell lymphoma with aggressive phenotype, including DLBCL, Burkitt’s lymphoma, and MCL.101 In DLBCL, BMI1 expression is also a predictive biomarker to distinguish GCB type versus ABC type, and its expression is associated with unfavorable clinical outcome in primary nodal DLBCL.102 These studies indicate the possible role of BMI1 in DLBCL and need for further validation using a large cohort of DLBCL clinical samples for BMI1 expression as a biomarker of this heterogeneous disease.
Multiple myeloma
Multiple myeloma (MM) is one of the most common hematologic malignancy that accounts for ~13% of all hematologic malignancies and 1% of overall cancer.103 It is a terminally differentiated neoplasm of immunoglobulin producing plasma cells (PCs). In vitro and in vivo studies suggest that BMI1 is critical for the maintenance of transformed phenotype of MM. RNA interference-mediated silencing of BMI1 in MM cell line RPMI-8226 triggered apoptosis and increased production of apoptotic marker cleaved caspase 3.104 However, BMI1 profiling and genetic analysis in MM patient sample is required to implicate it as potential biomarker and therapeutic target for this disease.
T-cell lymphoma
T-cell lymphoma is a diverse group of lymphoproliferative disorder of T-cell origin. Frequently BMI1 overexpression has been associated with T-cell lymhomagenesis through various mechanisms.105 Often, down-regulation of miRNA) that specifically targets BMI1 expression is implicated in T-cell lymphomagenesis. For instance, miRNA 16, a specific regulator of BMI1, is downregulated in primary cutaneous T-cell lymphoma (CTCL).106 Similarly, downregulation of miRNA 200c that represses BMI1 expression has been associated with split radiation-induced thymic lymphoma (RITL).107 Downregulation of miRNA-200c or upregulation of BMI1 can be a prognostic biomarker of RITL. BMI1 is overexpressed in cutaneous T-cell lymphoma, particularly sezary syndrome.108 Our studies further support this finding showing recurrent copy number gain of BMI1 in sezary syndrome. However, this finding needs further detail functional validation.109 These independent studies implicate the role of BMI1 in various types of T-cell lymphoma and require detailed clinical evaluations to establish whether BMI1 levels/activity can be exploited as a biomarker for T-cell lymphoma.
Conclusion and Future Perspectives
BMI1 deregulation has been associated with evolution and progression of several types of cancer mostly by providing proliferative advantage to transformed cells through various mechanisms. This review elucidates BMI1 abnormalities identified in hematologic malignancies and their implication in disease progression, early diagnosis, prognostic evaluations, and therapeutic targeting. In summary, discussed studies collectively provide new insight into molecular mechanisms underlying aberrant overexpression of BMI1 as potential contributor to the pathogenesis of hematologic malignancies. BMI1 deregulations are associated with hematologic malignancies of stem, progenitor, mature, and even terminally differentiated cells. Therefore, it becomes important to comprehensively examine the role of BMI1 in other hematologic malignancies that are not yet associated with its deregulations. Since, BMI1 does not possess its own enzymatic activity, the challenge in future lies in development of pharmacological modulator that specifically targets BMI1 expression and activity. Targeting BMI1 activity might offer more curative success for the hematologic malignancies associated with its aberrant activity.
Acknowledgments
I would like to thank Dr. Amogh A. Sahasrabuddhe, Dr. Delphine Rolland, and Dr. Thirunavukkarasu Velusamy for critical reading of this manuscript and their valuable inputs.
Footnotes
ACADEMIC EDITOR: Barbara Guinn, Editor in Chief
PEER REVIEW: One peer reviewers contributed to the peer review report. Reviewers’ reports totaled 323 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by Ramanujan Fellowship from the Department of Science and Technology, Government of India (SERB/F/4639/2015-16). The author confirms that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: The author discloses no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Provenance: the author was invited to submit this paper.
Author Contributions
AAS conceived the concept and wrote the manuscript. Author reviewed and approved of the final manuscript.
REFERENCES
- 1.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139(1):3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon JA, Kingston RE. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49(5):808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 6.Piunti A, Pasini D. Epigenetic factors in cancer development: polycomb group proteins. Future Oncol. 2011;7(1):57–75. doi: 10.2217/fon.10.157. [DOI] [PubMed] [Google Scholar]
- 7.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 9.Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19(2):150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16(11):643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14(12):853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 12.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20(10):1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 13.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8(4):263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 14.Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19(12):692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz YB, Pirrotta V. A new world of polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14(12):853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 16.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65(5):737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 17.Adorno M, Sikandar S, Mitra SS, et al. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature. 2013;501(7467):380–384. doi: 10.1038/nature12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allegra E, Trapasso S, Pisani D, Puzzo L. The role of BMI1 as a biomarker of cancer stem cells in head and neck cancer: a review. Oncology. 2014;86(4):199–205. doi: 10.1159/000358598. [DOI] [PubMed] [Google Scholar]
- 19.Brecqueville M, Adelaide J, Bertucci F, et al. Alterations of polycomb gene BMI1 in human myeloproliferative neoplasms. Cell Cycle. 2012;11(16):3141–3142. doi: 10.4161/cc.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24(2):117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy N. Metastasis: twisting BMI1. Nat Rev Cancer. 2010;10(10):666. doi: 10.1038/nrc2940. [DOI] [PubMed] [Google Scholar]
- 22.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118(4):409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Alkema MJ, Wiegant J, Raap AK, Berns A, van Lohuizen M. Characterization and chromosomal localization of the human proto-oncogene BMI-1. Hum Mol Genet. 1993;2(10):1597–1603. doi: 10.1093/hmg/2.10.1597. [DOI] [PubMed] [Google Scholar]
- 24.Ishida A, Asano H, Hasegawa M, et al. Cloning and chromosome mapping of the human Mel-18 gene which encodes a DNA-binding protein with a new ‘RING-finger’ motif. Gene. 1993;129(2):249–255. doi: 10.1016/0378-1119(93)90275-8. [DOI] [PubMed] [Google Scholar]
- 25.Wiederschain D, Chen L, Johnson B, et al. Contribution of polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol Cell Biol. 2007;27(13):4968–4979. doi: 10.1128/MCB.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18(2):536–546. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo WJ, Zeng MS, Yadav A, et al. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 2007;67(11):5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav AK, Sahasrabuddhe AA, Dimri M, Bommi PV, Sainger R, Dimri GP. Deletion analysis of BMI1 oncoprotein identifies its negative regulatory domain. Mol Cancer. 2010;9:158. doi: 10.1186/1476-4598-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi-1–Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281(29):20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 30.Balasubramanian S, Scharadin TM, Han B, Xu W, Eckert RL. The Bmi-1 helix-turn and ring finger domains are required for Bmi-1 antagonism of (-) epigallocatechin-3–gallate suppression of skin cancer cell survival. Cell Signal. 2015;27(7):1336–1344. doi: 10.1016/j.cellsig.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 31.Ginjala V, Nacerddine K, Kulkarni A, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31(10):1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itahana K, Zou Y, Itahana Y, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23(1):389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Beato M, Sanchez E, Gonzalez-Carrero J, et al. Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Mod Pathol. 2006;19(5):684–694. doi: 10.1038/modpathol.3800577. [DOI] [PubMed] [Google Scholar]
- 34.Huang R, Cheung NK, Vider J, et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. 2011;25(12):4138–4149. doi: 10.1096/fj.11-185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin A, Cano A. Tumorigenesis: twist1 links EMT to self-renewal. Nat Cell Biol. 2010;12(10):924–925. doi: 10.1038/ncb1010-924. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Zheng Y, Park HJ, et al. Targeting FoxM1 effectively retards p53–null lymphoma and sarcoma. Mol Cancer Ther. 2013;12(5):759–767. doi: 10.1158/1535-7163.MCT-12-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bommi PV, Dimri M, Sahasrabuddhe AA, Khandekar J, Dimri GP. The polycomb group protein BMI1 is a transcriptional target of HDAC inhibitors. Cell Cycle. 2010;9(13):2663–2673. doi: 10.4161/cc.9.13.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak K, Kerl K, Fehr D, et al. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 2006;34(6):1745–1754. doi: 10.1093/nar/gkl119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang HB, Liu GH, Zhang H, et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 2013;280(12):2929–2944. doi: 10.1111/febs.12299. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Chai L, Liu F, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci U S A. 2007;104(25):10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T, Chen X, Zhang W, et al. Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4: implications for colon cancer. J Biol Chem. 2012;287(6):3760–3768. doi: 10.1074/jbc.M111.316349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Sanchez-Tillo E, Lu X, et al. The ZEB1 transcription factor acts in a negative feedback loop with miR200 downstream of Ras and Rb1 to regulate Bmi1 expression. J Biol Chem. 2014;289(7):4116–4125. doi: 10.1074/jbc.M113.533505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X, Piao L, Cavey GS, et al. Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis. Oncogene. 2014;33(16):2040–2052. doi: 10.1038/onc.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang MH, Hsu DS, Wang HW, et al. Bmi1 is essential in Twist1–induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 45.Ismail IH, Gagne JP, Caron MC, et al. CBX4–mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40(12):5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voncken JW, Niessen H, Neufeld B, et al. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J Biol Chem. 2005;280(7):5178–5187. doi: 10.1074/jbc.M407155200. [DOI] [PubMed] [Google Scholar]
- 47.Nacerddine K, Beaudry JB, Ginjala V, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J Clin Invest. 2012;122(5):1920–1932. doi: 10.1172/JCI57477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Liu F, Yu H, et al. Akt phosphorylates the transcriptional repressor bmi1 to block its effects on the tumor-suppressing ink4a-arf locus. Sci Signal. 2012;5(247):ra77. doi: 10.1126/scisignal.2003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sahasrabuddhe AA, Dimri M, Bommi PV, Dimri GP. betaTrCP regulates BMI1 protein turnover via ubiquitination and degradation. Cell Cycle. 2011;10(8):1322–1330. doi: 10.4161/cc.10.8.15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya R, Mustafi SB, Street M, Dey A, Dwivedi SK. Bmi-1: at the crossroads of physiological and pathological biology. Genes Dis. 2015;2(3):225–239. doi: 10.1016/j.gendis.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Lugt NM, Domen J, Linders K, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 52.Ginjala V, Nacerddine K, Kulkarni A, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31(10):1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1–mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191(1):45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Cao L, Chen J, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459(7245):387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113(2):175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang L, Song L, Wu J, et al. Bmi-1 promotes glioma angiogenesis by activating NF-kappaB signaling. PLoS One. 2013;8(1):e55527. doi: 10.1371/journal.pone.0055527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen H, Zhou L, Wan G, Dou T, Tian J. BMI1 promotes the progression of laryngeal squamous cell carcinoma. Oral Oncol. 2011;47(6):472–481. doi: 10.1016/j.oraloncology.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Bednar F, Schofield HK, Collins MA, et al. Bmi1 is required for the initiation of pancreatic cancer through an Ink4a-independent mechanism. Carcinogenesis. 2015;36(7):730–738. doi: 10.1093/carcin/bgv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin T, Wei H, Leng Z, et al. Bmi-1 promotes the chemoresistance, invasion and tumorigenesis of pancreatic cancer cells. Chemotherapy. 2011;57(6):488–496. doi: 10.1159/000334103. [DOI] [PubMed] [Google Scholar]
- 60.Tong YQ, Liu B, Zheng HY, et al. Overexpression of BMI-1 is associated with poor prognosis in cervical cancer. Asia Pac J Clin Oncol. 2012;8(4):e55–e62. doi: 10.1111/j.1743-7563.2012.01564.x. [DOI] [PubMed] [Google Scholar]
- 61.Tong YQ, Liu B, Zheng HY, et al. BMI-1 autoantibody as a new potential biomarker for cervical carcinoma. PLoS One. 2011;6(11):e27804. doi: 10.1371/journal.pone.0027804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong P, Kaneuchi M, Watari H, et al. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer. 2011;10:99. doi: 10.1186/1476-4598-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren R, Liu W, Huang L, et al. Role of B lymphoma Mo-MLV insertion region 1 in the oncogenic behavior of retinoblastomas. Mol Vis. 2013;19:561–574. [PMC free article] [PubMed] [Google Scholar]
- 64.Kumamoto H, Ohki K. Detection of CD133, Bmi-1, and ABCG2 in ameloblastic tumors. J Oral Pathol Med. 2010;39(1):87–93. doi: 10.1111/j.1600-0714.2009.00807.x. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Li Y, Tan Y, Ma K, Cui J. Bmi-1 is critical for the proliferation and invasiveness of gastric carcinoma cells. J Gastroenterol Hepatol. 2010;25(3):568–575. doi: 10.1111/j.1440-1746.2009.06045.x. [DOI] [PubMed] [Google Scholar]
- 66.Liu JH, Song LB, Zhang X, et al. Bmi-1 expression predicts prognosis for patients with gastric carcinoma. J Surg Oncol. 2008;97(3):267–272. doi: 10.1002/jso.20934. [DOI] [PubMed] [Google Scholar]
- 67.Qin ZK, Yang JA, Ye YL, et al. Expression of Bmi-1 is a prognostic marker in bladder cancer. BMC Cancer. 2009;9:61. doi: 10.1186/1471-2407-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu WL, Guo XZ, Zhang LJ, et al. Prognostic relevance of Bmi-1 expression and autoantibodies in esophageal squamous cell carcinoma. BMC Cancer. 2010;10:467. doi: 10.1186/1471-2407-10-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Z, Min L, Chen D, et al. Overexpression of BMI-1 promotes cell growth and resistance to cisplatin treatment in osteosarcoma. PLoS One. 2011;6(2):e14648. doi: 10.1371/journal.pone.0014648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 71.Schuringa JJ, Vellenga E. Role of the polycomb group gene BMI1 in normal and leukemic hematopoietic stem and progenitor cells. Curr Opin Hematol. 2010;17(4):294–299. doi: 10.1097/MOH.0b013e328338c439. [DOI] [PubMed] [Google Scholar]
- 72.Mihara K, Takihara Y, Kimura A. Genetic and epigenetic alterations in myelodysplastic syndrome. Cytogenet Genome Res. 2007;118(2–4):297–303. doi: 10.1159/000108313. [DOI] [PubMed] [Google Scholar]
- 73.Mihara K, Chowdhury M, Nakaju N, et al. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood. 2006;107(1):305–308. doi: 10.1182/blood-2005-06-2393. [DOI] [PubMed] [Google Scholar]
- 74.Xu F, Yang R, Wu L, et al. Overexpression of BMI1 confers clonal cells resistance to apoptosis and contributes to adverse prognosis in myelodysplastic syndrome. Cancer Lett. 2012;317(1):33–40. doi: 10.1016/j.canlet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 75.Rizo A, Olthof S, Han L, Vellenga E, de Haan G, Schuringa JJ. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood. 2009;114(8):1498–1505. doi: 10.1182/blood-2009-03-209734. [DOI] [PubMed] [Google Scholar]
- 76.Sawa M, Yamamoto K, Yokozawa T, et al. BMI-1 is highly expressed in M0–subtype acute myeloid leukemia. Int J Hematol. 2005;82(1):42–47. doi: 10.1532/IJH97.05013. [DOI] [PubMed] [Google Scholar]
- 77.Chowdhury M, Mihara K, Yasunaga S, Ohtaki M, Takihara Y, Kimura A. Expression of Polycomb-group (PcG) protein BMI-1 predicts prognosis in patients with acute myeloid leukemia. Leukemia. 2007;21(5):1116–1122. doi: 10.1038/sj.leu.2404623. [DOI] [PubMed] [Google Scholar]
- 78.Nishida Y, Kojima K, Maeda A, et al. Prognostic impact and targeting of BMI-1 in acute myeloid leukemia; Paper presented at: Blood; San Francisco. December 6, 2014. [Google Scholar]
- 79.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 80.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 81.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 82.Rowley JD. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 83.Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315(6020):550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 84.Mohty M, Yong AS, Szydlo RM, Apperley JF, Melo JV. The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood. 2007;110(1):380–383. doi: 10.1182/blood-2006-12-065599. [DOI] [PubMed] [Google Scholar]
- 85.Saudy NS, Fawzy IM, Azmy E, Goda EF, Eneen A, Abdul Salam EM. BMI1 gene expression in myeloid leukemias and its impact on prognosis. Blood Cells Mol Dis. 2014;53(4):194–198. doi: 10.1016/j.bcmd.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 86.Datta S, Hoenerhoff MJ, Bommi P, et al. Bmi-1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer Res. 2007;67(21):10286–10295. doi: 10.1158/0008-5472.CAN-07-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoenerhoff MJ, Chu I, Barkan D, et al. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28(34):3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mourgues L, Imbert V, Nebout M, et al. The BMI1 polycomb protein represses cyclin G2–induced autophagy to support proliferation in chronic myeloid leukemia cells. Leukemia. 2015;29(10):1993–2002. doi: 10.1038/leu.2015.112. [DOI] [PubMed] [Google Scholar]
- 89.Sengupta A, Ficker AM, Dunn SK, Madhu M, Cancelas JA. Bmi1 reprograms CML B-lymphoid progenitors to become B-ALL-initiating cells. Blood. 2012;119(2):494–502. doi: 10.1182/blood-2011-06-359232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 91.Neumann M, Heesch S, Schlee C, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121(23):4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 92.Dik WA, Brahim W, Braun C, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19(11):1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 93.Strati P, Shanafelt TD. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood. 2015;126(4):454–462. doi: 10.1182/blood-2015-02-585059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rouhigharabaei L, Ferreiro JF, Put N, et al. BMI1, the polycomb-group gene, is recurrently targeted by genomic rearrangements in progressive B-cell leukemia/lymphoma. Genes Chromosomes Cancer. 2013;52(10):928–944. doi: 10.1002/gcc.22088. [DOI] [PubMed] [Google Scholar]
- 95.Bosch F, Jares P, Campo E, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood. 1994;84(8):2726–2732. [PubMed] [Google Scholar]
- 96.de Boer CJ, Schuuring E, Dreef E, et al. Cyclin D1 protein analysis in the diagnosis of mantle cell lymphoma. Blood. 1995;86(7):2715–2723. [PubMed] [Google Scholar]
- 97.Rimokh R, Berger F, Delsol G, et al. Detection of the chromosomal translocation t(11;14) by polymerase chain reaction in mantle cell lymphomas. Blood. 1994;83(7):1871–1875. [PubMed] [Google Scholar]
- 98.Bea S, Tort F, Pinyol M, et al. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61(6):2409–2412. [PubMed] [Google Scholar]
- 99.Teshima K, Nara M, Watanabe A, et al. Dysregulation of BMI1 and microRNA-16 collaborate to enhance an anti-apoptotic potential in the side population of refractory mantle cell lymphoma. Oncogene. 2014;33(17):2191–2203. doi: 10.1038/onc.2013.177. [DOI] [PubMed] [Google Scholar]
- 100.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Kemenade FJ, Raaphorst FM, Blokzijl T, et al. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97(12):3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- 102.van Galen JC, Muris JJ, Oudejans JJ, et al. Expression of the polycomb-group gene BMI1 is related to an unfavourable prognosis in primary nodal DLBCL. J Clin Pathol. 2007;60(2):167–172. doi: 10.1136/jcp.2006.038752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreau P, Attal M, Facon T. Frontline therapy of multiple myeloma. Blood. 2015;125(20):3076–3084. doi: 10.1182/blood-2014-09-568915. [DOI] [PubMed] [Google Scholar]
- 104.Jagani Z, Wiederschain D, Loo A, et al. The Polycomb group protein Bmi-1 is essential for the growth of multiple myeloma cells. Cancer Res. 2010;70(13):5528–5538. doi: 10.1158/0008-5472.CAN-09-4229. [DOI] [PubMed] [Google Scholar]
- 105.Abd Al, Kader L, Oka T, Takata K, et al. In aggressive variants of non-Hodgkin lymphomas, Ezh2 is strongly expressed and polycomb repressive complex PRC1.4 dominates over PRC1.2. Virchows Arch. 2013;463(5):697–711. doi: 10.1007/s00428-013-1428-y. [DOI] [PubMed] [Google Scholar]
- 106.Kitadate A, Ikeda S, Teshima K, et al. MicroRNA-16 mediates the regulation of a senescence-apoptosis switch in cutaneous T-cell and other non-Hodgkin lymphomas. Oncogene. 2015 doi: 10.1038/onc.2015.435. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 107.Cui J, Cheng Y, Zhang P, et al. Down regulation of miR200c promotes radiation-induced thymic lymphoma by targeting BMI1. J Cell Biochem. 2014;115(6):1033–1042. doi: 10.1002/jcb.24754. [DOI] [PubMed] [Google Scholar]
- 108.Zhang C, Toulev A, Kamarashev J, Qin JZ, Dummer R, Dobbeling U. Consequences of p16 tumor suppressor gene inactivation in mycosis fungoides and Sezary syndrome and role of the bmi-1 and ras oncogenes in disease progression. Hum Pathol. 2007;38(7):995–1002. doi: 10.1016/j.humpath.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 109.Kiel MJ, Sahasrabuddhe AA, Rolland DC, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.He Z, Xia Y, Pan C, et al. Up-regulation of MiR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37(1):387–398. doi: 10.1159/000430362. [DOI] [PubMed] [Google Scholar]
- 111.Chen T, Xu C, Chen J, et al. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol Lett. 2015;9(6):2639–2646. doi: 10.3892/ol.2015.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xi S, Xu H, Shan J, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest. 2013;123(3):1241–1261. doi: 10.1172/JCI61271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhu G, Wang Y, Mijiti M, Wang Z, Wu PF, Jiafu D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem Biophys Res Commun. 2015;465(2):194–199. doi: 10.1016/j.bbrc.2015.07.149. [DOI] [PubMed] [Google Scholar]
- 114.Cui JG, Zhao Y, Sethi P, et al. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98(3):297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 115.Fu J, Rodova M, Nanta R, et al. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013;15(6):691–706. doi: 10.1093/neuonc/not011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Peruzzi P, Bronisz A, Nowicki MO, et al. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro Oncol. 2013;15(9):1212–1224. doi: 10.1093/neuonc/not055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ye L, Yu G, Wang C, et al. MicroRNA128a, BMI1 polycomb ring finger oncogene, and reactive oxygen species inhibit the growth of U87 MG glioblastoma cells following exposure to Xray radiation. Mol Med Rep. 2015;12(4):6247–6254. doi: 10.3892/mmr.2015.4175. [DOI] [PubMed] [Google Scholar]
- 118.Tu Y, Gao X, Li G, et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013;73(19):6046–6055. doi: 10.1158/0008-5472.CAN-13-0358. [DOI] [PubMed] [Google Scholar]
- 119.Palumbo T, Faucz FR, Azevedo M, Xekouki P, Iliopoulos D, Stratakis CA. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene. 2013;32(13):1651–1659. doi: 10.1038/onc.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5(6):e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang YC, Jan CI, Peng CY, Lai YC, Hu FW, Yu CC. Activation of microRNA-494–targeting Bmi1 and ADAM10 by silibinin ablates cancer stemness and predicts favourable prognostic value in head and neck squamous cell carcinomas. Oncotarget. 2015;6(27):24002–24016. doi: 10.18632/oncotarget.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lo WL, Yu CC, Chiou GY, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol. 2011;223(4):482–495. doi: 10.1002/path.2826. [DOI] [PubMed] [Google Scholar]
- 123.Wang L, Liu JL, Yu L, et al. Downregulated miR-495 [corrected] inhibits the G1–S phase transition by targeting Bmi-1 in breast cancer. Medicine (Baltimore) 2015;94(21):e718. doi: 10.1097/MD.0000000000000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cho JH, Dimri M, Dimri GP. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J Biol Chem. 2015;290(16):10555–10567. doi: 10.1074/jbc.M114.624361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lugrin J, Parapanov R, Rosenblatt-Velin N, et al. Cutting edge: IL-1alpha is a crucial danger signal triggering acute myocardial inflammation during myocardial infarction. J Immunol. 2015;194(2):499–503. doi: 10.4049/jimmunol.1401948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song SJ, Poliseno L, Song MS, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154(2):311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kopp F, Oak PS, Wagner E, Roidl A. miR-200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression. PLoS One. 2012;7(11):e50469. doi: 10.1371/journal.pone.0050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yin J, Zheng G, Jia X, et al. A Bmi1–miRNAs cross-talk modulates chemotherapy response to 5–fluorouracil in breast cancer cells. PLoS One. 2013;8(9):e73268. doi: 10.1371/journal.pone.0073268. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Guo J, Li W, Shi H, et al. Synergistic effects of curcumin with emodin against the proliferation and invasion of breast cancer cells through upregulation of miR-34a. Mol Cell Biochem. 2013;382(1–2):103–111. doi: 10.1007/s11010-013-1723-6. [DOI] [PubMed] [Google Scholar]
- 131.Qian P, Banerjee A, Wu ZS, et al. Loss of SNAIL regulated miR-128–2 on chromosome 3p22.3 targets multiple stem cell factors to promote transformation of mammary epithelial cells. Cancer Res. 2012;72(22):6036–6050. doi: 10.1158/0008-5472.CAN-12-1507. [DOI] [PubMed] [Google Scholar]
- 132.Zhu Y, Yu F, Jiao Y, et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17(22):7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- 133.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci U S A. 2012;109(36):14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jones MF, Hara T, Francis P, et al. The CDX1–microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci U S A. 2015;112(13):E1550–E1558. doi: 10.1073/pnas.1503370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He X, Dong Y, Wu CW, et al. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med. 2013;18:1491–1498. doi: 10.2119/molmed.2012.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218–5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martinez-Fernandez M, Duenas M, Feber A, et al. A Polycomb-mir200 loop regulates clinical outcome in bladder cancer. Oncotarget. 2015;6(39):42258–42275. doi: 10.18632/oncotarget.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang X, Sun Y, Han S, Zhu W, Zhang H, Lian S. MiR-203 inhibits melanoma invasive and proliferative abilities by targeting the polycomb group gene BMI1. Biochem Biophys Res Commun. 2015;456(1):361–366. doi: 10.1016/j.bbrc.2014.11.087. [DOI] [PubMed] [Google Scholar]
- 139.Wei Y, Du Y, Chen X, et al. Expression patterns of microRNA-218 and its potential functions by targeting CIP2A and BMI1 genes in melanoma. Tumour Biol. 2014;35(8):8007–8015. doi: 10.1007/s13277-014-2079-6. [DOI] [PubMed] [Google Scholar]
- 140.Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012;181(5):1823–1835. doi: 10.1016/j.ajpath.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fu WM, Tang LP, Zhu X, et al. MiR-218–targeting-Bmi-1 mediates the suppressive effect of 1,6,7–trihydroxyxanthone on liver cancer cells. Apoptosis. 2015;20(1):75–82. doi: 10.1007/s10495-014-1047-3. [DOI] [PubMed] [Google Scholar]
- 142.Tu K, Li C, Zheng X, Yang W, Yao Y, Liu Q. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32(4):1571–1577. doi: 10.3892/or.2014.3386. [DOI] [PubMed] [Google Scholar]
- 143.Qi X, Li J, Zhou C, Lv C, Tian M. MicroRNA-320a inhibits cell proliferation, migration and invasion by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett. 2014;588(20):3732–3738. doi: 10.1016/j.febslet.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 144.Zhou L, Zhang WG, Wang DS, Tao KS, Song WJ, Dou KF. MicroRNA-183 is involved in cell proliferation, survival and poor prognosis in pancreatic ductal adenocarcinoma by regulating Bmi-1. Oncol Rep. 2014;32(4):1734–1740. doi: 10.3892/or.2014.3374. [DOI] [PubMed] [Google Scholar]
- 145.Dang Z, Xu WH, Lu P, et al. MicroRNA-135a inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma. Int J Biol Sci. 2014;10(7):733–745. doi: 10.7150/ijbs.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo S, Xu X, Tang Y, et al. miR-15a inhibits cell proliferation and epithelial to mesenchymal transition in pancreatic ductal adenocarcinoma by down-regulating Bmi-1 expression. Cancer Lett. 2014;344(1):40–46. doi: 10.1016/j.canlet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 147.Jin M, Zhang T, Liu C, et al. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014;74(15):4183–4195. doi: 10.1158/0008-5472.CAN-14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yu J, Lu Y, Cui D, et al. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol Rep. 2014;31(2):910–918. doi: 10.3892/or.2013.2897. [DOI] [PubMed] [Google Scholar]
- 149.Cui J, Cheng Y, Zhang P, et al. Down regulation of miR200c promotes radiation-induced thymic lymphoma by targeting BMI1. J Cell Biochem. 2014;115(6):1033–1042. doi: 10.1002/jcb.24754. [DOI] [PubMed] [Google Scholar]
- 150.Sugihara H, Ishimoto T, Watanabe M, et al. Identification of miR-30e* regulation of Bmi1 expression mediated by tumor-associated macrophages in gastrointestinal cancer. PLoS One. 2013;1839;8(11):e8. doi: 10.1371/journal.pone.0081839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu X, Jiang X, Li H, Guo L, Jiang W, Lu SH. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014;23(6):576–585. doi: 10.1089/scd.2013.0308. [DOI] [PubMed] [Google Scholar]
- 152.Bhattacharya R, Nicoloso M, Arvizo R, et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69(23):9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Saini S, Yamamura S, Majid S, et al. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71(19):6208–6219. doi: 10.1158/0008-5472.CAN-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun L, Yao Y, Liu B, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31(4):432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]