Abstract
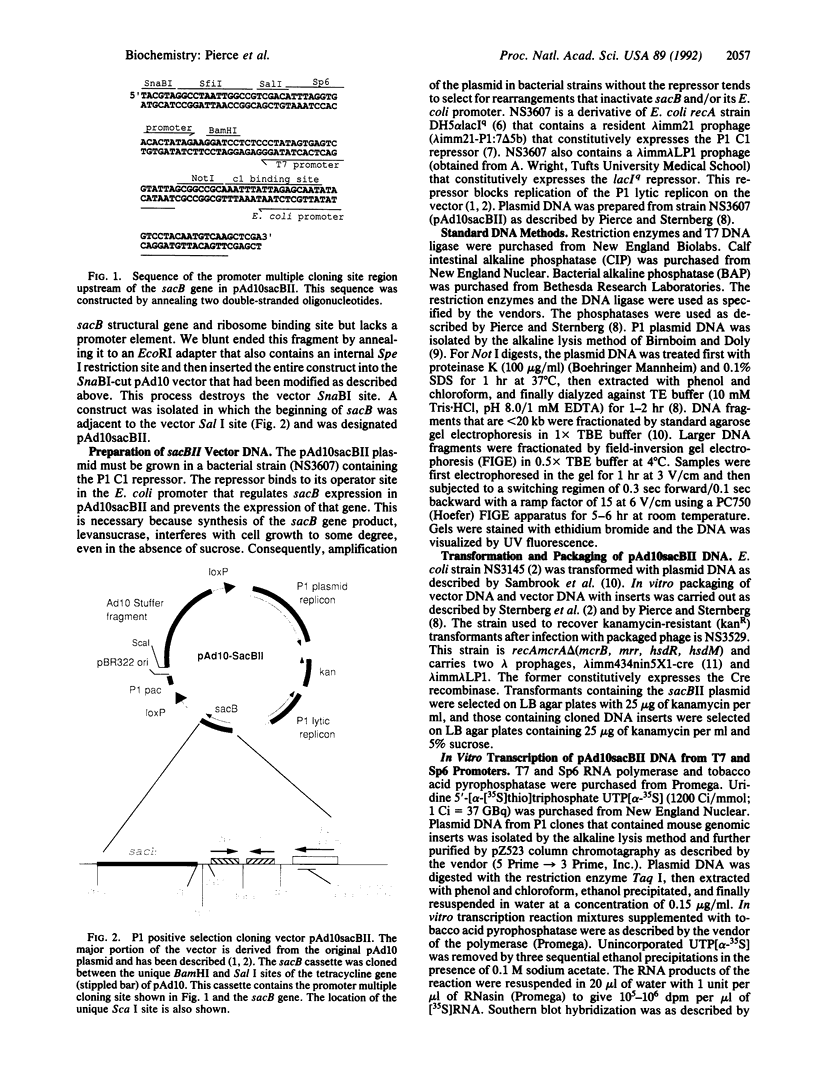
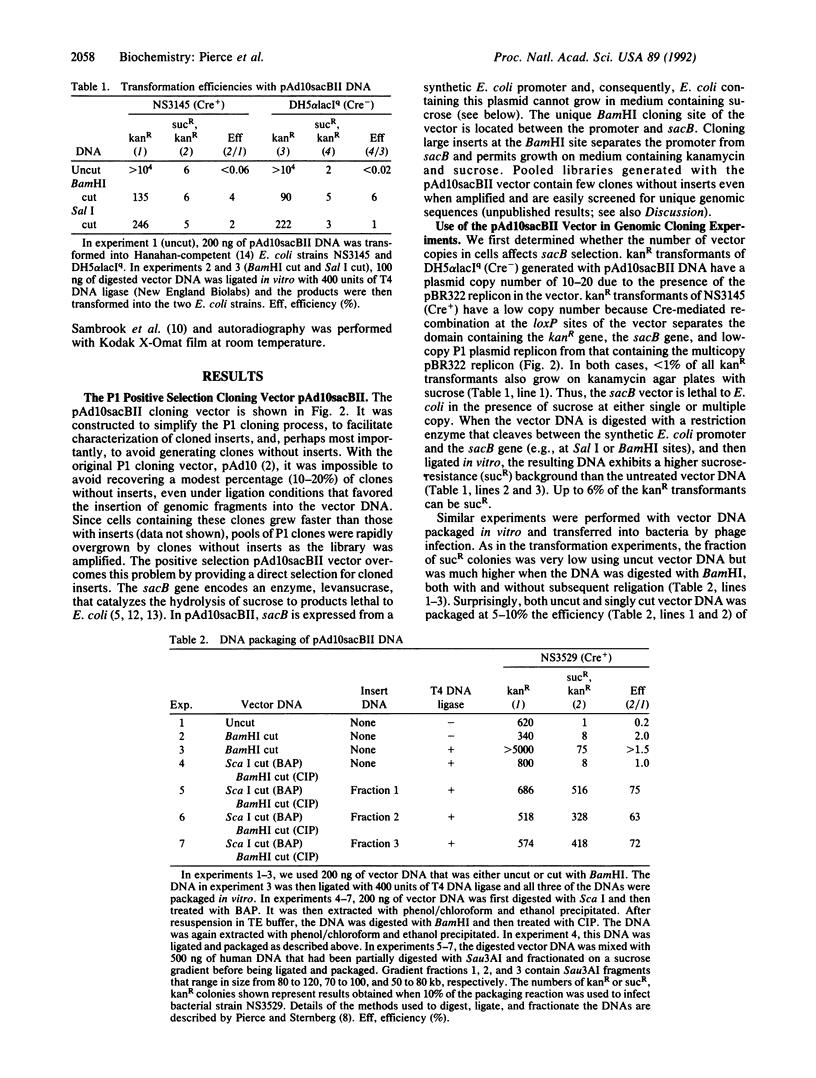
The bacteriophage P1 cloning system can package and propagate DNA inserts that are up to 95 kilobases. Clones are maintained in Escherichia coli by a low-copy replicon in the P1 cloning vector and can be amplified by inducing a second replicon in the vector with isopropyl beta-D-thiogalactopyranoside. To overcome the necessity of screening clones for DNA inserts, we have developed a P1 vector with a positive selection system that is based on the properties of the sacB gene from Bacillus amyloliquefaciens. Expression of that gene kills E. coli cells that are grown in the presence of sucrose. In the new P1 vector (pAd10sacBII) sacB expression is regulated by a synthetic E. coli promoter that also contains a P1 C1 repressor binding site. A unique BamHI cloning site is located between the promoter and the sacB structural gene. Cloning DNA fragments into the BamHI site interrupts sacB expression and permits growth of plasmid-containing cells in the presence of sucrose. We have also bordered the BamHI site with unique rare-cutting restriction sites Not I, Sal I, and Sfi I and with T7 and Sp6 promoter sequences to facilitate characterization and analysis of P1 clones. We describe here the use of Not I digestion to size the cloned DNA fragments and RNA probes to identify the ends of those fragments. The positive selection P1 vector provides a 65- to 75-fold discrimination of P1 clones that contain inserts from those that do not. It therefore permits generation of genomic libraries that are much easier to use for gene isolation and genome mapping than are our previous libraries. Also, the new vector makes it feasible to generate P1 libraries from small amounts of genomic insert DNA, such as from sorted chromosomes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliason J. L., Sternberg N. Characterization of the binding sites of c1 repressor of bacteriophage P1. Evidence for multiple asymmetric sites. J Mol Biol. 1987 Nov 20;198(2):281–293. doi: 10.1016/0022-2836(87)90313-5. [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Ferrari E., Hoch J. A. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983 Mar;153(3):1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene. 1988 Oct 30;70(2):331–341. doi: 10.1016/0378-1119(88)90205-3. [DOI] [PubMed] [Google Scholar]
- Smoller D. A., Petrov D., Hartl D. L. Characterization of bacteriophage P1 library containing inserts of Drosophila DNA of 75-100 kilobase pairs. Chromosoma. 1991 Sep;100(8):487–494. doi: 10.1007/BF00352199. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(1):103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Ruether J., deRiel K. Generation of a 50,000-member human DNA library with an average DNA insert size of 75-100 kbp in a bacteriophage P1 cloning vector. New Biol. 1990 Feb;2(2):151–162. [PubMed] [Google Scholar]
- Sternberg N., Sauer B., Hoess R., Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986 Jan 20;187(2):197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Emrich J., Stahl M. M. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A. 1967 Feb;57(2):292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L. B., Lenstra R., Borchert T. V., Nagarajan V. Isolation and characterization of levansucrase-encoding gene from Bacillus amyloliquefaciens. Gene. 1990 Nov 30;96(1):89–93. doi: 10.1016/0378-1119(90)90345-r. [DOI] [PubMed] [Google Scholar]