ABSTRACT
Chlamydia trachomatis is an obligate intracellular pathogen that is the etiological agent of a variety of human diseases, including blinding trachoma and sexually transmitted infections. Chlamydiae replicate within a membrane-bound compartment, termed an inclusion, which they extensively modify by the insertion of type III secreted proteins called Inc proteins. IncA is an inclusion membrane protein that encodes two coiled-coil domains that are homologous to eukaryotic SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) motifs. Recent biochemical evidence suggests that a functional core, composed of SNARE-like domain 1 (SLD-1) and part of SNARE-like domain 2 (SLD-2), is required for the characteristic homotypic fusion of C. trachomatis inclusions in multiply infected cells. To verify the importance of IncA in homotypic fusion in Chlamydia, we generated an incA::bla mutant. Insertional inactivation of incA resulted in the formation of nonfusogenic inclusions, a phenotype that was completely rescued by complementation with full-length IncA. Rescue of homotypic inclusion fusion was dependent on the presence of the functional core consisting of SLD-1 and part of SLD-2. Collectively, these results confirm in vitro membrane fusion assays identifying functional domains of IncA and expand the genetic tools available for identification of chlamydia with a method for complementation of site-specific mutants.
IMPORTANCE Chlamydia trachomatis replicates within a parasitophorous vacuole termed an inclusion. The chlamydial inclusions are nonfusogenic with vesicles in the endocytic pathway but, in multiply infected cells, fuse with each other to form a single large inclusion. This homotypic fusion is dependent upon the presence of a chlamydial inclusion membrane-localized protein, IncA. Specificity of membrane fusion in eukaryotic cells is regulated by SNARE (soluble N-ethylmaleimide sensitive factor attachment receptor) proteins on the cytosolic face of vesicles and target membranes. IncA contains two SNARE-like domains. Newly developed genetic tools for the complementation of targeted mutants in C. trachomatis are used to confirm the minimal requirement of SNARE-like motifs necessary to promote the homotypic fusion of inclusions.
INTRODUCTION
Chlamydiae are Gram-negative obligate intracellular bacteria that infect both humans and animals. All chlamydiae share a unique biphasic developmental cycle characterized by a metabolically inactive infectious elementary body (EB) and a metabolically active, replicative form termed the reticulate body (RB) (1). EBs bind specific receptors on a susceptible host cell, triggering internalization into a plasma membrane-derived vacuole that is rapidly modified by the bacteria to establish a replicative compartment termed the inclusion (2). Throughout the developmental cycle, the pathogen engages many host organelles, signaling networks, and the cytoskeleton to acquire host lipids, amino acids, and iron, all while suppressing activation of the immune response (3, 4). Starting at about 18 h, a proportion of RBs differentiate back to EBs which accumulate within the inclusion until they are released from the host cell by lysis or extrusion, which occurs between 48 and 72 h postinfection (5).
Chlamydiae encode a type III secretion system that is utilized to secrete bacterial effector proteins. A subset of these secreted effectors are inserted into the inclusion membrane and thus are poised to mediate crucial host-pathogen interactions (6). These inclusion membrane proteins, referred to as Incs, are defined by the presence of a bilobed hydrophobic domain of 40 amino acids (7). The presence of this domain has been used to predict up to 59 Incs in C. trachomatis (8–11); the number of Incs varies in other chlamydial species. Generation of specific antibodies (8, 9, 12, 13) and expression of epitope-tagged recombinant Incs in C. trachomatis (14, 15) have validated at least 37 of these as bona fide Incs; however, the function of most of these proteins is unknown.
In cells multiply infected with C. trachomatis, the inclusions fuse to form a single large vacuole (16, 17). This homotypic vesicle fusion is dependent on the presence of the IncA inclusion membrane protein. Microinjection of antibody to IncA into the cytosol of C. trachomatis-infected cells inhibits inclusion fusion (6). Clinical isolates deficient in IncA have been identified and also do not undergo inclusion fusion (18), supporting the idea of a role of IncA in this fusion event.
Eukaryotic SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) proteins play an integral role in membrane fusion through the assembly of multimeric complexes that facilitate or inhibit fusion of lipid bilayers (19–21). Given their crucial role in membrane fusion, SNARE proteins represent prime targets for bacterial effector proteins, particularly for those pathogens which occupy parasitophorous vacuoles. IncA, an ∼30-kDa inclusion membrane protein, possesses two putative α-helical SNARE-like domains, termed SNARE-like domain-1 (SLD-1) and SNARE-like domain 2 (SLD-2), that functionally mimic eukaryotic SNARE motifs (22–24). IncA is exposed on the cytosolic face of the inclusion membrane (6) and promotes homotypic fusion of inclusions but also inhibits SNARE-mediated membrane fusion, possibly to avoid fusion with endocytic compartments (6, 22–24). Recently, a functional domain, encompassing SLD-1 and part of SLD-2, has been identified (23, 24). However, it has not been determined whether this domain is necessary and sufficient for promoting inclusion fusion in C. trachomatis without IncA in a wild-type background (23, 24).
Until recently, a lack of genetic tools significantly hindered our ability to understand the molecular mechanisms of pathogenesis for this important obligate intracellular pathogen. Recent advances, including plasmid transformation (14, 25), generation of random mutants through chemical mutagenesis (26, 27), and adaption of a group II intron-based approach allowing selectable site-specific gene inactivation (28), have facilitated studies aimed at identifying and characterizing chlamydial virulence factors. In the current report, we highlight the importance of IncA in mediating homotypic inclusion fusion using these recently developed genetic approaches. Importantly, we implemented a complementation system that can be used to complement site-specific mutants. Through this system, we demonstrate the necessity of a functional core of IncA for promoting homotypic inclusion fusion in C. trachomatis.
MATERIALS AND METHODS
Bacterial and cell culture.
Chlamydia trachomatis serovar L2 (LGV 434/Bu) was propagated in HeLa 229 cells (American Type Culture Collection) and EBs were density gradient purified as previously described (29). HeLa and Vero CCL-81 cells (ATCC) were grown in RPMI 1640 media (Invitrogen) containing 10% fetal bovine serum (FBS) at 37°C and 5% CO2.
TargeTron construction.
All restriction enzymes and ligases, phosphatases, and DNA polymerases were purchased from New England BioLabs (NEB; Beverly, MA) unless otherwise specified. Oligonucleotides and primers used in this study were purchased from Integrated DNA Technologies (IDT; Skokie, IL) unless otherwise specified. All cloning was performed in Escherichia coli DH5α MAX Efficiency competent cells (Life Technologies, Carlsbad, CA).
The Targetron pACDK4-C plasmid was purchased from Sigma-Aldrich (Atlanta, GA) and modified for intron integration in chlamydia (Fig. 1A). The kanamycin–retrotransposition-activated marker cassette for postintegration selection was removed by digestion with MluI. The bla gene encoding penicillin resistance was PCR amplified from chlamydial plasmid pBOMB4 (14) using primers with 5′ MluI sites. The amplicon was digested with MluI and ligated into pACDK4-C to make pACDP4-C.
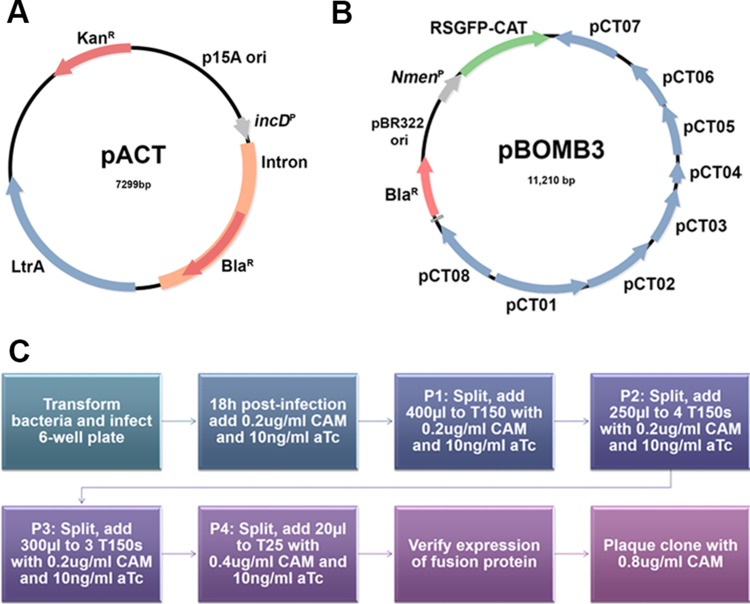
FIG 1.
Construction of pACT and pBOMB3 for mutation and complementation in C. trachomatis. (A) Map of pACT used for site-specific mutagenesis. (B) Map of pBOMB3 used for complementation. The pBOMB3 vector is similar to the previously reported pBOMB4 except that it contains the cat gene fused to the red-shifted green fluorescent protein gene, enabling selection for this vector with chloramphenicol. It also maintains the BamHI and PstI restriction sites within the native L2 plasmid backbone. (C) Flow chart depicting the methods used to transform and select for chloramphenicol (CAM)-resistant, complemented strains.
The chloramphenicol acetyltransferase resistance cassette (cat) located on the pACDK4-C plasmid backbone originally used for selection in E. coli was removed. The cat gene was removed by whole-plasmid PCR amplification of the pACDP4-C plasmid using primers pACD CATrmv F/R with incorporated 5′ XmaI sites. The primer sequences were specific to the up- and downstream regions immediately flanking cat in pACDP4-C, producing a linear DNA product of 6,486 bp. The DNA product was purified and then digested with XmaI. The aphA1 kanamycin resistance gene, an aminoglycoside 3-phosphotransferase suitable for selection in E. coli, was amplified from plasmid pMW1650 using primers pACD Kan Xma F/R containing 5′ XmaI restriction sites. pADP4-C and the purified aphA1 PCR product were digested with XmaI and ligated to form the pAP4-C kanamycin-resistant plasmid.
The T7 promoter was removed through digestion of pAP4-C with ClaI and HindIII and replaced with a polylinker containing three unique restriction sites for the modularity of promoters used to drive the group II intron. The polylinker contained (from 5′ to 3′) ClaI, DraIII, and HindIII sites and was inserted upstream of a 5′ exon by digestion of pAP4-C with ClaI and HindIII. The polylinker was ligated to the cut, phosphorylated pAP4-C vector to create pAPL-C. The upstream promoter region of chlamydial gene incD was amplified from C. trachomatis L2 genomic DNA using primers pACD incD-P F/R containing DraIII and HindIII sites incorporated into the forward and reverse primers, respectively. The PCR product was digested with DraIII and HindIII, purified, and then ligated to a DraIII/HindIII-digested pAPL-C vector to create pACT-C.
The intron was retargeted for C. trachomatis 434/Bu incA using the TargeTron computer algorithm (TargeTronics). Insertion sites with the highest score and the closest proximity to the 5′ ATG start codon were selected. Using the primers listed in Table 1, the intron was retargeted and amplified using a Qiagen core PCR kit (Qiagen). The PCR product was cloned into the BsrGI/HindIII site of pACT, and the ligated plasmid was transformed into methylation-deficient Escherichia coli K-12 ER2925 (New England BioLabs). The integrity of all constructs was verified by sequencing.
TABLE 1.
Primers used in this studya
| Primer name | Sequence | Use |
|---|---|---|
| CT119 24/25 IBS | AAAAAAGCTTATAATTATCCTTAGGCAACGGCTGCGTGCGCCCAGATAGGGTG | CT_119 KOa |
| CT119 24/25 EBS1 | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGGCTGCTATAACTTACCTTTCTTTGT | CT_119 KO |
| CT119 24/25 EBS2 | TGAACGCAAGTTTCTAATTTCGATTTTGCCTCGATAGAGGAAAGTGTCT | CT_119 KO |
| EBS Universal | AATTAGAAACTTGCGTTCAGTAAACACAACTTATAC | CT_119 KO |
| Tet Promoter Sacll F | CCCCGCGGATAATTTTAATTATATCACGGATCC | Complementation |
| Flag SalI R | GCGCGTCGACCTACTTGTCATCGTCATCCTTGTAGTC | Complementation |
| pACT Insert F seq | CAGATAAAATATTTCTAGCTAGATTTCAGTGC | Sequencing |
| pACT Insert R seq | CCAGTTAGTGTTAAGTCTTGGTAAATTCAG | Sequencing |
| pBOMB Tet F Seq | GGGTTGTTAAACCTTCGATTCCGACC | Sequencing |
| pBOMB R2 Seq | GCAAAAACAGGAAGGCAAAATGCCGC | Sequencing |
| CT119 NS NotI F | CCGCGGCCGCTTGATGGACAAAATTAAGAAAATAGC | Sequencing |
| CT119 NS Flag SalI R | CCGTCGACTTACTTATCGTCGTCATCCTTGTAATCGGAGCTTTTTGTAGAGGGTGAT | Sequencing |
KO, knockout.
Complementation vector construction.
The pBOMB3 construct was cloned using methods described by Bauler and Hackstadt (14) (Fig. 1B). Briefly, this vector contains regions from the pGFP::SW2 vector constructed by Wang et al. (25) but is modified to contain the entire L2 plasmid backbone and to remove any unnecessary regions, reducing vector size. There are two major differences between the pBOMB4 vector and the pBOMB3 vector. (i) The cat gene fused to the red-shifted green fluorescent protein-encoding gene was not removed from the pBOMB3 vector, enabling selection for this vector with chloramphenicol. (ii) the pBOMB3 vector maintains the BamHI and PstI restriction sites within the native L2 plasmid backbone, limiting the selection of enzymes available for use in the multiple-cloning site. To complement the incA::bla mutant, the tetracycline-inducible promoter and Flag-tagged incA were PCR amplified from pBOMB4-Tet-incA, pBOMB4-Tet-incA core, pBOMB4-Tet-incA 1-141, and pBOMB4-Tet-incA F/D 1-141 (23) using the primers listed in Table 1. All PCR products were cloned into the SacII/SalI site of pBOMB3 and transformed into E. coli K-12 ER2925.
Isolation of an incA::bla mutant.
C. trachomatis serovar L2 was transformed with pACT-incA as previously described (14, 15, 28). Plasmid DNA was transformed into C. trachomatis serovar L2 (LGV 434/Bu) density gradient-purified EBs using CaCl2 buffer (10 mM Tris [pH 7.5], 50 mM CaCl2). At 18 h postinfection, 0.1 U/ml penicillin G was added and cultures were incubated an additional 24 h. Following three passages, transformants were plaque cloned in Vero cells and individual plaques were picked and expanded. Disruption of incA was verified using PCR and sequencing of genomic DNA isolated from plaque-purified bacteria using a Qiagen blood and tissue kit (Qiagen).
Complementation of C. trachomatis incA::bla mutant.
To adapt a system of complementation for site-specific mutants, we focused on the incA::bla mutant because of the readily observed phenotype associated with disruption of IncA. A plaque-purified incA::bla mutant was expanded and EBs were density gradient purified as previously described (29). The transformation was conducted as previously described (30) with modifications outlined below and shown in Fig. 1C.
(i) A total of 107 purified EBs were mixed with 100 μl CaCl2 buffer and 3 μg DNA. Following a 30-min incubation at room temperature, 4.5 ml RPMI medium with 10% FBS was added, and 2 ml of the mixture was added to two wells of a HeLa monolayer in a 6-well plate. Cultures were centrifuged for 30 min at 2,400 rpm and were subsequently incubated at 37°C with 5% CO2.
(ii) At 18 h postinfection, 0.2 μg/ml chloramphenicol was added, IncA expression was induced with 10 ng/ml anhydrous tetracycline (aTc), and cultures were incubated an additional 24 h.
(iii) At 40 to 48 h postinfection, medium was removed and host cells were lysed in 1 ml sterile water/well. Host cell debris were pelleted for 5 min at 1,500 rpm, and 400 μl of supernatant was applied to a new HeLa monolayer in a T150 flask with RPMI 1640 plus 10% fetal bovine serum (FBS), 1 μg/ml cycloheximide, 0.2 μg/ml chloramphenicol, and 10 ng/ml aTc (passage 1 [P1]).
(iv) Following 40 to 48 h of incubation, cultures were split by lysing in 1 ml sterile water (total) and 250 μl was applied to 4 new HeLa monolayers (P2) in T150 flasks with RPMI 1640 plus 10% fetal bovine serum (FBS), 1 μg/ml cycloheximide, 0.2 μg/ml chloramphenicol, and 10 ng/ml aTc.
(v) At 40 to 48 h postinfection, the cultures were again split as described above and 300 μl of supernatant was applied to 3 new HeLa monolayers in a T150 flask containing RPMI 1640 plus 10% fetal bovine serum (FBS), 1 μg/ml cycloheximide, 0.2 μg/ml chloramphenicol, and 10 ng/ml aTc (P3).
(vi) At 40 to 48 h postinfection, the cultures were again split as described above and 20 μl of supernatant was applied to new HeLa monolayers in a T25 flask containing RPMI 1640 plus 10% fetal bovine serum (FBS), 1 μg/ml cycloheximide, 0.4 μg/ml chloramphenicol, and 10 ng/ml aTc (T3). The remaining supernatant was pelleted at 12,000 rpm for 20 min, and the pellet was resuspended in sucrose phosphate glycerol (SPG) buffer and stored at −80°C.
(vii) At 40 to 48 h postinfection, host cells were lysed as conducted above. The entire supernatant was pelleted at 12,000 rpm for 20 min and the pellet was resuspended in SPG buffer. Expression of the Flag-tagged fusion protein was verified using immunofluorescence microscopy.
(viii) Complemented mutants, expressing the fusion protein, were plaque cloned in Vero cells under selection with 0.8 μg/ml chloramphenicol, and individual plaques were picked and expanded.
Immunofluorescence.
HeLa cells were seeded at 105/ml onto glass coverslips and after 24 h were infected at an multiplicity of infection (MOI) of 1. IncA expression was induced with 10 ng/ml of aTc, and, 24 h postinfection, cells were fixed with 100% methanol and blocked using 1% BSA and DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). EBs were stained with anti-Momp antisera, and IncA was visualized using either anti-IncA antisera or an anti-Flag antibody (Sigma). Primary antibodies were observed by staining with goat anti-rabbit IgG–DyLight 594 and goat anti-mouse IgG–DyLight 488 (Jackson Laboratories) secondary antibodies. Images were captured on a Nikon Eclipse 80i fluorescence microscope and analyzed using Nikon Elements software. Data are representative of results of at least two independent experiments performed with at least 100 infected cells per experiment.
Growth curve analysis.
HeLa cells were infected on ice with an MOI of 1 with each bacterial strain, and expression of IncA was induced with 10 ng/ml aTc. After 30 min on ice, cultures were shifted to 37°C to stimulate bacterial uptake. At 0 h, 4 h, 12 h, 24 h, 36 h, and 48 h, postinfected cells were lysed in water and supernatants were applied to fresh HeLa monolayers to enumerate inclusion-forming units (IFUs).
Western blotting.
HeLa cells were infected with an MOI of 5, and expression of IncA was induced with 10 ng/ml aTc. At 24 h postinfection, cells were lysed in 1% SDS and analyzed by immunoblotting.
Southern blotting.
A 5-μg volume of genomic DNA from incA::bla L2 and wild-type C. trachomatis L2 was digested to completion with HindIII. As a positive control, 750 ng of purified pACT vector was also linearized by digestion with HindIII. Digested DNA was resolved in a 5-mm-thick 1% agarose gel, stained with ethidium bromide, and photographed in a UV cabinet. DNA ladder standards were marked, and the resolved DNA was transferred to a Hybond-N (GE Healthcare; Pittsburgh, PA) nylon membrane via capillary transfer with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (pH 7.0) overnight. Following transfer, the blot was UV cross-linked using a Stratalinker 1800 instrument (Stratagene, Los Angeles, CA) at 70,000 J/cm2. The Hybond-N membrane was dried and hybridized with an 866-bp probe specific to bla, an intron-specific gene encoding ß lactamase.
The probe was labeled with dCTP (α-32P) (PerkinElmer, Shelton, CT) (3,000 Ci/mmol, 10 mCi/ml) using a DecaPrime II labeling kit (Life Technologies) and then purified using Illustra microspin G-25 Sephadex columns (GE Healthcare, Pittsburgh, PA).
The membrane was prehybridized for 2 h at 42°C using 20 ml of hybridization solution (50% formamide, 6× SSC, 5× Denhardt's solution, 0.5% SDS) with 50 μg/ml denatured salmon sperm DNA. Following prehybridization, 20 ml of hybridization solution was combined with 10 μl of probe and the blot was hybridized with probe overnight at 42°C with shaking. The nylon membrane was washed, dried, and exposed to CL-Xposure film (Thermo Scientific; Atlanta, GA) for 24 h.
RESULTS
Generation of C. trachomatis incA::bla mutant.
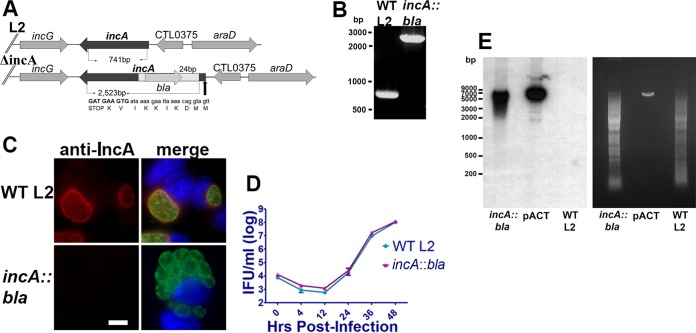
To disrupt IncA, the TargeTron pACD4K vector was modified for use in C. trachomatis (Fig. 1A) essentially as previously described (28), except the strong incD promoter was used to drive the intron in place of the CTL0665 promoter and kanamycin resistance was used for selection in E. coli. The intron was retargeted to incA using primer sequences obtained from TargeTronics. Primers that resulted in an insertion nearest the 5′ ATG and had the highest score were selected for retargeting. The resulting construct, pACT-incA24, was transformed into C. trachomatis serovar L2 (LGV 434/Bu), and transformants were expanded under conditions of penicillin G selection for four passages and were subsequently plaque purified. As previously described (28), viable inclusions were readily observed in passage 2 (P2) and nonfusogenic inclusions were apparent in P4 when the MOI exceeded 1. To verify disruption of IncA, genomic DNA was isolated from wild-type C. trachomatis serovar L2 (LGV 434/Bu) and from the L2 incA::bla mutant. PCR was employed to amplify incA using gene-specific primers. Sanger sequencing of incA::bla verified insertion of the intron in the antisense orientation relative to incA (Fig. 2A), as predicted by the TargeTronics algorithm. Agarose gel electrophoresis of the PCR product confirmed insertion of the group II intron as indicated by the 1.7-kB shift in the mutant band relative to the wild-type results (Fig. 2B). To confirm that loss of IncA expression was due to premature termination, immunofluorescence analysis of wild-type- and L2 incA::bla mutant-infected cells was conducted. As shown in Fig. 2C, IncA membrane staining was clearly evident on wild-type inclusions but was absent from incA::bla mutant-infected cells. Furthermore, nonfusogenic inclusions were present only in L2 incA::bla mutant-infected cells. No significant differences between the strains in growth rate were noted (Fig. 2D). Southern blotting was used to confirm a single intron insertion site (Fig. 2E). Collectively, these results indicate successful disruption of IncA.
FIG 2.
Generation of an incA::bla mutant. PCR was conducted on genomic DNA isolated from wild-type and incA::bla C. trachomatis L2. (A) Sanger sequencing of isolated DNA using gene-specific primers was conducted to verify orientation and insertion of the intron. The site of premature termination relative to the start site of C. trachomatis 434/Bu incA (YP_001654458) is shown. (B) PCR products were separated on a 1% agarose gel, and DNA was visualized using ethidium bromide staining. WT, wild type. (C) Immunofluorescence analysis was conducted on HeLa cells infected with an MOI of 1, and, after 24 h, cells were fixed with methanol and probed with anti-Momp (green) and anti-IncA (red) primary antibodies. Nuclei were stained using DAPI (blue). Data are representative of results of 3 independent experiments performed with at least 100 infected cells observed per experiment. Bar = 10 μm. (D) HeLa cells infected with wild-type L2 or incA::bla L2 were incubated for 0 h, 4 h, 12 h, 24 h, 36 h, or 48 h. Next, cells were lysed in water and replated on fresh HeLa monolayers to enumerate IFUs. Data are representative of results of 2 independent experiments. (E) Southern blot analysis of incA::bla L2, plasmid pACT containing bla, and parental C. trachomatis L2 (WT L2) using bla as a probe. A single insertion site is seen in incA::bla L2.
Generation of a complementation vector.
The pBOMB3 vector was constructed to facilitate complementation of genetic knockout mutants. It contains a chloramphenicol acetyltransferase gene (cat) to enable selection of the vector using chloramphenicol, an antibiotic that is not currently used for medical treatment of C. trachomatis infection and that has been shown to function for selection of plasmid maintenance in Chlamydia (30). The pBOMB3 vector contains the entire backbone of the native L2 plasmid; therefore, genetic knockout mutants complemented with this vector do not lack any additional genetic information formerly available on the native plasmid. Finally, the pBOMB3 vector encodes a red-shifted green fluorescent protein to visualize complemented organisms via immunofluorescence (Fig. 1B).
Complementation of incA::bla mutant rescues inclusion fusion.
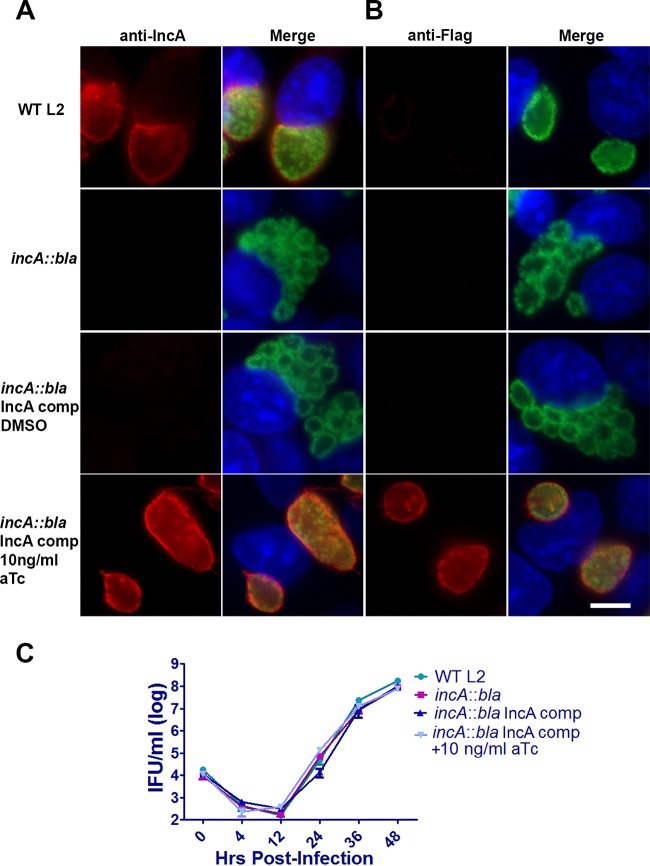
To verify that IncA is required for homotypic fusion of C. trachomatis inclusions, we expressed Flag-tagged IncA from a tetracycline-inducible promoter using the pBOMB3 vector. The resulting construct was transformed into infectious L2 incA::bla EBs that had been purified by Renografin density gradient centrifugation. Transformants were passaged and selected for by the use of 0.2 μg/ml chloramphenicol as outlined in Fig. 1C and as described in Materials and Methods. As shown in Fig. 3, induction of IncA expression by the addition of 10 ng/ml aTc resulted in IncA expression and localization to the inclusion membrane as evidenced by staining with anti-IncA (Fig. 3A) or anti-Flag (Fig. 3B) antibodies. In the absence of aTc induction, IncA expression and rescue of inclusion fusion were not observed in cells infected with the complemented strain. No growth difference was noted between the strains (Fig. 3C). Taken together, these results reinforce the idea of the importance of IncA in homotypic fusion and establish a system for complementing site-specific mutants in C. trachomatis.
FIG 3.
Complementation of incA::bla L2. (A and B) HeLa cells were infected at an MOI of 1 with wild-type L2 or incA::bla L2, and, after 24 h, cells were fixed with methanol and probed with (A) anti-IncA (red) and anti-Momp (green) or (B) anti-Flag (red) and anti-Momp (green) antibodies. Nuclei were visualized using DAPI (blue). Data are representative of results of 3 independent experiments performed with at least 100 infected cells per experiment. Bar = 10 μm. (C) For growth comparisons, HeLa cells were infected at an MOI of 1 with wild-type L2 or incA::bla L2. Infected cells were incubated for 0 h, 4 h, 12 h, 24 h, 36 h, or 48 h, at which point, cells were lysed in water and replated on fresh HeLa monolayers to enumerate IFUs. Data are representative of results of 2 independent experiments.
Homotypic fusion of inclusions requires a functional core of IncA.
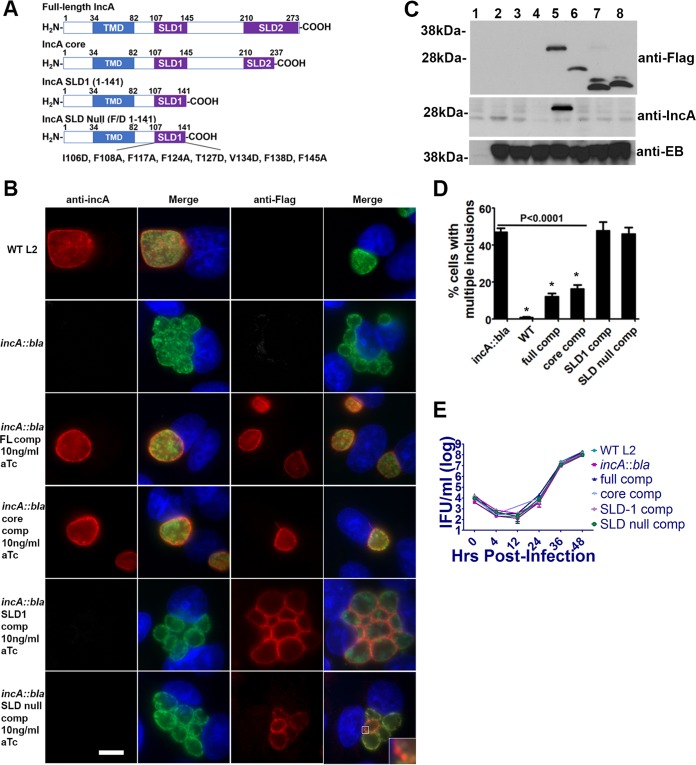
The ability to transform and overexpress Incs in wild-type chlamydiae has enabled experiments that assess the function of IncA during infection (23); however, the ability to generate site-specific mutants now allows more-direct evaluation of the function of IncA SNARE-like motifs in mediating inclusion fusion in the absence of a potentially confounding background of wild-type native IncA. IncA is composed of a bilobed hydrophobic domain (amino acids [aa] 34 to 82) and two SNARE-like domains termed SLD-1 (aa 107 to 145) and SLD-2 (aa 210 to 273) (Fig. 4A). Recent studies indicate that inclusion fusion is dependent on the presence of a functional core consisting of SLD-1 and part of SLD-2. To verify that inclusion fusion requires these domains, we expressed mutant IncA and full-length Flag-tagged IncA in the L2 incA::bla mutant under the control of a Tet promoter. HeLa cells were infected with C. trachomatis transformed with the various IncA constructs (Fig. 4A), and, 24 h postinfection, cells were stained using anti-IncA (Fig. 4B) or anti-Flag (Fig. 4C) antibodies. IncA staining at the inclusion membrane was evident for cells infected with C. trachomatis expressing either full-length IncA or the IncA core. Cells infected with the incA::bla mutant expressing IncA SLD-1 alone (pBOMB4-tet-incA 1-141) or an SLD-null construct (pBOMB4-tet-incA F/D 1-141) did not exhibit characteristic IncA staining. The IncA antibody used in this study is specific for the C terminus of IncA, confirming that the latter two strains lack the C terminus. Immunofluorescent staining of the recombinant chlamydia confirmed expression of each of the constructs and appropriate localization into the inclusion membrane of the full-length, core, and SLD1 constructs. The SLD-null (F/D 1-141) construct was not translocated to the inclusion membrane but appeared to be retained in association with the bacteria. Importantly, expression of the IncA core resulted in the formation of a single inclusion, comparably to wild-type C. trachomatis or cells complemented with full-length IncA. Multiple, nonfusogenic inclusions were observed in bacteria complemented with IncA SLD-1 or the SLD-null construct. To confirm the expression levels of the various constructs, immunoblotting was performed. As shown in Fig. 4C, expression levels of the induced constructs were equivalent as assessed by anti-Flag staining. Notably, expression levels of IncA under conditions of aTc induction were greater than that of IncA from the wild-type parental strain, as IncA was not expressed to levels detectable by anti-IncA in the parental L2 strain. The IncA antibody was produced against a domain near the C terminus of the protein and thus is not observed in the truncated versions of the protein. To better assess the fusogenicity of these inclusions, we scored cells infected with an MOI of 1 for the presence of multiple inclusions (more than 2 per cell). As shown in Fig. 4D, expression of the IncA core significantly rescued inclusion fusion, whereas mutants complemented with SLD-1 or the SLD-null construct exhibited defects comparable to those seen with the incA::bla mutation alone, with over half of the infected cells possessing multiple inclusions. Although complementation of the incA::bla mutant with either full-length IncA or the IncA core significantly restores inclusion fusion, the level of inclusion fusion for the IncA core was not equivalent to that seen with the parental L2 strain (P < 0.01). The failure of full-length or core IncA constructs to fully restore fusion to wild-type levels may be due to the abnormally high levels of IncA in the induced recombinant strains. Importantly, the differences in homotypic fusion were not due to differences in the replication rates, as all strains replicated with similar kinetics (Fig. 4E). Collectively, our results indicate that the functional core composed of SLD-1 and part of SLD-2 is necessary to promote IncA-mediated inclusion fusion.
FIG 4.
IncA-mediated homotypic fusion requires a functional core composed of SLD-1 and part of SLD-2. (A) IncA is composed of a 34-amino-acid N terminus, a bilobed transmembrane domain (amino acids 35 to 82), SLD-1 (amino acids 107 to 145), and SLD-2 (amino acids 210 to 273). (B) For immunofluorescence analysis, HeLa cells were infected at an MOI of 1 with each strain. At 24 h postinfection, cells were fixed with methanol and probed with anti-IncA (red) and anti-Momp (green) or anti-Flag (red) and anti-Momp (green) antibodies. Nuclei were visualized using DAPI (blue). The inset shows higher magnification of anti-Flag staining with the chlamydia expressing IncA SLD-null. Shown are wild-type L2 (WT L2); incA::bla L2; incA::bla L2 IncA full-length complemented (FL comp) induced; incA::bla L2 core complemented induced; incA::bla L2 SLD1 (1-141) complemented induced; and incA::bla L2 SLD null (F/D 1-141) complemented induced. Data are representative of results of 3 independent experiments performed with at least 100 infected cells per experiment. Bar = 10 μm. (C) HeLa cells, infected at an MOI of 2.5 for 24 h and subjected to immunoblotting using anti-Flag, anti-IncA, or anti-EB sera. The 40-kDa band representing Momp is shown. Samples are as follows: lane 1, uninfected cells; lane 2, wild-type L2; lane 3, incA::bla L2; lane 4, incA::bla L2 IncA full-length complemented uninduced; lane 5, incA::bla L2 IncA full-length complemented induced; lane 6, incA::bla L2 core complemented induced; lane 7, incA::bla L2 SLD1 (1-141) complemented induced; lane 8, incA::bla L2 SLD null (F/D 1-141) complemented induced. (D) Quantification of multiple inclusions (more than 2 per cell) was conducted by counting at least 200 infected cells in duplicate. Asterisks indicate differences from incA::bla L2 at a P level of <0.0001. Data are representative of results of 3 independent experiments. (E) Growth curve analyses were conducted with HeLa cells infected at an MOI of 1 with each strain. After 0 h, 4 h, 12 h, 24 h, 36 h, or 48 h, cells were lysed in water and replated on fresh HeLa monolayers to enumerate IFUs.
DISCUSSION
Until recently, the genetic intractability of chlamydiae significantly hindered our ability to understand this important pathogen. In the past decade, great strides have been made in applying modern molecular biology techniques to the study of chlamydiae. In 2011, Wang et al. (25) described a system for plasmid transformation of C. trachomatis EBs using calcium chloride and penicillin selection. Since that seminal study, the genetic toolbox available for use in C. trachomatis studies has been expanded to include expression of a variety of fluorescent tags (14, 31), epitope tags (14), inducible promoters (14), and alternative selectable markers (30, 32). Additionally, dendrimers have been used to silence gene expression (33, 34) and a library of random mutants has been generated using chemical mutagenesis (26, 27). Recently, a mobile group II intron was adapted for use in C. trachomatis that allows selectable insertional inactivation of target genes (28) and this technology has been extended to insertionally inactivate multiple genes (32). While complementation has been demonstrated for random chemical mutants, the ability to complement specific mutants harboring a selectable marker has been lacking. Here we adapted a chlamydial shuttle vector for complementation of mutants and used this system to verify that a functional core of IncA is required to mediate homotypic inclusion fusion.
To establish a method for complementation of C. trachomatis site-specific mutants, we focused on incA because of the readily observable phenotype associated with loss of incA. Naturally occurring IncA− mutants from clinical samples (18) and a incA::bla mutant generated by site-specific mutagenesis (35) exhibit a nonfusogenic phenotype. IncA is located in close proximity to other inc genes (incDEFG) but is regulated independently (13), suggesting that this phenotype is most likely not due to a polar effect. Here we used a modified chlamydial shuttle vector to express full-length Flag-tagged IncA under the control of an aTc-inducible promoter. Inducible expression of IncA rescued IncA localization to the inclusion membrane and homotypic fusion, confirming that IncA is necessary to promote the homotypic fusion of inclusions in C. trachomatis.
Previously, we showed that overexpression of recombinant, full-length IncA or IncA core in wild-type C. trachomatis has minimal effect on the characteristic homotypic fusion of the inclusions. In contrast, overexpression of either SLD-1 or SLD-2 acted in a dominant-negative fashion to block homotypic fusion with a concomitant increase in the percentage of cells exhibiting multiple, nonfused inclusions (23). Here, we used a group II intron mutant of IncA to evaluate IncA function in the absence of a background of wild-type IncA. While either wild-type IncA-flag or IncA core-flag substantially complemented homotypic fusion of the incA::bla mutant, the presence of IncA SLD-1 or the IncA SLD-null construct was insufficient to rescue homotypic fusion. Full-length IncA-flag, IncA-core, or IncA SLD-1 was localized appropriately to the inclusion membrane; however, the SLD-null construct was not observed on the inclusion membrane but appeared to remain associated with the bacteria. The phenylalanine/aspartic acid mutations introduced into the SLD-null IncA mutant (pBOMB4-tet-incA F/D 1-141) disrupted its alpha-helical structure, which was necessary to impair its function (21). This loss of secondary structure may account for the absence of secretion and its retention within the bacteria.
In addition to promoting homotypic fusion of C. trachomatis inclusions, wild-type IncA and IncA core also inhibit SNARE-mediated membrane fusion (23). The C. trachomatis L2 incA::bla mutant grew at a normal rate regardless of the presence or absence of IncA constructs that did or did not restore fusogenicity of the inclusion. The chlamydial inclusion is well known not to be fusogenic with endosomal/lysosomal compartments (36–39), and this avoidance of interactions with the endocytic pathway is evident quite early in infection (40), occurring before expression of IncA, which is not expressed until about 10 to 12 h in C. trachomatis L2 (9). It appears that IncA is not essential for avoidance of fusion with endocytic compartments, at least in vitro, and that there may be redundant mechanisms to avoid host defense mechanisms.
The number of molecular genetic tools for experimental manipulation of chlamydiae is increasing rapidly, and new tools promise to continue to strengthen the study of chlamydial pathogenesis. A robust means to complement mutants using a second, selectable marker such as chloramphenicol resistance adds a valuable tool for confirming the function of genes in chlamydia. As an example of how rapidly the field of chlamydia genetics is expanding, a means for allelic exchange by homologous recombination under conditions of beta-lactamase selection was recently described and with it a means for complementation using a separate plasmid expressing spectinomycin resistance (41). The availability of multiple selectable markers can only help accelerate the development of chlamydial genetic systems.
ACKNOWLEDGMENTS
We thank Tina Clark and Cheryl Dooley for excellent technical assistance.
This work was supported by the Intramural Research Program of the NIAID/NIH and by NIH grants AI073486 and AI116983 to F.P.
REFERENCES
- 1.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun 64:5366–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb Perspect Med 3:a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol 18:221–245. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- 5.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol 1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 7.Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol 2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun 76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 10.Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. 2011. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics 12:109. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutter EI, Martens C, Hackstadt T. 2012. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp Funct Genomics 2012:362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol 12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol 33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 14.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber MM, Bauler LD, Lam J, Hackstadt T. 2015. Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83:4710–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blyth WA, Taverne J. 1972. Some consequences of the multiple infection of cell cultures by TRIC organisms. J Hyg (Lond) 70:33–37. doi: 10.1017/S0022172400022063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridderhof JC, Barnes RC. 1989. Fusion of inclusions following superinfection of HeLa cells by two serovars of Chlamydia trachomatis. Infect Immun 57:3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun 68:360–367. doi: 10.1128/IAI.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahn R, Scheller RH. 2006. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 20.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. 1998. SNAREpins: minimal machinery for membrane fusion. Cell 92:759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- 21.Varlamov O, Volchuk A, Rahimian V, Doege CA, Paumet F, Eng WS, Arango N, Parlati F, Ravazzola M, Orci L, Sollner TH, Rothman JE. 2004. i-SNAREs: inhibitory SNAREs that fine-tune the specificity of membrane fusion. J Cell Biol 164:79–88. doi: 10.1083/jcb.200307066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delevoye C, Nilges M, Dehoux P, Paumet F, Perrinet S, Dautry-Varsat A, Subtil A. 2008. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog 4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronzone E, Wesolowski J, Bauler LD, Bhardwaj A, Hackstadt T, Paumet F. 2014. An alpha-helical core encodes the dual functions of the chlamydial protein IncA. J Biol Chem 289:33469–33480. doi: 10.1074/jbc.M114.592063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronzone E, Paumet F. 2013. Two coiled-coil domains of Chlamydia trachomatis IncA affect membrane fusion events during infection. PLoS One 8:e69769. doi: 10.1371/journal.pone.0069769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen BD, Valdivia R. 2012. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A 109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu S, Battaglia L, Bao X, Fan H. 2013. Chloramphenicol acetyltransferase as a selection marker for chlamydial transformation. BMC Res Notes 6:377. doi: 10.1186/1756-0500-6-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agaisse H, Derré I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ. 2015. Use of aminoglycoside 3′ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes 8:570. doi: 10.1186/s13104-015-1542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gérard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Kannan RM, Hudson AP. 2013. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9:996–1008. doi: 10.1016/j.nano.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Kannan RM, Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Hudson AP. 2013. Dendrimer-enabled transformation of Chlamydia trachomatis. Microb Pathog 65:29–35. doi: 10.1016/j.micpath.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KA, Lee JK, Chen AL, Tan M, Sutterlin C. 2015. Induction and inhibition of CPAF activity during analysis of Chlamydia-infected cells. Pathog Dis 73:1–8. doi: 10.1093/femspd/ftv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friis RR. 1972. Interaction of L cells and Chlamydia psittaci: entry of the parasite and host responses to its development. J Bacteriol 110:706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eissenberg LG, Wyrick PB, Davis CH, Rumpp JW. 1983. Chlamydia psittaci elementary body envelopes: ingestion and inhibition of phagolysosome fusion. Infect Immun 40:741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taraska T, Ward DM, Ajioka RS, Wyrick PB, Davis-Kaplan SR, Davis CH, Kaplan J. 1996. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun 64:3713–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scidmore MA, Fischer E, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller KE, Wolf K, Fields KA. 2016. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio 7:e01817-15. doi: 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]