ABSTRACT
The general stress response (GSR) system of the intracellular pathogen Brucella abortus controls the transcription of approximately 100 genes in response to a range of stress cues. The core genetic regulatory components of the GSR are required for B. abortus survival under nonoptimal growth conditions in vitro and for maintenance of chronic infection in an in vivo mouse model. The functions of the majority of the genes in the GSR transcriptional regulon remain undefined. bab1_1070 is among the most highly regulated genes in this regulon: its transcription is activated 20- to 30-fold by the GSR system under oxidative conditions in vitro. We have solved crystal structures of Bab1_1070 and demonstrate that it forms a homotetrameric complex that resembles those of WrbA-type NADH:quinone oxidoreductases, which are members of the flavodoxin protein family. However, B. abortus WrbA-related protein (WrpA) does not bind flavin cofactors with a high affinity and does not function as an NADH:quinone oxidoreductase in vitro. Soaking crystals with flavin mononucleotide (FMN) revealed a likely low-affinity binding site adjacent to the canonical WrbA flavin binding site. Deletion of wrpA (ΔwrpA) does not compromise cell survival under acute oxidative stress in vitro or attenuate infection in cell-based or mouse models. However, a ΔwrpA strain does elicit increased splenomegaly in a mouse model, suggesting that WrpA modulates B. abortus interaction with its mammalian host. Despite high structural homology with canonical WrbA proteins, we propose that B. abortus WrpA represents a functionally distinct member of the diverse flavodoxin family.
IMPORTANCE Brucella abortus is an etiological agent of brucellosis, which is among the most common zoonotic diseases worldwide. The general stress response (GSR) regulatory system of B. abortus controls the transcription of approximately 100 genes and is required for maintenance of chronic infection in a murine model; the majority of GSR-regulated genes remain uncharacterized. We present in vitro and in vivo functional and structural analyses of WrpA, whose expression is strongly induced by GSR under oxidative conditions. Though WrpA is structurally related to NADH:quinone oxidoreductases, it does not bind redox cofactors in solution, nor does it exhibit oxidoreductase activity in vitro. However, WrpA does affect spleen inflammation in a murine infection model. Our data provide evidence that WrpA forms a new functional class of WrbA/flavodoxin family proteins.
INTRODUCTION
Bacterial pathogens face fluctuating chemical and physical challenges in the host environment, including low pH, antimicrobial peptides, nutrient sequestration, and oxidative burst. These conditions can damage essential cellular components, including nucleic acids, proteins, and lipids. As such, bacteria encode a variety of protection mechanisms, including antioxidant enzymes, DNA damage repair systems, and efflux systems (1–4), which enable the cell to resist attacks by the host immune system (5–7).
The diverse flavodoxin family, which is composed of enzymes that noncovalently bind a flavin cofactor and transfer electrons between a variety of substrates, can help to maintain cellular redox balance and confer resistance to oxidative stress (8–10). Flavodoxins and flavodoxin-like proteins are also known to promote pathogen virulence and to play a role in host infection and colonization (8, 11–18), in addition to important roles in enzyme activation and metabolism (15). The flavodoxin-like protein WrbA (tryptophan [W] repressor binding protein) has been described for several microbial species, in which it is proposed to facilitate adaptation to varied chemical changes in the environment (13, 19–31). Though WrbA was initially reported as a TrpR (trp repressor) binding protein (24), data supporting its function as a regulator of TrpR are not conclusive (32). In Escherichia coli, wrbA transcription is activated by σS during stationary phase or when the organism is challenged with acid, salt, H2O2, or diauxic stress (23–30) and is repressed when the quinone pool is reduced (22) or by fumarate and nitrate reductase (FNR) proteins under anaerobic conditions (21). WrbA-like proteins have also been reported to confer oxidative stress protection in Pseudomonas aeruginosa (13, 31) and to promote persistence in a mouse model of Yersinia pseudotuberculosis infection (20).
WrbA binds a flavin mononucleotide (FMN) cofactor and adopts a characteristic β-α-β fold with a five-stranded, parallel β-sheet surrounded by five α-helices (30, 31, 33–38). Three conserved insertions, in β-strand 5 (loop 2), before α-helix 5 (loop 3), and between β2 and α2b (loop 1; contains α2a), supplement this core topology (31, 33, 35, 38). Insertion loop 1 distinguishes WrbA from classic flavodoxins (15). WrbA is tetrameric in solution (30, 32, 39, 40) and in crystal structures (31, 33, 36, 38), which is distinct from classic flavodoxins, which are monomeric/dimeric (15, 16, 32, 41–44). The WrbA tetramer presents four active sites, each with a bound flavin cofactor. Each active site can accommodate a nicotinamide cofactor as an electron donor or a quinone as an electron acceptor but not both simultaneously (45). This structural feature enables serial flavin reduction and oxidation by its substrates during electron transfer reactions (33).
This study focuses on a functional and structural characterization of a WrbA-related protein (WrpA) encoded by the intracellular mammalian pathogen Brucella abortus (46, 47). Analogous to σS activation of wrbA expression in E. coli, transcription of wrpA and its homologs is strongly activated by the general stress response (GSR) sigma factor, σE1/σEcfG (48, 49), in B. abortus and related alphaproteobacteria (19). Considering that σE1 enables growth and survival under a range of nonoptimal growth conditions, including several experienced by B. abortus cells in the mammalian host (5–7, 50, 51), we tested the hypothesis that WrpA functions as an enzyme that facilitates cellular adaptation to particular stress conditions in vitro or in vivo. Unlike strains lacking the primary general stress regulators (σE1, PhyR, or LovhK) (48, 49), a B. abortus ΔwrpA strain was not deficient in a hydrogen peroxide stress survival assay, nor was it deficient in growth under 1,920 metabolic and drug/chemical conditions we assayed in vitro. Moreover, the ΔwrpA strain was not attenuated in a macrophage infection model and had no colonization deficit in the spleen in a BALB/c mouse infection model. However, mice infected with a ΔwrpA null strain did display increased splenomegaly compared to mice infected with wild-type (WT) B. abortus at 4 weeks postinfection, suggesting that WrpA affects the inflammatory capacity of B. abortus in an animal host. These results provide evidence that wrpA is not a major downstream genetic contributor to the stress survival and animal infection deficits observed in strains harboring mutations in the GSR regulatory genes rpoE1, phyR, and lovhK.
Given the results of our in vitro and in vivo analyses of WrpA function, we further tested whether the biochemical and structural properties of WrpA differ from those of closely related WrbA proteins characterized for other bacterial species. We solved high-resolution structures of B. abortus WrpA in two crystal forms and characterized its biochemical properties. WrpA is a tetramer in crystals and in solution and possesses a WrbA/flavodoxin-type fold, but it failed to bind a flavin cofactor in solution and did not exhibit NADH:quinone oxidoreductase activity in vitro, even in the presence of high concentrations of flavin and nicotinamide cofactors. Soaking one of our two crystal forms of WrpA with high concentrations of FMN revealed a clear electron density for FMN at a site adjacent to the FMN binding pocket described for E. coli WrbA, though the functional relevance of this binding site remains undefined. Our data provide evidence that WrpA, although structurally homologous to WrbA NADH:quinone oxidoreductase enzymes, is part of a functionally distinct class of protein.
MATERIALS AND METHODS
All experiments on live B. abortus cells were performed at the University of Chicago Howard Taylor Ricketts Laboratory under biosafety level 3 (BSL3) conditions per CDC select agent regulations.
Chromosomal deletion of wrpA in B. abortus.
The B. abortus wrpA deletion strain was built by use of a double-recombination strategy. Briefly, the ΔwrpA in-frame deletion allele (carrying 5′- and 3′-flanking sequences of B. abortus locus tag bab1_1070/BAB_RS21040) was cloned into the suicide plasmid pNPTS138 (M. R. K. Alley, unpublished data), which carries the nptI gene for initial selection and the sacB gene for counterselection on sucrose. Wild-type B. abortus strain 2308 was transformed with pNPTS138-ΔwrpA by electroporation, and single-crossover integrants were selected on Schaedler blood agar (SBA) plates supplemented with kanamycin (Kan; 25 μg/ml). Counterselection for the second crossover was carried out by growing Kan-resistant colonies overnight under nonselective conditions and by plating the overnight cultures on SBA plates supplemented with sucrose (150 mM) (52, 53). We performed PCR screening to identify colonies in which the wild-type allele was replaced with the deletion allele. Genetic complementation of the deletion strain was carried out by transforming the B. abortus ΔwrpA strain with the pNPTS138-wrpA plasmid, carrying the wild-type wrpA allele. Primer and strain information is available in Tables S1 and S2 in the supplemental material.
Oxidative stress assay.
B. abortus wild-type, ΔwrpA, and ΔrpoE1 strains (strain information is available in Table S2 in the supplemental material) grown on SBA plates for 48 h were harvested and resuspended in Gerhardt's minimal medium, pH 6.8 (GMM). Each cell sample was then adjusted to a final cell density of 1 × 108 CFU/ml in 2 ml of GMM. The test group was subjected to oxidative stress by the addition of 5 mM H2O2 (final concentration); the control group was mock treated with sterile water. After 1 h of incubation in a shaking incubator at 37°C, cultures were serially diluted and plated on tryptic soy agar (TSA) for counting of viable CFU. Each experiment was performed at least three times for each strain.
Identification of compounds that support B. abortus metabolism in axenic culture.
Bacterial phenotype microarray (PM) assays (Biolog) were carried out according to the supplier's protocols. Briefly, B. abortus strains (wild-type, ΔwrpA, and ΔrpoE1 strains; strain information is available in Table S2 in the supplemental material) were streaked on an SBA plate, cultivated at 37°C and 5% CO2 for 48 h, restreaked, and grown for another 48 h. Cells were scraped off the plate and resuspended in 1× IF-0a medium (Biolog) at a final density equivalent to 5% transmittance (optical density [OD]) at 600 nm. PM inoculating fluids (PM1 to PM9+) were prepared from 0.2-μm-filter-sterilized stock solutions at the following final concentrations: for PM1 and -2, 2 mM MgCl2, 1 mM CaCl2, 25 μM l-arginine, 50 μM l-glutamic acid, 5 μM β-NAD, 25 μM hypoxanthine, 25 μM 5′-UMP, 25 μM l-cystine (pH 8.5), 0.005% yeast extract, and 0.005% Tween 40; for PM3, -6, -7, and -8, 20 mM tricarballylic acid (pH 7.1), 2 mM MgCl2, 1 mM CaCl2, 5 μM β-NAD, 25 μM hypoxanthine, 25 μM 5′-UMP, 25 μM l-cystine (pH 8.5), 0.005% yeast extract, 0.005% Tween 40, 0.625 mM d-glucose, and 5 mM pyruvate; for PM4, 20 mM tricarballylic acid (pH 7.1), 2 mM MgCl2, 1 mM CaCl2, 25 μM l-arginine, 50 μM l-glutamic acid, 0.005% yeast extract, 0.005% Tween 40, 0.625 mM d-glucose, and 5 mM pyruvate; for PM5, 20 mM tricarballylic acid (pH 7.1), 2 mM MgCl2, 1 mM CaCl2, 0.625 mM d-glucose, and 5 mM pyruvate; and for PM9+, 2 mM MgCl2, 1 mM CaCl2, 0.005% yeast extract, 0.005% Tween 40, 2.5 mM d-glucose, and 5 mM pyruvate. Inoculating fluids PM1 to -5 were prepared in 1× IF-0a GN/GP medium (final concentration; Biolog), while PM9+ inoculating fluid was prepared in 1× IF-10b GN/GP medium (final concentration; Biolog). All solutions had dye mix G (Biolog) added to a final concentration of 1×. A 1:13.6 dilution of 5% transmittance Brucella suspension was added to each inoculating fluid. One hundred microliters of Brucella-containing inoculating fluid per well was dispensed into each PM plate. PM plates were incubated at 37°C and 5% CO2, and the absorbance at 630 nm was measured every 12 to 24 h for up to 5 days, using a Tecan Infinite 200 Pro microplate reader. Each screening experiment was performed two times for each strain.
Cell culture and macrophage infection assay.
Human monocytic THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% heat-treated fetal bovine serum (FBS) and 2 mM l-glutamine and were differentiated into inactivated macrophages by the addition of 40 ng/ml phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) for 48 h at 37°C in a 5% CO2 atmosphere. For differentiation into activated macrophages, 40 ng/ml of PMA and 150 ng/ml of gamma interferon (IFN-γ; PrepoTech) were added to the THP-1 cell cultures for 48 h at 37°C in a 5% CO2 atmosphere. For differentiation into immature dendritic cells, 100 ng/ml of recombinant human interleukin-4 (rhIL-4; Sigma-Aldrich) and 100 ng/ml of recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; Sigma-Aldrich) were added to the THP-1 cell cultures for 48 h at 37°C in a 5% CO2 atmosphere.
Prior to infection, bacteria were harvested from freshly plated SBA plates and resuspended in sterile RPMI medium, and cell densities were adjusted. For infection assays, 5 × 104 cells were infected with 5 × 106 brucellae (wild-type or ΔwrpA strain [see Table S2 in the supplemental material for strain information]) to achieve a multiplicity of infection (MOI) of 1:100 in 96-well plates for 1 h in triplicate. Following infection, extracellular bacteria were removed by treatment with gentamicin (50 μg/ml) for 20 min. To determine the numbers of intracellular bacteria at 1 h, 24 h, 48 h, and 72 h postinfection, cells (with the exception of immature dendritic cells) were washed three times with phosphate-buffered saline (PBS) and lysed with 0.1 ml of 0.1% Triton X-100. The lysate was serially diluted and plated on TSA plates for CFU counting. Each infection experiment was performed in triplicate.
Mouse infection assay.
Bacteria (WT, ΔwrpA, and ΔwrpA::wrpA strains [see Table S2 in the supplemental material for strain information]) harvested from freshly plated SBA plates were resuspended in sterile PBS and diluted to a final concentration of 5 × 105 CFU/ml. One hundred microliters of each bacterial suspension was injected intraperitoneally into 6-week-old female BALB/c mice (Harlan). At weeks 1, 4, 8, and 12 postinfection, 5 mice per strain were sacrificed, and spleens were removed for weighing and CFU counting. For bacterial enumeration, each spleen was homogenized in 5 ml of sterile PBS supplemented with 0.1% Tween 20. Each homogenized spleen solution was then serially diluted, and 10-μl aliquots were plated on TSA plates for CFU counting. All mouse studies were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC).
Recombinant expression plasmids.
Primers used for E. coli wrbA and B. abortus wrpA and wrbA(Y80F) PCR amplification are listed in Table S1 in the supplemental material. After purification, the different PCR products were either treated with T4 polymerase (NEB) and directly inserted into the pMCSG68 expression plasmid by a ligation-independent cloning technique or digested with NdeI-XhoI and ligated into the pET28c expression plasmid. Plasmids were then transformed into E. coli Top10 cells. After sequencing, plasmids with the different inserts were purified and transformed into E. coli Rosetta (DE3)pLysS for protein expression (see Table S2 for strain information).
Protein expression and purification.
A 100-ml overnight Luria broth (LB; Fisher) culture inoculated with the protein expression strains (see Table S2 in the supplemental material for strain information) was used to inoculate 1 liter of LB medium supplemented with the appropriate antibiotics. Overexpression of His-tagged WrbA and WrpA (used for P4222 crystal form) proteins was induced at an OD600 of ∼0.8 (37°C, 220 rpm) by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; GoldBio). After 5 h of induction, cells were harvested by centrifugation at 11,000 × g for 20 min at 4°C. Cell pellets were resuspended in 20 ml of 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM imidazole buffer supplemented with 5 μg/ml of DNase I and half a tablet of complete protease inhibitor cocktail (Roche). Cells were disrupted by one passage in a microfluidizer (Microfluidics LV1). The resulting cell lysate was clarified by centrifugation at 39,000 × g for 20 min at 4°C.
Purification of His-tagged proteins was performed using nickel affinity chromatography (nitrilotriacetic acid [NTA] resin; GE Healthcare). After binding of the clarified lysate samples to the column, three washing steps were performed, using Tris-NaCl buffers containing 10 mM, 30 mM, and 75 mM imidazole, followed by elution with Tris-NaCl buffer containing 500 mM imidazole. All purification steps were carried out at 4°C. The protein purity of the different samples was assessed by 14% SDS-PAGE and staining with Coomassie blue. B. abortus WrpA strongly precipitates if imidazole is removed from the buffer. As such, the B. abortus and E. coli purified proteins were dialyzed against Tris-NaCl buffer (Tris-HCl, pH 7.4, 150 mM NaCl) containing 200 mM imidazole.
For purification of the protein used for crystallization of the WrpA P43212 crystal form, a 1-liter culture of M9 medium was grown at 37°C (190 rpm). At an OD600 of ∼1, the culture was cooled to 4°C and supplied with 90 mg of l-selenomethionine (Se-Met) and 25 mg each of methionine biosynthetic inhibitory amino acids (l-valine, l-isoleucine, l-leucine, l-lysine, l-threonine, and l-phenylalanine). Protein expression was induced overnight at 18°C with 0.5 mM IPTG. Cell pellets were resuspended in lysis buffer (500 mM NaCl, 5% [vol/vol] glycerol, 50 mM HEPES, pH 8.0, 20 mM imidazole, and 10 mM β-mercaptoethanol) and treated with lysozyme (1 mg/ml). Debris was removed by centrifugation. Se-Met WrpA was purified via Ni-NTA affinity chromatography, using an AKTAxpress system (GE Health Systems). The protein was washed and eluted using a linear imidazole gradient (10 to 250 mM). Immediately after purification, the His tag was cleaved at 4°C for 24 to 48 h, using a recombinant His-tagged tobacco etch virus (TEV) protease, resulting in an untagged WrpA protein with an N-terminal Ser-Asn-Ala peptide. An additional Ni-NTA purification step was performed to remove the protease, uncut protein, and affinity tag. The purified protein was then dialyzed against a 20 mM HEPES, pH 8.0, 250 mM NaCl, and 2 mM dithiothreitol (DTT) buffer.
For every construct, protein concentrations were estimated by UV absorption spectroscopy (280 nm) using a NanoDrop 1000 spectrophotometer (Thermo Scientific) or by use of a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). When necessary, purified proteins were concentrated using a centrifugal filter unit (3-kDa molecular mass cutoff; Amicon-Millipore).
UV-visible spectrophotometry.
Immediately after purification, UV-visible spectra were measured for E. coli WrbA and B. abortus WrpA. One milliliter (200 μM) of each protein was loaded into a 1-cm-path-length quartz cuvette, and the absorbance was measured from 200 to 700 nm, using a Shimadzu UV-1650pc spectrophotometer.
Size exclusion chromatography.
Protein complexes were analyzed by size exclusion chromatography after affinity purification. Concentrated samples (300 μl of a 10-mg/ml protein sample, with or without 1 mM FMN) were injected onto a GE Healthcare Superdex 200 10/300 GL column (flow rate, 0.5 ml/min). Elution profiles were measured at two different wavelengths (280 and 446 nm), and fractions of 500 μl were collected during the run; the same dialyzing buffer as that described above for protein purification was used for all runs. Protein standards (blue dextran [2,000 kDa], aldolase [158 kDa], conalbumin [75 kDa], and ovalbumin [43 kDa]) were also injected into the column, and the corresponding calibration curve was use for molecular size estimation of the different complexes (see Fig. S1 in the supplemental material).
Enzyme activity assays.
The enzymatic activities of purified E. coli WrbA and B. abortus WrpA in the presence of FMN or flavin adenine dinucleotide (FAD) were assayed by monitoring the oxidation of NADH at 340 nm. Each purified protein (100 nM) was mixed with 100 μM NADH and 250 μM benzoquinone in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl buffer. Reactions were started by the addition of the purified proteins. WrbA was assayed immediately after purification, without addition of FMN. WrpA was preincubated with the FMN/FAD cofactors (1 mM) for 1 h on ice, and activity was then assayed. Assays were performed in a 1-cm-path-length quartz cuvette at room temperature with a total volume of 1 ml and were monitored for 120 s by use of a Shimadzu UV-1650pc spectrophotometer.
Crystallization of B. abortus WrpA. (i) P4222 crystal form.
Initial crystallization screening was carried out using the hanging-drop vapor diffusion technique in 96-well microplates (Nunc). Trays were prepared using a Mosquito robot (TTP LabTech) and commercial crystallization kits (Nextal-Qiagen). The drops were set up by mixing equal volumes (0.1 μl) of the purified protein and the precipitant solutions equilibrated against 75 μl of crystallization solution. The protein concentration used was 17 mg/ml. P4222 crystals appeared in approximately 5 days under condition 83 of the Cryos Suite crystallization kit (Qiagen). After manual refinement of the crystallization conditions, the best crystals were obtained at 19°C with the following crystallization solution: 100 mM sodium acetate, pH 4.6, 300 mM ammonium sulfate, 20% polyethylene glycol 2000 (PEG 2000) MME. Prior to flash freezing in liquid nitrogen, crystals were cryo-protected by soaking in crystallization solution containing 20% glycerol.
(ii) P43212 crystal form.
Initial crystallization screening was carried out using the sitting-drop vapor diffusion technique in 96-well CrystalQuick plates (Greiner Bio-One). Trays were prepared using a Mosquito robot (TTP LabTech) and commercial crystallization kits (MCSG-1 to -4; Microlytic Inc.). The drops were set up by mixing equal volumes (0.4 μl) of the purified protein and the precipitant solutions equilibrated against 140 μl of crystallization solution. The protein concentration used was 12.7 mg/ml. P43212 crystals appeared within a week under condition 8 of the MSCG-2 crystallization kit (Qiagen). After manual refinement of the crystallization conditions, the best crystals were obtained at 16°C with the following crystallization solution: 0.2 M potassium formate, pH 7.3, 20% (wt/vol) PEG 3350. For FMN binding studies, crystals were quickly soaked (10 min) in 0.1 M FMN with 0.2 M potassium formate, pH 7.3, 20% (wt/vol) PEG 3350, and 10% glycerol. Prior to flash freezing in liquid nitrogen, crystals were cryo-protected by soaking in crystallization solution containing 20% glycerol.
Crystallographic data collection and data processing.
For the apo-WrpA P4222 crystal form, crystal diffraction was measured at 100 K, using a 1-degree oscillation range, on beamline 21-ID-F (LS-CAT; Advanced Photon Source, Argonne, IL). Diffraction images were collected on a MAR Mosaic 300 detector (LS-CAT) and processed using the −3d approach in xia2 (54–59). Geometric refinement and examination of the scaled amplitudes revealed that the WrpA crystals belonged to tetragonal space group P4222, with the following cell dimensions: a = 61.28, b = 61.28, and c = 128.34 (see Table S3 in the supplemental material).
Diffraction from a single WrpA protein crystal was measured to 2.5 Å at 12.66 keV (0.979 Å). The structure was solved by molecular replacement in PHENIX (60), using a WrpA search model based on the E. coli WrbA structure (PDB ID 3B6I) generated with the Expasy SWISS-Model server (61). The number of WrpA molecules in the asymmetric unit was estimated using the Matthews Probability Calculator (http://www.ruppweb.org/mattprob/). We found one molecule in the asymmetric unit for the P4222 form. The initial structural models were manually examined and corrected; ions and water molecules were added, and refinement of the structure was conducted iteratively using COOT and PHENIX.refine (60). The final structural model was refined to an Rwork value of 23.9% and an Rfree value of 26.8%. Crystallographic data and refined model statistics are presented in Table S3 in the supplemental material. The diffraction data set for this crystal form has been uploaded to the Structural Biology Data Grid (62) and is available for download at http://dx.doi.org/10.15785/SBGRID/203.
For the WrpA P43212 crystal form, Se-Met crystal diffraction was measured at 100 K, using a 1-degree oscillation range, on beamline 19-ID (SBC-CAT; Advanced Photon Source, Argonne, IL). Diffraction images were collected on an ADSC Q315r detector (SBC-CAT) and processed using the HKL3000 suite (63). Diffraction data measured to 2.50 Å for both the apo-form and the FMN-bound form were collected with an X-ray wavelength near the selenium edge of 12.66 keV (0.979 Å). Geometric refinement and examination of the scaled amplitudes revealed that the WrpA crystals belonged to tetragonal space group P43212, with the following cell dimensions: a = 63.58, b = 63.58, and c = 188.53 (see Table S3 in the supplemental material). The diffraction data set for this crystal form has been uploaded to the Structural Biology Data Grid (62) and is available for download at http://dx.doi.org/10.15785/SBGRID/202.
The structure of the apo-form was determined independently by single-wavelength anomalous diffraction (SAD)phasing and initial automatic protein model building with resolve, buccaneer, or Arp/Warp, all as implemented in the HKL3000 software package. The initial model for the apo-form built using COOT was used to determine the structure of the FMN-bound form by molecular replacement using HKL3000. The structure of the FMN-bound form was then iteratively refined using COOT, PHENIX, and/or REFMAC (60); FMN cofactors, ions, and water molecules were also added. The final structural model was refined to an Rwork value of 23.03% and an Rfree value of 26.90%. Crystallographic data and refined model statistics are presented in Table S3 in the supplemental material.
Protein sequence and structure alignments.
Amino acid sequences were aligned using the T-COFFEE multiple-sequence alignment server (64) and shaded using BoxShade. Structural alignments and root mean square deviation (RMSD) calculations were carried out using FATCAT (65) or PyMOL (PyMOL molecular graphics system, version 1.7.4; Schrödinger, LLC). PyMOL was used to visualize hydrophobic and electrostatic surfaces. The ConSurf server (66) was used to visualize conserved regions. The XtalPred server (67) and the Dali server (68) were used to find proteins with the highest structural and sequence homologies. A structural model of WrpA based on the E. coli WrbA structure (PDB ID 3B6J) was generated using the ExPASy SWISS-Model server (61) and used to model the missing loops.
Protein structure accession numbers.
The coordinates of the B. abortus WrpA P4222 structure have been deposited in the Protein Data Bank under PDB ID 5F51, and the coordinates for B. abortus WrpA bound to FMN have been deposited under PDB ID 5F4B.
RESULTS
wrpA transcription is strongly activated by the general stress response (GSR) system under oxidative conditions, but wrpA is not necessary for resistance to acute oxidative stress in vitro.
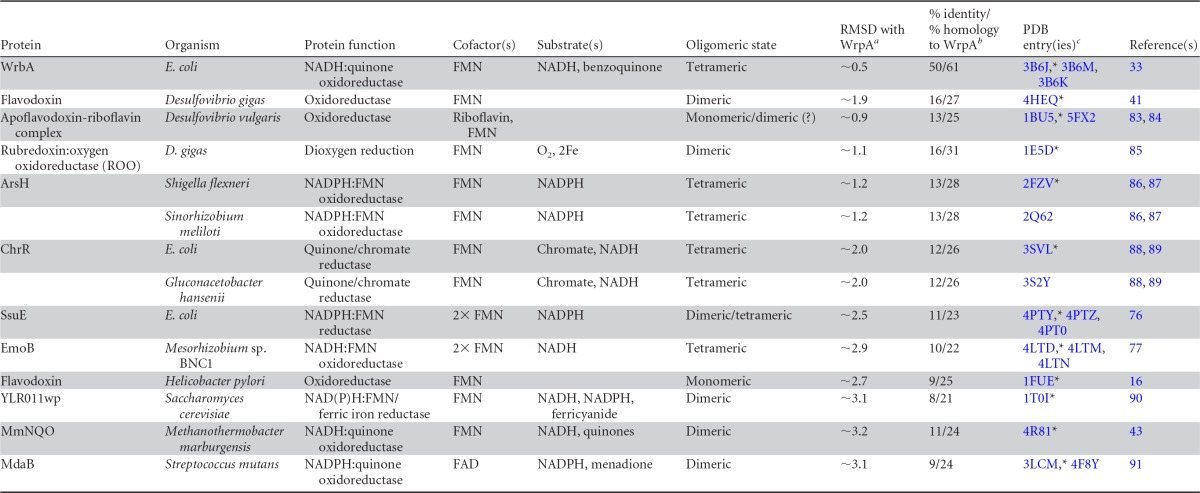
Wild-type B. abortus and strains harboring in-frame deletions of the GSR regulator genes rpoE1 and lovhK were previously challenged with oxidative stress in an effort to characterize the in vitro stress survival deficit of GSR regulatory mutants and to define the GSR transcriptional regulon (48, 49). We found that σE1 and LovhK mediated a 30-fold increase in transcription of the bab1_1070 gene (wrpA) under oxidative conditions (Fig. 1A). wrpA contains a canonical σE1 binding site in its promoter and is among the most strongly activated genes in the GSR regulon (Fig. 1B). WrpA is 40 to 55% identical at the primary structure level to known WrbA NADH:quinone oxidoreductases, which, as outlined in the introduction, have been implicated as resistance factors in response to oxidative stress in other bacterial species. As such, we investigated the role of WrpA in B. abortus oxidative stress survival in vitro.
FIG 1.
Regulated expression of wrpA (bab1_1070). (A) Differential expression of wrpA under oxidative stress in B. abortus ΔrpoE1 and ΔlovhK backgrounds relative to the wild type (WT) as measured by transcriptome sequencing (RNA-seq) (49). (B) Cartoon representation of the wrpA promoter region, with the consensus σE1 binding site at the −35 and −10 regions. (C) Oxidative stress (H2O2) resistance of the B. abortus 2308 wild-type (WT) strain, the ΔwrpA strain, and the ΔrpoE1 strain. CFU ratios between the treated and untreated strains (mock) are presented on a log scale. Experiments were performed in triplicate; error bars represent standard deviations.
We treated a B. abortus strain harboring an in-frame deletion of wrpA (ΔwrpA) with 5 mM H2O2 for 1 h and plated it on TSA plates to enumerate viable CFU. Loss of ΔwrpA cell viability after treatment was statistically equivalent to that of the wild-type strain subjected to the same treatment (Fig. 1C). As a positive control, we also subjected the ΔrpoE1 strain to the same oxidative stress treatment. As expected (48), the ΔrpoE1 strain exhibited a >1-log loss in cell viability under this condition.
wrpA is not a major genetic contributor to GSR-mediated cell survival.
While we observed no deficit in viability of the ΔwrpA strain relative to that of the wild type in the presence of 5 mM H2O2, we considered that WrpA may confer a growth or survival advantage under other conditions. As such, we implemented the Biolog phenotyping platform (69) to assay the growth of B. abortus wild-type and ΔwrpA strains under 1,920 distinct metabolic and chemical/drug conditions. As a positive control, we also screened a strain harboring an in-frame deletion of the known GSR regulator gene rpoE1.
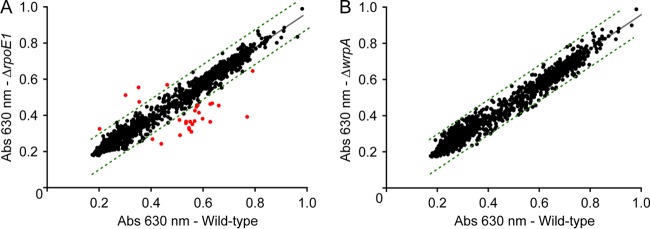
Wild-type B. abortus growth in the Biolog platform identified molecules that are catabolized as sole carbon, nitrogen, sulfur, and phosphorus sources. These measurements captured known metabolic traits of B. abortus (70), including utilization of erythritol, which has previously been described for Brucella (71, 72) and related Rhizobiales (73–75). B. abortus exhibited growth on several hexose (d-fructose, d-glucose, d-galactose, and d-fucose) and pentose (l-arabinose, d-xylose, d-ribose, and adonitol) sugars, amino acids, and dipeptides. While a B. abortus rpoE1 null mutant exhibited growth defects under a number of metabolic and stress conditions (Fig. 2A; see Table S4 in the supplemental material), the ΔwrpA strain did not exhibit comparable growth defects under any assayed condition (Fig. 2B).
FIG 2.
Bacterial phenotype microarray analysis. (A) Comparison of cell growth between wild-type B. abortus 2308 (WT) and the B. abortus ΔrpoE1 strain across a range of distinct metabolic and stress conditions. The plot shows tetrazolium reduction (measured at 630 nm) after 72 h in the WT strain versus the ΔrpoE1 strain. Dotted lines show the 99% confidence prediction band; red dots correspond to conditions where the ΔrpoE1 strain showed a difference in growth relative to that of the WT, outside the 99% confidence prediction band, after 72 h. The full list of conditions in which the ΔrpoE1 strain differed from the WT (with data collected at multiple points on the growth curve) is presented in Table S4 in the supplemental material. (B) Comparison of cell growth between wild-type B. abortus 2308 (WT) and the B. abortus ΔwrpA strain across the same set of conditions after 72 h. We detected no significant, repeatable differences between strains after 72 h or at any other time points measured. Each full screen of 1,920 conditions was performed in duplicate for each strain, and results were averaged.
The ΔwrpA strain is not attenuated in cell or animal infection models, and wrpA affects the magnitude of splenomegaly in infected mice.
We next tested whether deletion of wrpA affects B. abortus infection and replication in cell-based and animal infection models. Inactive and IFN-γ-activated human macrophage-like cells and immature dendritic cells were derived from a THP-1 monocytic cell line and infected with either the wild-type or ΔwrpA B. abortus strain. Infected mammalian cells were lysed and plated on TSA for CFU enumeration at 1, 24, 48, and 72 h postinfection. We observed no difference in the number of CFU between the wild-type strain and the ΔwrpA strain for any cell type, providing evidence that deletion of wrpA does not affect the potential of B. abortus to infect and replicate in the host in these in vitro infection models (Fig. 3A). We further evaluated the capacity of the ΔwrpA strain to infect, colonize, and persist in the mouse spleen over a 3-month time course. Mice were injected intraperitoneally with 5 × 104 CFU of the wild-type, ΔwrpA, or complemented ΔwrpA (ΔwrpA::wrpA) strain and sacrificed at 1, 4, 8, and 12 weeks postinfection. For each time point, spleens were harvested and plated for CFU enumeration. There was no difference in the number of B. abortus organisms per mouse spleen between strains at any time point (Fig. 3B). However, we did observe a statistically significant difference in spleen weight at 4 weeks postinfection: mice infected with the ΔwrpA strain had elevated splenomegaly at this time point relative to mice infected with the wild type (P < 0.05) (Fig. 3B). This result suggests that WrpA modulates the inflammatory capacity of B. abortus in a mouse model. Adding a wild-type copy of wrpA to the ΔwrpA null strain genetically complemented this splenomegaly phenotype.
FIG 3.
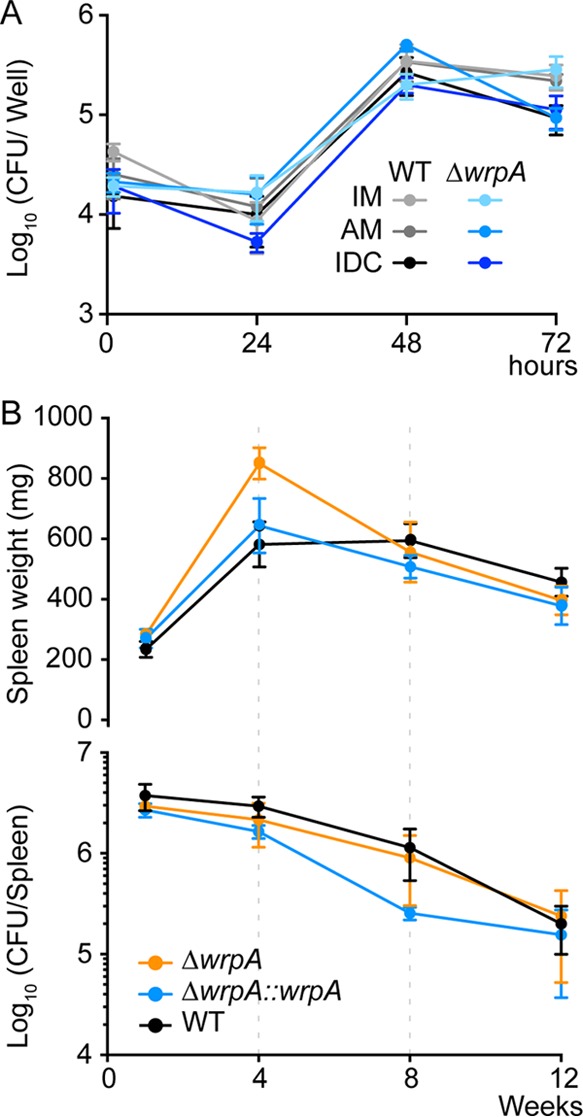
In vitro and in vivo infection assays. (A) Infection of differentiated THP-1 cells with wild-type B. abortus 2308 and ΔwrpA strains. The infected cells were inactivated macrophages (IM), activated macrophages (AM), or immature dendritic cells (IDC). The numbers of B. abortus CFU recovered from different THP-1 cell variants at 1, 12, 24, and 72 h postinfection were plotted. Each infection was performed in triplicate; error bars represent standard errors of the means (SEM). (B) Mice (n = 5 per strain per time point) were injected intraperitoneally with 5 × 104 brucellae (wild-type, ΔwrpA, and ΔwrpA::wrpA strains). Spleens were harvested, weighed (top), and plated for CFU enumeration (bottom) at 1, 4, 8, and 12 weeks postinfection. Error bars represent SEM. Data were analyzed by one-way analysis of variance (ANOVA) followed by Dunnett's posttest (for spleen weight at 4 weeks, P is <0.05).
WrpA forms a stable tetrameric protein complex that does not bind FMN in solution.
The results of our in vitro and in vivo functional analyses of B. abortus wrpA suggested that this gene, though homologous to the well-characterized wrbA family genes, may not encode a flavin-binding oxidoreductase involved in the oxidative stress response. As such, we sought to biochemically and structurally characterize the B. abortus WrpA protein. To this end, we expressed and purified recombinant WrpA; as a positive control, we also expressed and purified a recombinant version of the E. coli WrbA NADH:quinone oxidoreductase.
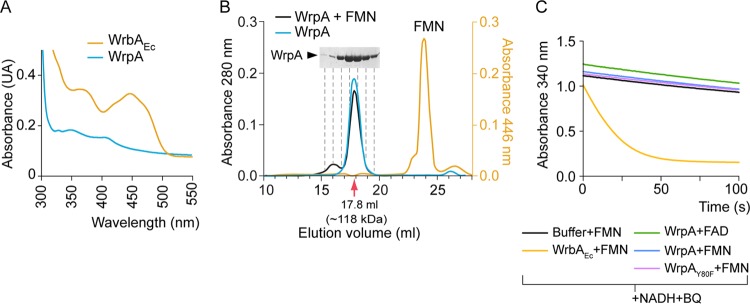
After Ni2+ affinity purification, WrpA appeared colorless at visible wavelengths, suggesting that no flavin cofactor was bound to the protein. Using the same purification protocol, E. coli WrbA appeared bright yellow, indicating the presence of a bound FMN cofactor. Indeed, a UV-visible (200 to 700 nm) absorption scan of purified E. coli WrbA clearly shows a spectrum characteristic of flavoproteins, consistent with its previous description as an FMN-binding enzyme. We observed no flavin absorption spectrum for purified B. abortus WrpA (Fig. 4A).
FIG 4.
Biochemical characterization of Brucella abortus WrpA. (A) UV-visible spectra of purified E. coli WrbA (WrbAEc) and B. abortus WrpA. (B) Size exclusion chromatography of purified WrpA incubated with or without FMN. The corresponding standard curve used for molecular size calculation is presented in Fig. S1 in the supplemental material. (C) Representative NADH:quinone oxidoreductase assay. Purified WrbAEc and WrpA with the FAD or FMN cofactor were subjected to an enzymatic activity assay in which oxidation of NADH in the presence of benzoquinone (BQ) was monitored spectrophotometrically at 340 nm.
Though WrpA did not copurify with a flavin cofactor, we considered the possibility that the protein formed a low-affinity interaction with FMN that was disrupted during affinity purification. To test this hypothesis, we mixed 420 μM WrpA with 1 mM FMN in Tris-HCl, pH 7.4, 150 mM NaCl buffer and performed size exclusion chromatography. The elution profile of this mixture was monitored at 280 nm and 446 nm and compared to that of WrpA alone. WrpA alone eluted at a volume with an apparent molecular mass of 118 kDa, which is consistent with a tetramer or pentamer (23.6 kDa per monomer) (Fig. 4B; see Fig. S1 in the supplemental material). WrpA injected in the presence of 1 mM FMN had an identical elution volume and no absorption signal at 446 nm. A large peak at 446 nm eluted after 23.8 ml, which corresponds to the full column volume and is consistent with a small molecule, such as FMN (Fig. 4B). This result provides evidence that WrpA is a multimer in solution that does not interact with FMN at the concentrations tested, though we cannot exclude the possibility that an FMN-WrpA association occurs but is rapidly reversible. These results also provide evidence that the presence of FMN does not affect the oligomeric state of WrpA at the assessed concentrations, unlike what has been described previously for E. coli WrbA (39) and other flavodoxin-like proteins (44, 76).
WrpA is not an active NADH:quinone oxidoreductase in vitro.
While our biochemical analysis of purified WrpA failed to identify binding between the protein and FMN, it remained possible that these molecules formed a transient but functionally relevant interaction. Therefore, we tested the ability of WrpA to oxidize NADH in the presence of FMN or FAD and benzoquinone. In this assay, we mixed 100 nM WrpA preincubated with 1 mM flavin with 250 μM NADH and 500 μM benzoquinone. We then monitored change in absorbance at 340 nm, which provides a colorimetric readout of NADH oxidation into NAD+. Our positive control, E. coli WrbA (36), exhibited clear NADH:quinone oxidoreductase activity, with full oxidation of NADH within 1 min. Purified B. abortus WrpA had no NADH:quinone oxidoreductase activity above that of the buffer control in the presence of either FMN or FAD (Fig. 4C). We note that for some flavodoxins, the enzymatic activity depends on the protein's oligomeric state (44). Our data show that B. abortus WrpA is a stable oligomer in the presence and absence of flavin cofactors but do not exclude the possibility that other proteins or cofactors are required to elicit a change in oligomeric state and/or enzymatic activity.
WrpA crystal structures reveal a WrbA-like fold that can interact with FMN in a noncanonical fashion.
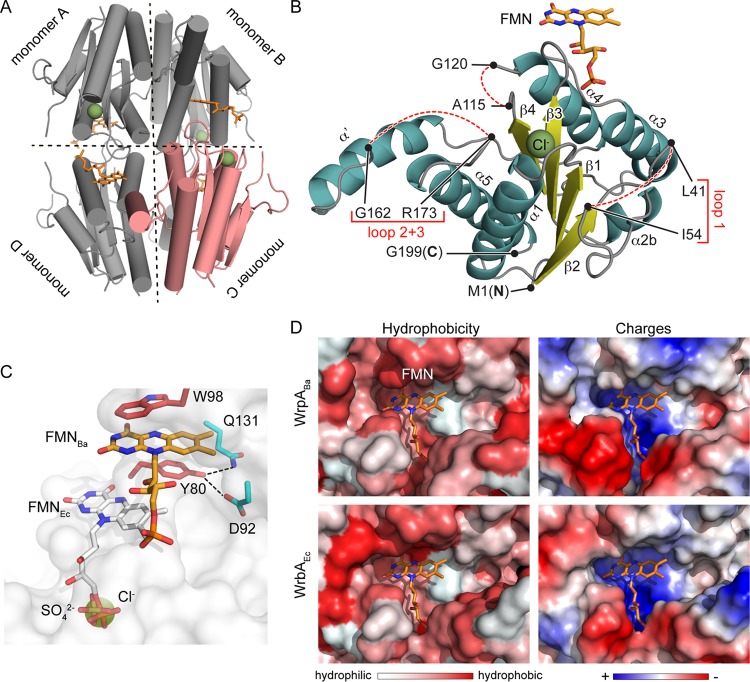
To address the question of whether particular structural features of WrpA could explain the absence of flavin cofactor binding and oxidoreductase activity in vitro, we solved structures of B. abortus WrpA in two tetragonal crystal forms: P43212, with two monomers in the asymmetric unit (ASU), and P4222, with one monomer in the ASU. In both crystal forms, WrpA monomers are arranged about crystallographic symmetry axes in the same tetrameric configuration (Fig. 5A). This oligomeric organization is consistent with the biological assembly described for WrbA-like proteins of other species (31, 33, 38) and is congruent with our size exclusion chromatography data (Fig. 4B). Monomers from both crystal forms are highly similar (average RMSD of the C-α atomic coordinates = 0.35 Å) (see Fig. S2A in the supplemental material) and are further described below. Crystallographic data and refinement statistics are presented in Table S3.
FIG 5.
Structure of B. abortus WrpA. (A) WrpA tetramer with one monomer colored pink. Chloride ions and FMNs are colored green and orange, respectively. (B) Ribbon diagram of a WrpA monomer (from the P43212 crystal form), with α-helices shown in cyan, β-strands in yellow, a chloride ion as a green sphere, and the FMN cofactor in orange. Missing regions of polypeptide are represented with dashed lines. (C) Zoomed view of the WrpA FMN binding site. The model is a surface-rendered model of WrpA (white) with the FMN cofactor from E. coli WrbA (FMNEc; PDB ID 3B6M) modeled at its expected position into the open B. abortus WrpA cavity. The sulfate ion from the P4222 crystal form is modeled as yellow sticks, and the chloride ion from the P43212 crystal form is modeled as a green sphere. Chloride and sulfate ions from the two WrpA crystal forms overlap the terminal phosphate of FMNEc. The FMN cofactor (FMNBa; orange) from the FMN-soaked WrpA crystal structure is stacked between the W98 and Y80 side chains (red); Y80 hydrogen bonds with residues D92 and Q131 (cyan) are shown as dotted lines. (D) Zoomed views of B. abortus WrpA (WrpABa) and E. coli WrbA (WrbAEc) active site cavities. The WrbAEc FMN cofactor (orange; PDB ID 3B6M) (33) is modeled into the WrbAEc and WrpABa active sites to show the similarity in the physicochemical properties of these regions. (Left) Surface representation of active site hydrophobicity. (Right) Surface representation of the electrostatic potentials of the active sites. Missing regions of the WrpABa structure described to participate in active site formation in E. coli WrbA were modeled using the SWISS-Model server (using the structure under PDB ID 3B6J as the template).
The overall tertiary structure of the WrpA monomer is most closely related to that of E. coli WrbA (PDB ID 3B6M; ∼50% sequence identity; C-α RMSD = 0.5 Å). B. abortus WrpA is composed of 4 parallel β-strands organized into a β-sheet with 5 surrounding α-helices (Fig. 5B). An additional α-helix (α′; Y144 to G157) is present in the P43212 crystal form (Fig. 5B; see Fig. S2A in the supplemental material). In the P4222 form, polypeptides spanning residues A115 to G119 and A145 to S172 are missing electron density, likely due to disorder, and are therefore not modeled. The electron density for the polypeptide spanning V42 to K53 is also poor (see Fig. S2A). The FMN binding site in this crystal form does not contain a flavin cofactor. However, a sulfate ion occupies the position corresponding to the 5′ phosphate of FMN in the E. coli holo-WrbA structure (Fig. 5C; see Fig. S2B).
The P43212 crystal form also has missing density for polypeptides spanning V42 to K53, A115 to G119, and M163 to S172 (Fig. 5B; see Fig. S2A in the supplemental material). After soaking the crystals in a solution containing FMN, we observed single bound FMN molecules at the same position in each WrpA monomer (Fig. 5A and B). However, the position of FMN was distinct from that in E. coli WrbA: the flavin isoalloxazine ring was stacked between the tryptophan (W98) side chain of one monomer and the tyrosine (Y80) side chain of the opposite monomer, approximately 11 Å away from the FMN binding site in E. coli WrbA (Fig. 5C). Residue W98 has previously been described for E. coli WrbA to be part of the hydrophobic active site, with the aromatic substrates being stacked between the side chain of the tryptophan and the FMN ring (33). In addition to this π-stacking interaction, FMN also makes hydrogen bonds with the side chains of H134 and E148, present in the opposite monomer and the adjacent monomer, respectively (see Fig. S2C). A simulated annealing composite omit map (contoured at 1σ) of this FMN molecule is presented in Fig. S2C.
The location of FMN positions the terminal phosphate of its ribityl tail outside the usual binding pocket. This pocket is instead occupied by a Cl− ion in this WrpA crystal form, which is in the same position as the SO42− ion in the P4222 (apo) crystal form (Fig. 5C; see Fig. S2D in the supplemental material). A chloride ion has previously been described for the same position in the E. coli apo-WrbA structure (PDB ID 2RG1) (38). Residue Y80, which forms π-stacking interactions with the FMN ring, also makes close hydrogen bonds with residues D92 and Q131 of the opposite monomer (Fig. 5C). A phenylalanine is present at the same position in enzymatically active WrbA-like proteins (Fig. 6C). To test whether interaction of the Y80 hydroxyl moiety with D92 and Q131 may constrain FMN binding in WrpA, we generated a Y80F mutant. However, WrpA(Y80F) also failed to bind FMN or to exhibit oxidoreductase activity (Fig. 4C).
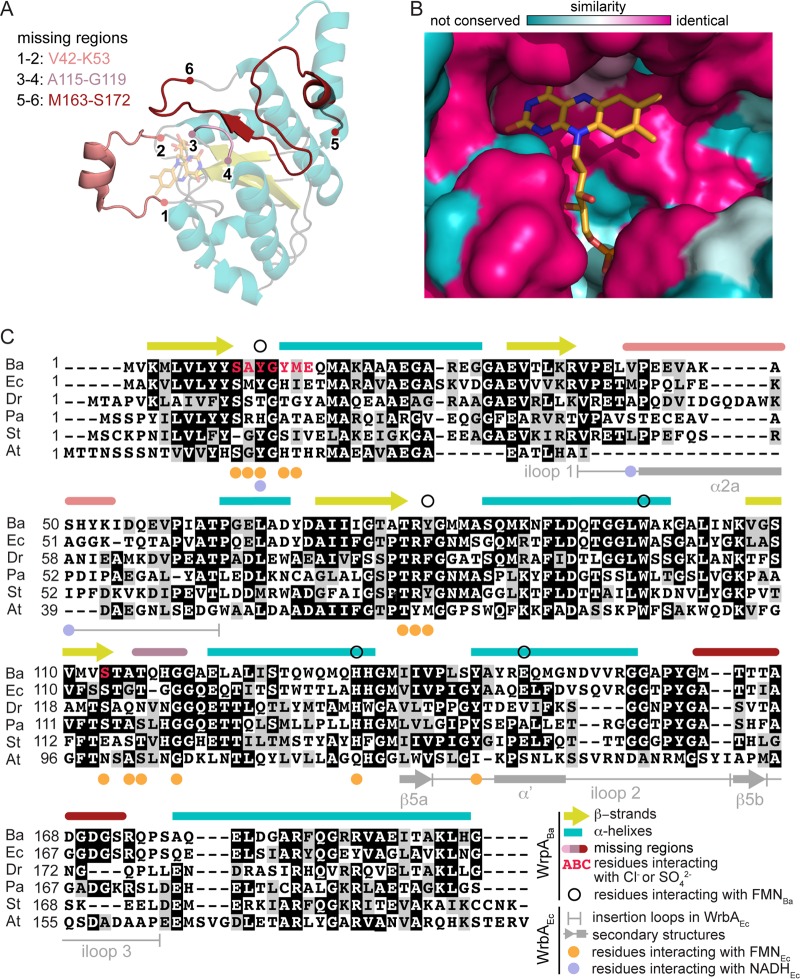
FIG 6.
Comparison of B. abortus WrpA structure and that of classic WrbA family proteins. (A) WrpA monomer (transparent) with missing regions modeled using the E. coli WrbAEc structure (PDB ID 3B6J) as a template. (B) Conservation of WrpA and WrbAEc active sites. The WrbAEc FMN cofactor (orange) from PDB structure 3B6J (33) was modeled into the WrpA active site. (C) WrpA sequence alignment to WrbA-like proteins from E. coli (Ec) (33), Deinococcus radiodurans (Dr) (31), P. aeruginosa (Pa) (31), Sulfolobus tokodaii (St) (PDB ID 2ZKI), and Agrobacterium tumefaciens (At) (PDB ID 3D7N). The WrpA secondary structure is reported above the alignment, with the β-strands shown in yellow and α-helices shown in cyan. Unmodeled regions are delimited as indicated in panel A. Residues reported to interact with FMN and NADH in WrbAEc structures (PDB IDs 3B6J, 3B6K, and 3B6M) (33) are marked below the alignment iloop, insertion loop.
The phosphate group of FMN bound to the P43212 crystal form interacts with the side chain of Y12 (see Fig. S2C in the supplemental material), which has been described to participate in a nonproductive NADH interaction in E. coli WrbA (PDB ID 3B6J) (33). However, an alternative view of NADH binding has been reported for the flavodoxin-like EmoB protein (PDB ID 4LTN) (77). In this structure, FMN and NADH molecules are stacked through their isoalloxazine and nicotinamide rings; modeling of these two molecules into the binding cavity suggests that B. abortus WrpA could accommodate both FMN and NADH simultaneously (see Fig. S3A). With this stated, however, the actual position of FMN that we observed in our crystal structure is clearly not compatible with NADH (see Fig. S3B) or benzoquinone (see Fig. S3C) binding.
The two crystal structures of B. abortus WrpA presented here are highly homologous to those of WrbA from E. coli and other species and do not easily explain why WrpA fails to bind FMN in solution. Indeed, the cofactor binding pockets of WrpA and WrbA have equivalent volumes and very similar hydrophobic and electrostatic characteristics, and it is clearly possible to model FMN into the WrpA cofactor binding pocket (Fig. 5D) with an NADH or benzoquinone substrate (see Fig. S3A in the supplemental material). We note that unusual FMN interactions in flavodoxin-like proteins have previously been reported in the literature: in the E. coli SsuE (PDB ID 4PTZ) and Mesorhizobium spp. EmoB (PDB ID 4LTM) flavoproteins, two stacked FMN molecules are present in the active site. The first FMN is in the typical binding site; the second FMN occupies a position closer to the FMN binding position we observed for WrpA (see Fig. S3D). However, in these proteins, the second FMN is displaced by an NAD(P)H molecule during the enzymatic reaction (76, 77).
The disordered regions of WrpA interact with the FMN cofactor and substrates in WrbA.
As discussed above, several regions of polypeptide corresponding to loops that connect elements of secondary structure in E. coli WrbA (31, 33, 38) are missing in both WrpA crystal forms, presumably due to disorder (Fig. 6A). However, if we model the missing regions in the WrpA active site and compare it to WrbA-like protein active sites, both sites appear to be very similar (Fig. 6B). The loop containing V42 to K53 (connecting β2 and α2b) (Fig. 6A and C) has previously been described for WrbA family proteins (31, 33, 38) and contains a helical segment (α2a) that helps to form a hydrophobic groove that accommodates the mispositioned NADH in E. coli WrbA (33). Electron density for this loop is absent in the P43212 crystal form and is poorly modeled in the P4222 form, suggesting that it is dynamic (see Fig. S2A in the supplemental material). The hydrophobic loop containing A115 to G119, which connects β4 to α4 and makes important contacts with the FMN isoalloxazine ring in WrbA (31, 33, 38), is also absent in both of our structures (Fig. 6A and C). Lastly, the region spanning β5a-β5b-α5 in E. coli WrbA (33, 38) is partially missing in our P4222 (from A145 to S172) and P43212 (from M163 to S172) structures (Fig. 6A and C). This region makes important contacts with FMN in WrbA (31, 33, 38). Notably, the WrpA primary structure (Y144 to G157) corresponding to insertion loop 2 in E. coli WrbA (Fig. 6C) is mostly helical (α′) in the WrpA P43212 crystal form and protrudes outside the tetramer (Fig. 6A and C; see Fig. S2A). This conformation is not favorable with the β5-strand formation and may explain the absence of β5 in our structural models. Overall, the regions of missing electron density in the B. abortus WrpA structures presented here are similar to regions missing from the E. coli apo-WrbA structure (PDB ID 2RG1) (38), suggesting that the interactions with the cofactor and substrates stabilize these portions of polypeptide.
DISCUSSION
PhyR phosphorylation by LovhK frees σE1 from an inhibiting interaction with NepR and promotes the transcription of genes involved in B. abortus stress adaptation and chronic infection (48, 49). B. abortus wrpA contains a consensus σE1 binding site, and its transcription is strongly activated by σE1 under oxidative conditions (Fig. 1A and B) (48). Crystal structures of WrpA reveal a flavodoxin-like protein that is closely related to known WrbA NADH:quinone oxidoreductases (Fig. 5 and 6). While the transcriptional and structural data suggest a role for WrpA in adaptation to oxidative stress, deletion of wrpA had no significant effect on the ability of B. abortus to survive acute peroxide stress in vitro, nor did deletion of wrpA compromise infection and replication of B. abortus in cell-based or animal infection models (Fig. 1C and 3A and B). However, mice infected with the ΔwrpA strain did exhibit elevated splenomegaly at week 4 (Fig. 3B), suggesting that WrpA can modulate B. abortus interaction with the host. Notably, increased splenomegaly did not correlate with an increase in the number of CFU per spleen, providing evidence that the ΔwrpA strain elicits a stronger inflammatory response than that seen with the wild type. The basis of this inflammatory response is not known at this time, though it is possibly related to levels of IFN-γ, IL-12, or cyclic beta-1,2-glucan in the host, which are correlated with the development of splenomegaly (78, 79).
The lack of an in vitro stress phenotype or attenuation of infection for the ΔwrpA strain could also be due to functional redundancy with other redox enzymes. Such cases have been noted for P. aeruginosa, where flavodoxin-like proteins have no apparent protective effect unless the katA catalase gene is also deleted (13).
Our functional analysis of purified WrpA revealed surprising biochemical properties. Specifically, WrpA does not form a stable complex with a flavin cofactor in solution (Fig. 4A and B), despite high sequence and structural homologies with E. coli WrbA (Fig. 6C). While soaking one crystal form of WrpA with FMN did reveal an atypical, and likely low-affinity, binding site (Fig. 5B and C), we failed to observe NADH:quinone oxidoreductase activity under several assay conditions (Fig. 4C). Notably, the FMN-protein interaction we observed in the P43212 crystal form is analogous to that of classic flavodoxins, with each side of the isoalloxazine ring π-stacking with tryptophan and tyrosine side chains (see Fig. S3E in the supplemental material) (14–16, 92), but is not compatible with NADH or benzoquinone binding (see Fig. S3B and C). While this cofactor interaction in WrpA is distal from the established WrbA functional FMN binding site, we cannot exclude the possibility that the FMN binding we observe in soaked crystals is functionally relevant in the cell. The atypical FMN position in WrpA evokes the second bound FMN cofactor that has been reported outside the FMN binding site in EmoB and SsuE (see Fig. S3D), though both of these proteins exhibit strong oxidoreductase activity in vitro (76, 77, 80–82).
A careful comparison of the sequences and structures of WrpA and related WrbA proteins failed to reveal differences that could easily explain why WrpA does not bind a flavin cofactor in the typical location (Fig. 5 and 6). Certainly, the archetypal FMN binding pocket of WrpA is fully intact and closely resembles that defined in E. coli and related WrbA crystal structures (Fig. 6; see Fig. S3A in the supplemental material). Indeed, expression and purification of E. coli WrbA by use of the same purification protocol (as a control for our enzymatic and binding assays) revealed a clearly visible absorption signal for FMN and corresponding NADH:quinone oxidoreductase activity (Fig. 4A and C). With this stated, it is important that two crystal forms of B. abortus WrpA grown in two distinct buffer conditions showed several shared regions of missing electron density (Fig. 5B and 6A and C; see Fig. S2A). These regions of protein structure are presumed to be disordered under the assayed conditions and may have positive or negative effects on the binding of the FMN cofactor or other redox substrates. WrpA activity may also require association with other accessory proteins that modulate the structure of these regions, and hence the ability of WrpA to function as an enzyme. As we present in Table 1 and as presented in previous studies (35, 76, 77), flavodoxin-like proteins have very conserved three-dimensional folds but strongly differ at the amino acid sequence level and in their oligomeric state in solution. These primary and quaternary structural differences define the nature of the cofactors and substrates utilized by these proteins. Thus, despite its clear homology to WrbA-like proteins, we cannot exclude the possibility that WrpA uses different cofactors and substrates and has a different enzymatic function from those predicted using sequence-based and structural bioinformatic approaches.
TABLE 1.
Diversity of flavodoxin and flavodoxin-like proteins
RMSD corresponding to monomer-monomer structural alignment of main-chain alpha carbons.
Based on alignment of the X-ray structures (using the FMN-bound WrpA structure).
*, structure used for RMSD and % identity/% homology calculations.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Crosson lab for helpful discussions and Lauriane Quenee and Lois Zitzow for guidance in animal experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00982-15.
REFERENCES
- 1.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra A, Mishra KP. 2015. Bacterial resistance mechanism against oxidative stress. J Med Pharm Innov 2:1–9. [Google Scholar]
- 3.Poole K. 2008. Bacterial multidrug efflux pumps serve other functions. Microbe 3:179–185. [Google Scholar]
- 4.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 5.Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 6.Slauch JM. 2011. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol 80:580–583. doi: 10.1111/j.1365-2958.2011.07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forman HJ, Torres M. 2002. Reactive oxygen species and cell signaling: respiratory burst in macrophage signaling. Am J Respir Crit Care Med 166:S4–S8. doi: 10.1164/rccm.2206007. [DOI] [PubMed] [Google Scholar]
- 8.Coba de la Pena T, Redondo FJ, Fillat MF, Lucas MM, Pueyo JJ. 2013. Flavodoxin overexpression confers tolerance to oxidative stress in beneficial soil bacteria and improves survival in the presence of the herbicides paraquat and atrazine. J Appl Microbiol 115:236–246. doi: 10.1111/jam.12224. [DOI] [PubMed] [Google Scholar]
- 9.Pueyo JJ, Gomez-Moreno C, Mayhew SG. 1991. Oxidation-reduction potentials of ferredoxin-NADP+ reductase and flavodoxin from Anabaena PCC 7119 and their electrostatic and covalent complexes. Eur J Biochem 202:1065–1071. doi: 10.1111/j.1432-1033.1991.tb16471.x. [DOI] [PubMed] [Google Scholar]
- 10.Liochev SI, Hausladen A, Beyer WF Jr, Fridovich I. 1994. NADPH: ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci U S A 91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Maier RJ. 2004. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect Immun 72:1391–1396. doi: 10.1128/IAI.72.3.1391-1396.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez CF, Ackerley DF, Lynch SV, Matin A. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–22595. doi: 10.1074/jbc.M501654200. [DOI] [PubMed] [Google Scholar]
- 13.Green LK, La Flamme AC, Ackerley DF. 2014. Pseudomonas aeruginosa MdaB and WrbA are water-soluble two-electron quinone oxidoreductases with the potential to defend against oxidative stress. J Microbiol 52:771–777. doi: 10.1007/s12275-014-4208-8. [DOI] [PubMed] [Google Scholar]
- 14.Moyano AJ, Tobares RA, Rizzi YS, Krapp AR, Mondotte JA, Bocco JL, Saleh MC, Carrillo N, Smania AM. 2014. A long-chain flavodoxin protects Pseudomonas aeruginosa from oxidative stress and host bacterial clearance. PLoS Genet 10:e1004163. doi: 10.1371/journal.pgen.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho J. 2006. Flavodoxins: sequence, folding, binding, function and beyond. Cell Mol Life Sci 63:855–864. doi: 10.1007/s00018-005-5514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freigang J, Diederichs K, Schafer KP, Welte W, Paul R. 2002. Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori. Protein Sci 11:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Naseem S, Sharma S, Konopka JB. 2015. Flavodoxin-like proteins protect Candida albicans from oxidative stress and promote virulence. PLoS Pathog 11:e1005147. doi: 10.1371/journal.ppat.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev 12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourion B, Rossignol M, Vorholt JA. 2006. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci U S A 103:13186–13191. doi: 10.1073/pnas.0603530103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avican K, Fahlgren A, Huss M, Heroven AK, Beckstette M, Dersch P, Fallman M. 2015. Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog 11:e1004600. doi: 10.1371/journal.ppat.1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem 279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 23.Lacour S, Landini P. 2004. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol 186:7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, Ni L, Somerville RL. 1993. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc Natl Acad Sci U S A 90:5796–5800. doi: 10.1073/pnas.90.12.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. 2003. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res 13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang DE, Smalley DJ, Conway T. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol Microbiol 45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirkpatrick C, Maurer LM, Oyelakin NE, Yoncheva YN, Maurer R, Slonczewski JL. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J Bacteriol 183:6466–6477. doi: 10.1128/JB.183.21.6466-6477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phadtare S, Kato I, Inouye M. 2002. DNA microarray analysis of the expression profile of Escherichia coli in response to treatment with 4,5-dihydroxy-2-cyclopenten-1-one. J Bacteriol 184:6725–6729. doi: 10.1128/JB.184.23.6725-6729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patridge EV, Ferry JG. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J Bacteriol 188:3498–3506. doi: 10.1128/JB.188.10.3498-3506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman J, Shapiro L. 2005. Crystal structures of the tryptophan repressor binding protein WrbA and complexes with flavin mononucleotide. Protein Sci 14:3004–3012. doi: 10.1110/ps.051680805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grandori R, Khalifah P, Boice JA, Fairman R, Giovanielli K, Carey J. 1998. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins. J Biol Chem 273:20960–20966. doi: 10.1074/jbc.273.33.20960. [DOI] [PubMed] [Google Scholar]
- 33.Andrade SL, Patridge EV, Ferry JG, Einsle O. 2007. Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J Bacteriol 189:9101–9107. doi: 10.1128/JB.01336-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carey J, Brynda J, Wolfova J, Grandori R, Gustavsson T, Ettrich R, Smatanova IK. 2007. WrbA bridges bacterial flavodoxins and eukaryotic NAD(P)H:quinone oxidoreductases. Protein Sci 16:2301–2305. doi: 10.1110/ps.073018907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grandori R, Carey J. 1994. Six new candidate members of the alpha/beta twisted open-sheet family detected by sequence similarity to flavodoxin. Protein Sci 3:2185–2193. doi: 10.1002/pro.5560031204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishko I, Carey J, Reha D, Brynda J, Winkler R, Harish B, Guerra R, Ettrichova O, Kukacka Z, Sheryemyetyeva O, Novak P, Kuty M, Kuta Smatanova I, Ettrich R, Lapkouski M. 2013. 1.2 A resolution crystal structure of Escherichia coli WrbA holoprotein. Acta Crystallogr D Biol Crystallogr 69:1748–1757. doi: 10.1107/S0907444913017162. [DOI] [PubMed] [Google Scholar]
- 37.Wolfova J, Mesters JR, Brynda J, Grandori R, Natalello A, Carey J, Kuta Smatanova I. 2007. Crystallization and preliminary diffraction analysis of Escherichia coli WrbA in complex with its cofactor flavin mononucleotide. Acta Crystallogr Sect F Struct Biol Cryst Commun 63:571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfova J, Smatanova IK, Brynda J, Mesters JR, Lapkouski M, Kuty M, Natalello A, Chatterjee N, Chern SY, Ebbel E, Ricci A, Grandori R, Ettrich R, Carey J. 2009. Structural organization of WrbA in apo- and holoprotein crystals. Biochim Biophys Acta 1794:1288–1298. doi: 10.1016/j.bbapap.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Natalello A, Doglia SM, Carey J, Grandori R. 2007. Role of flavin mononucleotide in the thermostability and oligomerization of Escherichia coli stress-defense protein WrbA. Biochemistry 46:543–553. doi: 10.1021/bi061769c. [DOI] [PubMed] [Google Scholar]
- 40.Reha D, Harish B, Sinha D, Kukacka Z, McSally J, Ettrichova O, Novak P, Carey J, Ettrich R. 2014. Molecular dynamics comparison of E. coli WrbA apoprotein and holoprotein. J Mol Model 20:2400. doi: 10.1007/s00894-014-2400-8. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh YC, Chia TS, Fun HK, Chen CJ. 2013. Crystal structure of dimeric flavodoxin from Desulfovibrio gigas suggests a potential binding region for the electron-transferring partner. Int J Mol Sci 14:1667–1683. doi: 10.3390/ijms14011667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Dorado I, Bortolotti A, Cortez N, Hermoso JA. 2013. Structural and phylogenetic analysis of Rhodobacter capsulatus NifF: uncovering general features of nitrogen-fixation (nif)-flavodoxins. Int J Mol Sci 14:1152–1163. doi: 10.3390/ijms14011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullmann E, Tan TC, Gundinger T, Herwig C, Divne C, Spadiut O. 2014. A novel cytosolic NADH:quinone oxidoreductase from Methanothermobacter marburgensis. Biosci Rep 34:e00167. doi: 10.1042/BSR20140143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoch DC. 1975. Dimerization of Azotobacter vinelandii flavodoxin (azotoflavin). Arch Biochem Biophys 170:326–333. doi: 10.1016/0003-9861(75)90124-1. [DOI] [PubMed] [Google Scholar]
- 45.Kishko I, Harish B, Zayats V, Reha D, Tenner B, Beri D, Gustavsson T, Ettrich R, Carey J. 2012. Biphasic kinetic behavior of E. coli WrbA, an FMN-dependent NAD(P)H:quinone oxidoreductase. PLoS One 7:e43902. doi: 10.1371/journal.pone.0043902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect Dis 6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 47.Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. 2011. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol 65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- 48.Kim HS, Caswell CC, Foreman R, Roop RM II, Crosson S. 2013. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J Biol Chem 288:13906–13916. doi: 10.1074/jbc.M113.459305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HS, Willett JW, Jain-Gupta N, Fiebig A, Crosson S. 2014. The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol Microbiol 94:913–925. doi: 10.1111/mmi.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossi F, Zabucchi G, Dri P, Bellavite P, Berton G. 1979. O2- and H2O2 production during the respiratory burst in alveolar macrophages. Adv Exp Med Biol 121:53–74. [DOI] [PubMed] [Google Scholar]
- 51.Sadikot RT, Blackwell TS, Christman JW, Prince AS. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ried JL, Collmer A. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 53.Fiebig A, Castro Rojas CM, Siegal-Gaskins D, Crosson S. 2010. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol Microbiol 77:236–251. doi: 10.1111/j.1365-2958.2010.07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauter NK, Grosse-Kunstleve RW, Adams PD. 2004. Robust indexing for automatic data collection. J Appl Crystallogr 37:399–409. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winter G. 2010. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr 43:186–190. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 59.Zhang Z, Sauter NK, van den Bedem H, Snell G, Deacon AM. 2006. Automated diffraction image analysis and spot searching for high-throughput crystal screening. J Appl Crystallogr 39:112–119. doi: 10.1107/S0021889805040677. [DOI] [Google Scholar]
- 60.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer PA, Socias S, Key J, Ransey E, Tjon EC, Buschiazzo A et al. , 2016. Data publication with the structural biology data grid supports live analysis. Nat Commun 7:10882. doi: 10.1038/ncomms10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 64.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 65.Ye Y, Godzik A. 2003. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19(Suppl 2):ii246–ii255. [DOI] [PubMed] [Google Scholar]
- 66.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A. 2007. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23:3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 68.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bochner BR. 2009. Global phenotypic characterization of bacteria. FEMS Microbiol Rev 33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al Dahouk S, Scholz HC, Tomaso H, Bahn P, Gollner C, Karges W, Appel B, Hensel A, Neubauer H, Nockler K. 2010. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol 10:269. doi: 10.1186/1471-2180-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson JD, Smith H. 1965. Metabolism of erythritol by Brucella abortus. J Gen Microbiol 38:109–124. doi: 10.1099/00221287-38-1-109. [DOI] [PubMed] [Google Scholar]
- 72.McCullough NB, Beal GA. 1951. Growth and manometric studies on carbohydrate utilization of Brucella. J Infect Dis 89:266–271. doi: 10.1093/infdis/89.3.266. [DOI] [PubMed] [Google Scholar]
- 73.Barbier T, Collard F, Zuniga-Ripa A, Moriyon I, Godard T, Becker J, Wittmann C, Van Schaftingen E, Letesson JJ. 2014. Erythritol feeds the pentose phosphate pathway via three new isomerases leading to d-erythrose-4-phosphate in Brucella. Proc Natl Acad Sci U S A 111:17815–17820. doi: 10.1073/pnas.1414622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geddes BA, Hausner G, Oresnik IJ. 2013. Phylogenetic analysis of erythritol catabolic loci within the Rhizobiales and proteobacteria. BMC Microbiol 13:46. doi: 10.1186/1471-2180-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yost CK, Rath AM, Noel TC, Hynes MF. 2006. Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152:2061–2074. doi: 10.1099/mic.0.28938-0. [DOI] [PubMed] [Google Scholar]
- 76.Driggers CM, Dayal PV, Ellis HR, Karplus PA. 2014. Crystal structure of Escherichia coli SsuE: defining a general catalytic cycle for FMN reductases of the flavodoxin-like superfamily. Biochemistry 53:3509–3519. doi: 10.1021/bi500314f. [DOI] [PubMed] [Google Scholar]
- 77.Nissen MS, Youn BY, Knowles BD, Ballinger JW, Jun SY, Belchik SM, Xun LY, Kang CH. 2008. Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis. J Biol Chem 283:28710–28720. doi: 10.1074/jbc.M804535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grillo MJ, Blasco JM, Gorvel JP, Moriyon I, Moreno E. 2012. What have we learned from brucellosis in the mouse model? Vet Res 43:29. doi: 10.1186/1297-9716-43-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roset MS, Ibanez AE, de Souza Filho JA, Spera JM, Minatel L, Oliveira SC, Giambartolomei GH, Cassataro J, Briones G. 2014. Brucella cyclic beta-1,2-glucan plays a critical role in the induction of splenomegaly in mice. PLoS One 9:e101279. doi: 10.1371/journal.pone.0101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao B, Ellis HR. 2007. Mechanism of flavin reduction in the alkanesulfonate monooxygenase system. Biochim Biophys Acta 1774:359–367. doi: 10.1016/j.bbapap.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 81.Bohuslavek J, Payne JW, Liu Y, Bolton H Jr, Xun L. 2001. Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1. Appl Environ Microbiol 67:688–695. doi: 10.1128/AEM.67.2.688-695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao B, Ellis HR. 2005. Altered mechanism of the alkanesulfonate FMN reductase with the monooxygenase enzyme. Biochem Biophys Res Commun 331:1137–1145. doi: 10.1016/j.bbrc.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 83.Walsh MA, McCarthy A, O'Farrell PA, McArdle P, Cunningham PD, Mayhew SG, Higgins TM. 1998. X-ray crystal structure of the Desulfovibrio vulgaris (Hildenborough) apoflavodoxin-riboflavin complex. Eur J Biochem 258:362–371. doi: 10.1046/j.1432-1327.1998.2580362.x. [DOI] [PubMed] [Google Scholar]
- 84.Watt W, Tulinsky A, Swenson RP, Watenpaugh KD. 1991. Comparison of the crystal structures of a flavodoxin in its three oxidation states at cryogenic temperatures. J Mol Biol 218:195–208. doi: 10.1016/0022-2836(91)90884-9. [DOI] [PubMed] [Google Scholar]
- 85.Frazao C, Silva G, Gomes CM, Matias P, Coelho R, Sieker L, Macedo S, Liu MY, Oliveira S, Teixeira M, Xavier AV, Rodrigues-Pousada C, Carrondo MA, Le Gall J. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat Struct Biol 7:1041–1045. doi: 10.1038/80961. [DOI] [PubMed] [Google Scholar]
- 86.Ye J, Yang HC, Rosen BP, Bhattacharjee H. 2007. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Lett 581:3996–4000. doi: 10.1016/j.febslet.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vorontsov II, Minasov G, Brunzelle JS, Shuvalova L, Kiryukhina O, Collart FR, Anderson WF. 2007. Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity. Protein Sci 16:2483–2490. doi: 10.1110/ps.073029607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eswaramoorthy S, Poulain S, Hienerwadel R, Bremond N, Sylvester MD, Zhang YB, Berthomieu C, Van Der Lelie D, Matin A. 2012. Crystal structure of ChrR—a quinone reductase with the capacity to reduce chromate. PLoS One 7:e36017. doi: 10.1371/journal.pone.0036017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jin H, Zhang Y, Buchko GW, Varnum SM, Robinson H, Squier TC, Long PE. 2012. Structure determination and functional analysis of a chromate reductase from Gluconacetobacter hansenii. PLoS One 7:e42432. doi: 10.1371/journal.pone.0042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, van Tilbeurgh H. 2004. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J Biol Chem 279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Li L, Dong YH, Su XD. 2014. Structural and biochemical characterization of MdaB from cariogenic Streptococcus mutans reveals an NADPH-specific quinone oxidoreductase. Acta Crystallogr D Biol Crystallogr 70:912–921. doi: 10.1107/S1399004713033749. [DOI] [PubMed] [Google Scholar]
- 92.Rao ST, Shaffie F, Yu C, Satyshur KA, Stockman BJ, Markley JL, Sundarlingam M. 1992. Structure of the oxidized long-chain flavodoxin from Anabaena 7120 at 2 A resolution. Protein Sci 1:1413–1427. doi: 10.1002/pro.5560011103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.