ABSTRACT
Conjugation in bacteria is the contact-dependent transfer of DNA from one cell to another via donor-encoded conjugation machinery. It is a major type of horizontal gene transfer between bacteria. Conjugation of the integrative and conjugative element ICEBs1 into Bacillus subtilis is affected by the composition of phospholipids in the cell membranes of the donor and recipient. We found that reduction (or elimination) of lysyl-phosphatidylglycerol caused by loss of mprF caused a decrease in conjugation efficiency. Conversely, alterations that caused an increase in lysyl-phosphatidylglycerol, including loss of ugtP or overproduction of mprF, caused an increase in conjugation efficiency. In addition, we found that mutations that alter production of other phospholipids, e.g., loss of clsA and yfnI, also affected conjugation, apparently without substantively altering levels of lysyl-phosphatidylglycerol, indicating that there are multiple pathways by which changes to the cell envelope affect conjugation. We found that the contribution of mprF to conjugation was affected by the chemical environment. Wild-type cells were generally more responsive to addition of anions that enhanced conjugation, whereas mprF mutant cells were more sensitive to combinations of anions that inhibited conjugation at pH 7. Our results indicate that mprF and lysyl-phosphatidylglycerol allow cells to maintain relatively consistent conjugation efficiencies under a variety of ionic conditions.
IMPORTANCE Horizontal gene transfer is a driving force in microbial evolution, enabling cells that receive DNA to acquire new genes and phenotypes. Conjugation, the contact-dependent transfer of DNA from a donor to a recipient by a donor-encoded secretion machine, is a prevalent type of horizontal gene transfer. Although critically important, it is not well understood how the recipient influences the success of conjugation. We found that the composition of phospholipids in the membranes of donors and recipients influences the success of transfer of the integrative and conjugative element ICEBs1 in Bacillus subtilis. Specifically, the presence of lysyl-phosphatidylglycerol enables relatively constant conjugation efficiencies in a range of diverse chemical environments.
INTRODUCTION
Conjugation is one of several processes bacteria use to acquire new genes. During conjugation, a donor bacterium transfers DNA directly to a recipient bacterium in a contact-dependent manner. The conjugation machinery is typically encoded by a mobile genetic element, which itself is frequently transferred during conjugation. Conjugation can also deliver genes that are not directly involved in the conjugation process but that are located on the mobile genetic element or on other DNA elements that are transferred. These genes are known to confer a wide variety of phenotypes to cells, and their transfer can allow recipients to rapidly acquire new characteristics. For example, conjugative elements are widely involved in the spread of antibiotic resistances (reviewed in references 1, 2, and 3).
ICEBs1 is an integrative and conjugative element (ICE) found in Bacillus subtilis (4, 5). ICEs are widespread and found in many bacterial species (6). Unlike conjugative plasmids, ICEs integrate into the host chromosome, where they are maintained during chromosomal replication, segregation, and cell division, much like a transposon or phage lysogen (reviewed in references 3 and 7). Under certain circumstances, ICEs can excise from the chromosome, forming a plasmid intermediate that can then be transferred to recipient cells by the element-encoded conjugation machinery.
ICEBs1 is found integrated in the trn-leu2 gene in the B. subtilis chromosome and becomes activated in response to extracellular signaling, starvation, or DNA damage (4). The regulatory genes of ICEBs1 involved in cell-cell signaling (rapI and phrI) have been defined (4, 8). Overexpression of RapI leads to excision of ICEBs1 in >90% of cells in a growing population, allowing a high frequency of experimentally induced conjugation (4, 8, 9). ICEBs1 encodes a type IV secretion system that transfers DNA from the donor to a recipient. Type IV secretion systems are found in other ICEs and conjugative plasmids in both Gram-positive and Gram-negative bacteria (10).
During conjugation, DNA is transferred from the cytoplasm of the donor to that of the recipient, crossing the envelope of each to generate a transconjugant. The composition of the cell envelopes of both the donor and recipient influences the success of conjugation. For example, in Gram-negative bacteria, the outer membrane protein OmpR and the lipopolysaccharide are important for formation of mating pairs (11–16). In Enterococcus faecalis, lipoteichoic acids may be important for mating pair formation (17–19). Recently, we found that in B. subtilis, the phospholipid head groups of the membrane bilayer make important contributions to conjugation (20).
The cell envelopes of B. subtilis and other Gram-positive bacteria contain a single lipid bilayer. The lipids of this membrane vary in the composition of their fatty acid tails and their head groups (reviewed in reference 21). The most abundant phospholipids in the membrane of B. subtilis are the negatively charged phosphatidylglycerol, zwitterionic phosphatidylethanolamine and neutral glycolipids, negatively charged cardiolipin, and positively charged lysyl-phosphatidylglycerol (reviewed in reference 22). The membrane of B. subtilis carries a net negative charge.
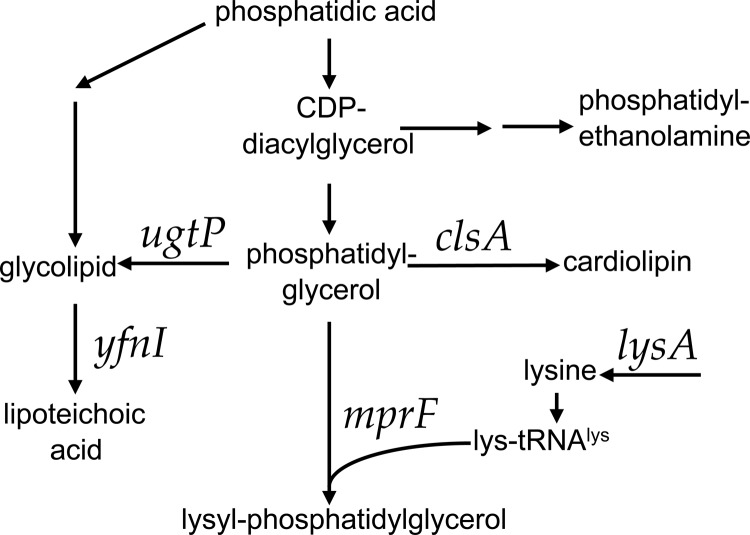
Although the conjugation machinery is encoded by the conjugative element, host genes, in both the donor and recipient, are also important for successful transfer of conjugative DNA. Previously, we used transposon insertions and deep sequencing (Tn-seq) to identify genes in recipients that affect the frequency of conjugation (20). We found that deletion of genes involved in the synthesis of various phospholipids has distinct effects on the ability of B. subtilis to act as a recipient in conjugation. Several of the mutations (in ugtP, yfnI, and mprF) that affect conjugation affect consumption of the phospholipid phosphatidylglycerol (Fig. 1).
FIG 1.
Pathways of phospholipid biosynthesis that affect conjugation of ICEBs1. Some of the pathways involved in phospholipid biosynthesis are shown. Genes relevant to this work are indicated above the arrows.
Here, we analyzed these mutants to evaluate the effects of phospholipids on conjugation. We used double mutant analysis to determine epistasis between several of the phospholipid mutations. Our results indicate that lysyl-phosphatidylglycerol stimulates conjugation and that other phospholipids are also important for conjugation, independently of lysyl-phosphatidylglycerol. We also found that the phenotype caused by loss of mprF (needed for production of lysyl-phosphatidylglycerol) was enhanced by some environmental conditions and suppressed by others. Our results indicate that the ability of cells to function in conjugation is buffered against some chemical variations in the environment by lysyl-phosphatidylglycerol.
MATERIALS AND METHODS
Media and growth conditions.
Escherichia coli cells were grown at 37°C in LB medium. B. subtilis cells were grown at 37°C in LB medium or S750 defined minimal medium with 0.1% glutamate and 40 μg/ml required amino acids (23). Arabinose (1%, wt/vol) was used as a carbon source. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was used to induce expression from the LacI-repressible promoter Pspank(hy) (24). Xylose (1%, wt/vol) was used to induce expression of Pxyl-rapI. Ampicillin was used at 100 μg/ml for E. coli. Antibiotics were used at the following concentrations for B. subtilis: spectinomycin at 100 μg/ml, kanamycin at 5 μg/ml, chloramphenicol at 5 μg/ml, and a combination of erythromycin at 0.5 μg/ml and lincomycin at 12.5 μg/ml to select for macrolide-lincosamide-streptogramin (MLS) resistance.
Strains and alleles.
B. subtilis strains are listed in Table 1. Strains with trp phe alleles are derived from JH642 (trpC2 pheA1). rapI was overexpressed from the xylose-inducible fusion Pxyl-rapI (integrated in the chromosome at amyE) to activate ICEBs1 in donor cells. The Δ(rapI-phrI)160::cat allele was constructed with the same genomic boundaries as the Δ(rapI-phrI)342::kan allele (4). Upstream and downstream genomic DNA fragments and the chloramphenicol resistance gene cat were amplified by PCR and joined together by isothermal (Gibson) assembly (25). This product was used to transform naturally competent B. subtilis cells, a chloramphenicol-resistant isolate was selected, and the allelic exchange was verified by PCR.
TABLE 1.
B. subtilis strains used
| Strain | Genotype (reference) |
|---|---|
| CAL89 | trp phe str-84 comK::spc (4) |
| CMJ44 | trp phe ICEBs10 ΔyfnI44::spc (20) |
| CMJ83 | trp phe ICEBs10 ugtP::mls amyE::[lacI spc] (20) |
| CMJ86 | trp phe ICEBs10 clsA::cat amyE::[lacI spc] |
| CMJ124 | trp phe ICEBs10 ΔmprF124::mls (20) |
| CMJ127 | trp phe amyE::[Pxyl-rapI cat] Δ(rapI-phrI)342::kan ΔmprF124::mls (20) |
| CMJ132 | trp phe ICEBs10 ΔyfnI44::spc ΔmprF124::mls |
| CMJ161 | trp phe ICEBs10 amyE::spc (20) |
| CMJ162 | trp phe ICEBs10 ΔmprF162::spc (20) |
| CMJ222 | trp phe ICEBs10 ΔmprF162::spc lacA::[Pspank(hy)-mprF lacI mls] |
| CMJ248 | trp phe ΔmprF162::spc lacA::[Pspank(hy)-mprF lacI mls] amyE::[Pxyl-rapI cat] Δ(rapI-phrI)342::kan |
| CMJ332 | trp phe ICEBs10 clsA::cat ΔmprF162::spc |
| CMJ333 | trp phe ICEBs10 ugtP::mls ΔmprF162::spc |
| CMJ335 | trp phe ICEBs10 ΔlysA73::mls amyE::[lacI spc] (20) |
| CMJ336 | trp phe ICEBs10 ΔlysA73::mls ΔmprF162::spc |
| CMJ337 | trp phe ICEBs10 ΔmprF162::spc lacA::[lacI mls] |
| CMJ348 | trp phe amyE::[Pxyl-rapI mls] Δ(rapI-phrI)160::cat |
| CMJ459 | trp phe ICEBs10 ΔmprF459::lox-cat |
| CMJ476 | trp phe amyE::[Pxyl-rapI mls] Δ(rapI-phrI)160::cat ΔmprF459 (unmarked) |
| HB5362 | clsA::cat (27) |
| JMA222 | trp phe ICEBs10 (4) |
| KM250 | trp phe amyE::[Pxyl-rapI cat] Δ(rapI-phrI)342::kan (52) |
The unmarked ΔmprF459 allele was constructed by replacing mprF with cat flanked by lox sites to generate strain CMJ459 and then recombining out the lox-cat allele using Cre recombinase expressed from pDR244, as previously described (20, 26). The genomic boundaries of this allele are the same as for the ΔmprF125::mls and ΔmprF162::spc alleles.
For MprF overexpression studies, mprF was cloned into a plasmid that carried the IPTG-inducible promoter Pspank(hy) (24), lacI, and mls situated between genomic sequence from lacA. mprF was placed under the control of the promoter Pspank(hy). This plasmid was transformed into naturally competent B. subtilis cells and Pspank(hy)-mprF, lacI, and mls introduced by double crossover at lacA. Expression of mprF was induced by the addition of 1 mM IPTG.
Thin-layer chromatography.
Lipids were extracted from cells using a modified Bligh-Dyer method (27). We grew cells in minimal medium to an optical density at 600nm (OD600) of ∼1, sampled 1 ml of culture, pelleted the cells, removed the supernatant, resuspended in 1 ml water, pelleted the cells, resuspended in 100 μl 1 M perchloric acid, and incubated for 30 min on ice. Lipids were extracted by adding 1 ml of 12:6:2 methanol-chloroform-water and 0.625 μg of a phosphatidylserine standard (Sigma-Aldrich) to each sample and incubating at 4°C overnight on a rocking platform. Lipids were recovered by adding 300 μl H2O and 300 μl chloroform, incubating the samples for 30 min at −20°C, and then centrifuging for 5 min at 720 × g. The organic (bottom) phase was recovered and dried under nitrogen, and the extracted lipids were resuspended in 12 μl of 2:1 chloroform-methanol.
The total volume of each sample was spotted on silica 60 plates (Angela) along with lysyl-phosphatidylglycerol (0.63 μg to 2.5 μg) (Avanti Polar Lipids) and phosphatidylserine (0.25 μg to 1 μg) standards and developed in a thin-layer chromatography chamber with 60:35:5 chloroform-methanol-water. The plates were dried, stained with ninhydrin (1.5 mg/ml ninhydrin in water-saturated butanol with 3% [vol/vol] acetic acid), and charred. The plates were scanned on a flat-bed scanner and analyzed with ImageJ (28). Standard curves were generated for lysyl-phosphatidylglycerol (0.16 μg to 5 μg) and phosphatidylserine (0.25 μg to 1 μg) to ensure that the amount of each phospholipid in the samples was within the linear range of the assay.
Mating assays.
Mating assays were performed on filters as previously described (9, 20). Briefly, donor and recipient cells were grown separately in minimal medium with 1% arabinose as a carbon source. Donors were induced with 1% (wt/vol) xylose for 2 h to induce expression of Pxyl-rapI, thereby activating ICEBs1 gene expression. An equal number of donors and recipients was mixed, collected on a mating filter, and placed on a mating support consisting of 1.5% agar with a buffered salt solution (see below) for 90 min. Mating filters were typically placed on Spizizen's minimal salts (SMS) agar. SMS agar contains 15 mM ammonium sulfate, 80 mM dibasic potassium phosphate, 44 mM monobasic potassium phosphate, 3.4 mM trisodium citrate, 0.8 mM magnesium sulfate, and 1.5% agar at pH 7.0 (29) unless otherwise specified. TSS agar contains 37 mM ammonium chloride, 2 mM dibasic potassium phosphate, 50 mM Tris base, 1 mM magnesium sulfate, 0.004% iron(III) chloride, 0.004% trisodium citrate, and 1.5% agar at pH 7.5 (29) and was used as an alternate buffer in the mating support in some experiments. TSS was further amended in some experiments, as noted in Results. Cells were then rinsed off the filter, diluted, and spread on LB plates with selective antibiotics to determine the numbers of transconjugants, donors, and/or recipients.
RESULTS
Effects of genes involved in phospholipid biosynthesis on conjugation.
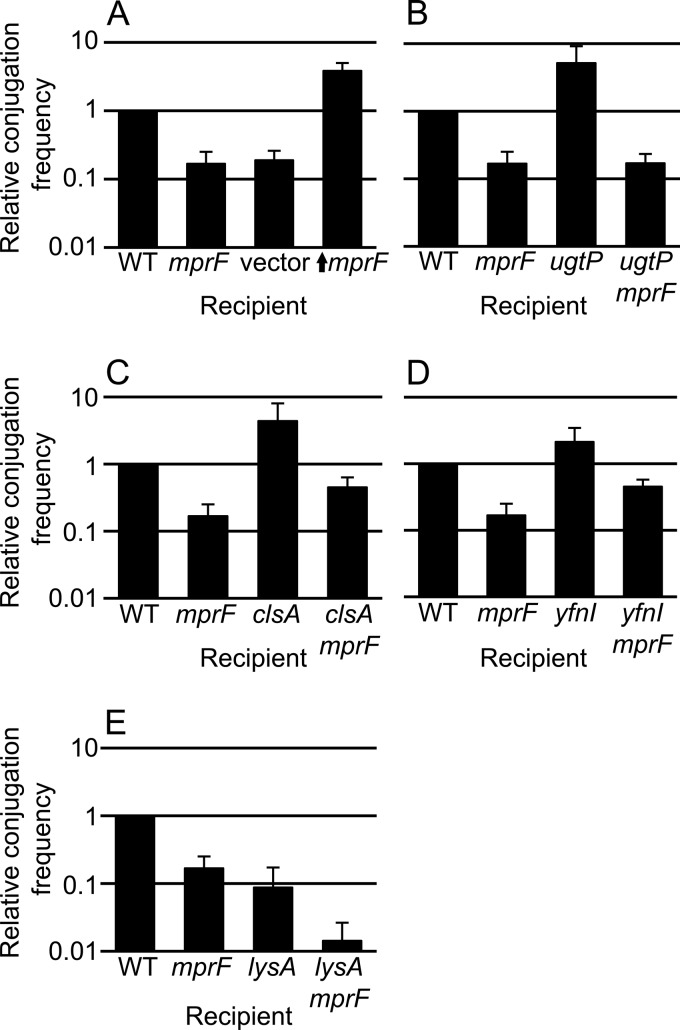
Previously, we found that mprF and several other genes involved in the synthesis of phospholipids affect the efficiency of conjugation (20). MprF catalyzes addition of a lysyl group from Lys-tRNALys to phosphatidylglycerol to form lysyl-phosphatidylglycerol (Fig. 1) (27, 30, 31). Loss of mprF in donors and/or recipients causes a decrease in conjugation of ICEBs1 (Fig. 2) (20), indicating that elimination of lysyl-phosphatidylglycerol is detrimental for conjugation.
FIG 2.
Effects of mutations in recipients on acquisition of ICEBs1. The relative conjugation frequency (y axis) is shown for each of the indicated recipients (x axis). The same donor strain (KM250) was used for all experiments, and ICEBs1 was induced in the donor by overproduction of the activator RapI (see Materials and Methods). The relative conjugation frequency (y axis) is the number of transconjugants per donor crossed to the indicated recipient strain, normalized to that of the wild-type (WT) recipient (CMJ161) in each experiment. The wild-type conjugation efficiency was approximately 4% transconjugants per donor in these experiments. Conjugation frequencies measured with recipients that are null for mprF, ugtP, yfnI, and lysA are similar to those previously reported (20) and were included in these experiments to allow direct comparison with the appropriate double mutants. The graph shows means and standard deviation from ≥3 experiments. The conjugation efficiency for each single mutant is statistically different from that for the wild type (P < 0.05). Data for the wild type (CMJ161) and an mprF null mutant recipient (CMJ162) are included in all panels for comparison. (A) The vector (CMJ337) contains Pspank(hy) with no insert; ↑mprF (CMJ222) indicates an mprF null mutant with Pspank(hy) driving expression of mprF. (B) ugtP (CMJ83) and ugtP mprF double mutant (CMJ333) (P < 0.05 versus ugtP). (C) clsA (CMJ86) and clsA mprF double mutant (CMJ332) (P < 0.05 versus clsA and mprF). (D) yfnI (CMJ44) and yfnI mprF double mutant (CMJ132) (P < 0.05 versus yfnI and mprF). (E) lysA (CMJ335) and lysA mprF double mutant (CMJ336) (P < 0.05 versus lysA and mprF). These strains were grown with 40 μg/ml lysine.
In contrast to the loss of mprF, we found that overexpression of mprF in recipients caused an increase in the acquisition of ICEBs1 via conjugation. We fused mprF to the LacI-repressible, IPTG-inducible promoter Pspank(hy) at an ectopic location (amyE) on the chromosome in a mutant missing the normal copy of mprF. We found that expression of Pspank(hy)-mprF in recipients caused an increase in mating efficiency (Fig. 2A). When mprF was similarly overexpressed in the donor (strain CMJ248) and mated to a wild-type recipient (CAL89), the mating efficiency was 7- to 8-fold greater than that of the wild-type donor (KM250) mated to the same recipient. Together with previous findings on the effects of loss of mprF on conjugation (20), our results indicate that both loss and overproduction of mprF affect conjugation efficiencies. Since the only known role of mprF in B. subtilis is in the production of lysyl-phosphatidylglycerol from phosphatidylglycerol and charged lysyl-tRNA, our results indicate that the amount of lysyl-phosphatidylglycerol, or of other compounds derived from phosphatidylglycerol, affects conjugation. If these effects are due to lysyl-phosphatidylglycerol, then this phospholipid appears to stimulate conjugation.
Other genes affecting phospholipid biosynthesis that were previously identified as having an effect on conjugation include lysA, ugtP, and yfnI (Fig. 1) (20). Similar to loss of mprF, loss of lysA in either the donor or the recipient inhibits conjugation (Fig. 2E) (20). lysA encodes diaminopimelate decarboxylase, which catalyzes synthesis of l-lysine from meso-diaminopimelate (32). lysA is essential for synthesis of lysine, used in the production of lysyl-phosphatidylglycerol, so lysA mutations might affect conjugation by altering lysyl-phosphatidylglycerol production. In contrast, loss of ugtP or yfnI enhances the ability of cells to act as recipients in conjugation (Fig. 2) (20). ugtP is involved in synthesis of glycolipid, a component of the membrane that also acts as a precursor in the synthesis of lipoteichoic acids (Fig. 1) (33). yfnI is one of four genes with overlapping roles in lipoteichoic acid synthesis in B. subtilis (34). Like MprF, the products of ugtP and yfnI consume phosphatidylglycerol.
Based on the functions of the genes described above and their consumption of phosphatidylglycerol, we decided to test the effects of clsA on conjugation. The clsA gene product, cardiolipin synthetase, consumes phosphatidylglycerol during the synthesis of cardiolipin, another phospholipid of the membrane bilayer. clsA was not identified previously in our mutant hunt because the apparent effect on conjugation was below the cutoff used to identify candidate genes (20).
We found that loss of clsA in recipients caused an increase in the acquisition of ICEBs1 via conjugation (Fig. 2C). This increase was similar to that caused by a ugtP null mutation. Together, these results indicate that phosphatidylglycerol or derivatives of phosphatidylglycerol can stimulate and/or inhibit the efficiency of conjugation.
Double mutant analysis of phospholipid biosynthesis mutants.
Deletion of individual genes encoding phospholipid synthetases that consume phosphatidylglycerol (Fig. 1) resulted in opposite affects on conjugation efficiency, depending on which gene was deleted. For example, deletion of mprF caused a decrease in conjugation, and deletion of ugtP, yfnI, or clsA caused an increase in conjugation. There are two simple models to explain these effects. (i) Lysyl-phosphatidylglycerol might enhance conjugation. In this model, loss of mprF (which is needed to make lysyl-phosphatidylglycerol) causes a decrease in conjugation because of loss of lysyl-phosphatidylglycerol. In addition, loss of ugtP, yfnI, and clsA might cause an increase in phosphatidylglycerol (substrate for MprF) and a subsequent increase in lysyl-phosphatidylglycerol, thereby causing an increase in conjugation. (ii) Alternatively (or in addition), cardiolipin, glycolipids, and lipoteichoic acids might act individually or together to inhibit conjugation. For example, phospholipids and teichoic acids can interfere with hydrolase activity (35–40) and might inhibit the cell wall hydrolase CwlT, which is encoded by and needed for transfer of ICEBs1. Loss of clsA (cardiolipin) and yfnI (lipoteichoic acids), and perhaps ugtP (glycolipids), relieves this inhibition, causing an increase in conjugation. In this model, loss of mprF leads to an increase in phosphatidylglycerol and a possible increase in the inhibitory molecule(s) and thus a decrease in conjugation. To test these models, we generated strains in which multiple phospholipid synthetases were inactivated and tested them as recipients in conjugation experiments (Fig. 2). The results described below indicate that lysyl-phosphatidylglycerol enhances conjugation.
We found that the decrease in conjugation frequency caused by loss of mprF was epistatic to the increase in conjugation frequency due to loss of ugtP (Fig. 2B). We measured the conjugation efficiencies using standard mating assays between a wild-type donor (KM250) and recipients carrying the mutation(s) of interest. An mprF ugtP double mutant recipient had essentially the same phenotype as the mprF single mutant recipient (Fig. 2B). This result indicates that mprF is needed for the increase in conjugation caused by loss of ugtP and that the ugtP phenotype is likely due to an increase in the level of lysyl-phosphatidylglycerol.
We also made double mutants between mprF and clsA (CMJ332), yfnI (CMJ132), and lysA (CMJ336). We used the double mutants as recipients in conjugation experiments and directly compared the results to those for the single mutants. The efficiency of conjugation of ICEBs1 into the mprF clsA double mutant was about half (0.45) of that into wild-type recipients. This appeared to be partly (mostly) additive between the conjugation efficiencies of the single mutants: an approximately 6-fold reduction (0.17) and an approximately 4-fold increase (4.4) for mprF and clsA, respectively (expect: 0.17 × 4.4 = 0.73) (Fig. 2C). The conjugation efficiency of the mprF yfnI double mutant was also about half (0.45) that of wild-type recipients, indicative of additive effects of the 6-fold decrease (0.17) and 2-fold increase (2.1) in the mprF and yfnI single mutants (expected, 0.17 × 2.1 = 0.36) (Fig. 2D). The conjugation efficiency of the mprF lysA double mutant was decreased 70-fold (0.014) and appeared to be fully additive between the effects of each of the single mutants, i.e., 6-fold (0.17) and ∼11-fold (0.087) decrease of the mprF and lysA mutants (expected, 0.17 × 0.087 = 0.014) (Fig. 2E). Although it is difficult to determine if the phenotypes of the double mutants are precisely additive, the data clearly indicate that mprF is epistatic to ugtP and not to clsA, yfnI, and lysA.
Together, the results of the double mutant analyses indicate that (i) loss of ugtP and mprF likely affects conjugation by affecting levels of lysyl-phosphatidylglycerol and (ii) loss of clsA, yfnI, and lysA probably does not affect levels of lysyl-phosphatidylglycerol, and their effects on conjugation are likely by altering other components of the cell membrane.
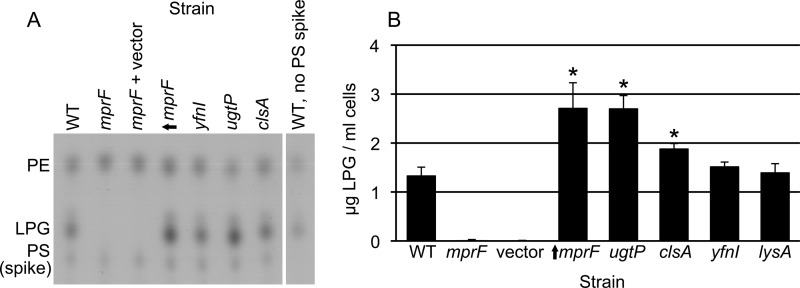
Analysis of lysyl-phosphatidylglycerol levels in mutant cells.
To test the inferences from the genetic analyses described above, we measured the amount of lysyl-phosphatidylglycerol in each of the different phospholipid synthesis mutants (Fig. 3). We grew cells in defined minimal medium, extracted phospholipids, and used thin-layer chromatography to measure lysyl-phosphatidylglycerol (Fig. 3). As expected (27), there was no detectable lysyl-phosphatidylglycerol in the mprF mutant (Fig. 3). In contrast, overproduction of MprF caused an increase in the amount of lysyl-phosphatidylglycerol above that found in otherwise wild-type cells (Fig. 3). We found that the ugtP null mutation, and to a lesser extent the clsA null mutation, also caused an increase in the amount of lysyl-phosphatidylglycerol (Fig. 3). The simplest interpretation of these results is that the increase in lysyl-phosphatidylglycerol in the ugtP mutant, and perhaps the clsA mutant, likely causes the increase in conjugation efficiency. However, the double mutant analysis described above demonstrated that mprF was epistatic to ugtP and apparently additive with clsA. The smaller effect of clsA than of ugtP on the level of lysyl-phosphatidylglycerol and the double mutant phenotypes indicates that the conjugation phenotype of ugtP, but not that of clsA, was due to an increase in lysyl-phosphatidylglycerol.
FIG 3.
Effects of mutations on the level of lysyl-phosphatidylglycerol. The amount of lysyl-phosphatidylglycerol (LPG) recovered from a 1-ml culture of cells at an OD600 of 1 was determined for the indicated strains: CMJ161 (wild type [WT]), CMJ162 (mprF), CMJ337 (mprF plus vector), CMJ222 {↑mprF [mprF null with Pspank(hy) driving expression of mprF]}, CMJ44 (yfnI), CMJ83 (ugtP), CMJ86 (clsA), and CMJ335 (lysA) grown with 40 μg/ml lysine (in panel B only). (A) LPG was extracted from cell membranes and examined using thin-layer chromatography (see Materials and Methods). LPG and phosphatidylethanolamine (PE) standards were used to identify the LPG and PE bands. Phosphatidylserine (PS) was added to samples as an internal standard. The locations of the LPG, PE, and PS bands are indicated. The last part of the panel shows the wild-type sample with no added PS. (B) The LPG content of each strain was quantified from ≥3 experiments. Asterisks indicate a significant difference in the amount of LPG recovered compared to that from the wild-type strain (P < 0.05, t test).
In contrast to the mutations that affected levels of lysyl-phosphatidylglycerol, yfnI or lysA null mutations caused no detectable change in levels of lysyl-phosphatidylglycerol (Fig. 3). The results of the conjugation and thin-layer chromatography experiments are summarized in Table 2. Together with the analysis of double mutants (Fig. 2; Table 2), these results indicate that the conjugation phenotypes caused by mutations in mprF and ugtP are likely due to changes in levels of lysyl-phosphatidylglycerol and that the conjugation phenotypes caused by mutations in clsA, yfnI, and lysA are most likely not due to changes in levels of lysyl-phosphatidylglycerol.
TABLE 2.
Summary of mutations affecting conjugation and phospholipid synthesis
| Mutationa | Phospholipid biosynthesisb | Matingc | LPGd | Phenotype with mprFe |
|---|---|---|---|---|
| mprF | Lysyl-phosphatidylglycerol | Decreased | None | |
| ↑mprF | Lysyl-phosphatidylglycerol | Increased | Increased | |
| ugtP | Glycolipid | Increased | Increased | Epistatic |
| clsA | Cardiolipin | Increased | (Increased) | Additive |
| yfnI | Lipoteichoic acid | Increased | WT | Additive |
| lysA | Lysyl-phosphatidylglycerol | Decreased | WT | Additive |
All are null mutations except ↑mprF, which indicates overexpression of mprF.
Phospholipid whose synthesis depends on the indicated gene (Fig. 1).
Effect of the mutation on conjugation.
Amount of lysyl-phosphatidylglycerol (LPG) produced in cells with the indicated mutation relative to the amount produced cells with the indicated mutation relative to the amount produced in wild-type (WT) cells. “None” indicates that there was no detectable LPG. Parentheses indicates a possible effect but on the edge of statistical significance (Fig. 3).
Phenotype of the double mutant (with mprF) with respect to conjugation. Epistatic indicates that the phenotype of the double mutant is the same as that of the mprF single mutant (Fig. 2).
The lysA mutant requires addition of exogenous lysine to the medium in order to grow. This lysine is evidently enough to support wild-type levels of lysyl-phosphatidylglycerol production but not wild-type levels of conjugation. We suspect that the effects of loss of lysA on conjugation are complex. Synthesis and enzymatic activity of aspartokinase II (the lysC gene product) are regulated by lysine. Aspartokinase II synthesizes l-aspartate 4-phosphate, which is a precursor for synthesis of cell wall peptidoglycan as well as the amino acids lysine, methionine, isoleucine, and threonine (reviewed in reference 41). Deleting lysA and providing exogenous lysine might perturb this regulatory feedback pathway and alter several cellular processes, including peptidoglycan synthesis.
The mating defect of mprF mutants is affected by the chemical environment.
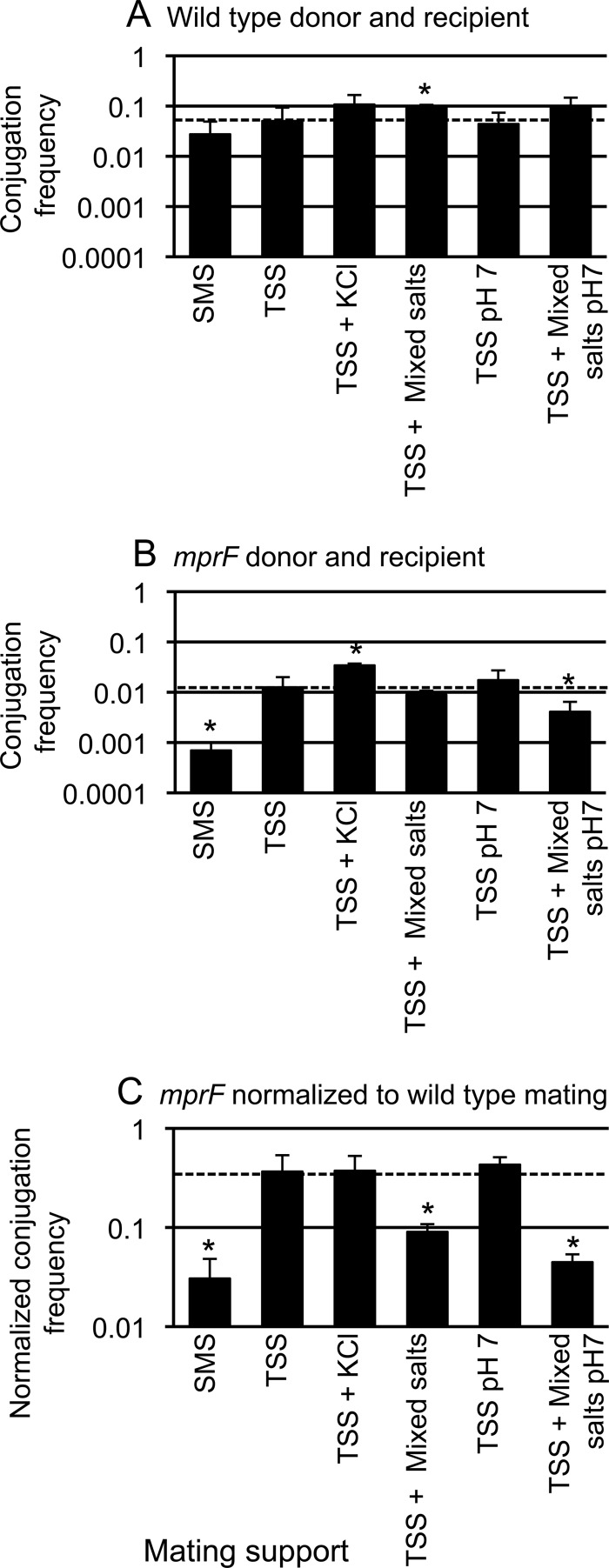
During the course of our investigations, we noticed that the composition of the agar surface on which the filter paper for mating was placed (the mating support) influenced the magnitude of the conjugation phenotype caused by loss of mprF. Specifically, loss of mprF from both donors (CMJ476) and recipients (CMJ162) caused a pronounced conjugation defect (0.031, ∼30-fold) compared to a cross between wild-type donors (CMJ348) and recipients (CMJ161), similar to previously reported results (20). This drop in conjugation was observed when matings were performed on SMS agar.
In contrast to the ∼30-fold decrease in conjugation between mprF mutants on SMS agar, there was a much smaller effect when matings were done on agar containing Spizizen's salts and Tris (TSS agar) (Fig. 4). TSS agar, compared to SMS agar, is buffered with Tris instead of potassium phosphate to pH 7.5 instead of 7.0 and contains a lower total concentration of salts (29). Under these conditions, the conjugation frequency of mprF mutant cells was reduced by approximately 3-fold (0.37) compared to that of wild-type donors and recipients. We ruled out the possibility that production of lysyl-phosphatidylglycerol was restored in the mprF mutant on the TSS agar support; there was no detectable lysyl-phosphatidylglycerol under these conditions in the mutant. These findings indicate that there is something about TSS that suppresses or something about SMS that exacerbates the conjugation defect of the mprF mutant.
FIG 4.
The chemical composition of the mating support affects conjugation. Standard filter matings were performed on supports with different chemical compositions. Donor and recipient cells were mixed in equal numbers and then collected on a filter that was placed on a mating support with the indicated composition. KCl was added to 125 mM. Mixed salts contained 106 mM sodium phosphate, 14 mM sodium sulfate, and 3 mM trisodium citrate. The dashed horizontal line in each panel marks the value for mating on TSS. The mean and standard deviation from ≥3 experiments for each condition are shown. Asterisks indicate that the difference in conjugation frequency on the given support compared to conjugation frequency on TSS is statistically significant (P < 0.05, t test). (A and B) The conjugation frequency is shown as transconjugants per donor for a wild-type donor (CMJ348) and recipient (CMJ161) (A) and for an mprF null mutant donor (CMJ476) and recipient (CMJ162) (B). (C) The conjugation frequencies obtained from panels A and B are directly compared. The ratio of the conjugation frequencies of the mprF mutant (B) and the wild-type strain (A) under each of the indicated conditions is shown.
We investigated what aspect of the different mating supports accounted for the magnitude of the mprF mutant phenotype. Since mating in the mprF mutants was much lower in SMS than TSS, we postulated that the lower pH and/or some of the additional ions in SMS were inhibiting conjugation of mprF mutants. There are several differences between TSS and SMS. Notably, SMS contains a higher total concentration of different salts than TSS and a lower pH (7 versus 7.5). SMS has higher concentrations of potassium (204 mM versus 4 mM), phosphate (124 mM versus 2 mM), sulfate (16 mM versus 1 mM), and citrate (3 mM versus 0.1 mM).
We measured mating efficiencies on TSS agar as the base support with additions to make it more closely resemble SMS. Addition of potassium chloride (125 mM) or mixed salts (106 mM sodium phosphate, 14 mM sodium sulfate, and 3 mM trisodium citrate) increased the conjugation frequency in matings between wild-type cells (Fig. 4A). Adjustment of the pH to 7.0 (without other changes) had little or no detectable effect and had no additional effect in the presence of mixed salts (Fig. 4A).
As with wild-type cells, the addition of potassium chloride also increased the conjugation frequency in matings between mprF mutant cells (Fig. 4B), and adjustment of the pH to 7.0 had little or no effect (Fig. 4B). However, unlike the effect on wild-type cells, addition of mixed salts did not cause an increase in the conjugation efficiency in matings between mprF mutants at either pH (Fig. 4B).
Direct comparison of the conjugation frequencies for wild-type cells (Fig. 4A) and mprF cells (Fig. 4B) showed that mprF caused a more severe phenotype when matings were performed on TSS with mixed salts (at pH 7.5 and pH 7) than when they were performed on TSS (Fig. 4C). Based on these results, we conclude that the salts found in SMS contributed to the defect in mating caused by loss of mprF, particularly at pH 7.
Ion-specific effects on conjugation and effects of mprF.
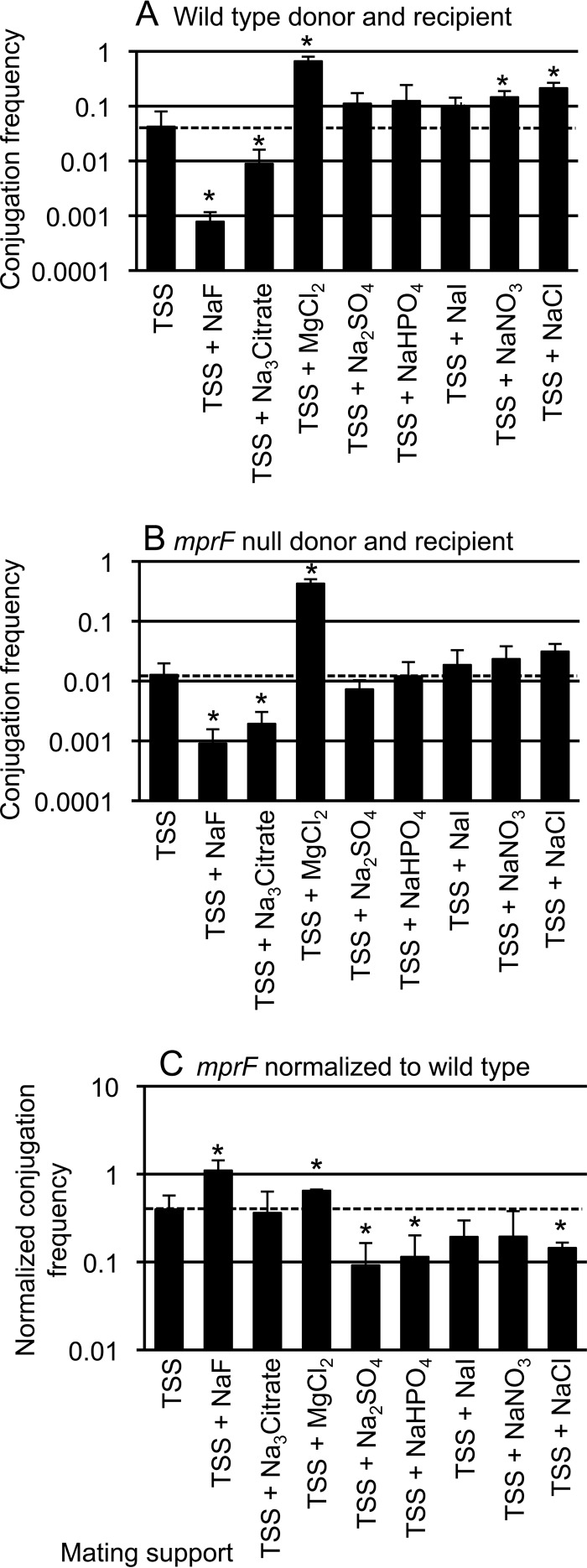
Based on the above results, we wondered if other salts might affect wild-type and mprF mutant strains differently. To test this, we used TSS agar as our base medium and supplemented it with a 125 mM concentration of different salts, including sodium fluoride, trisodium citrate, magnesium chloride, sodium sulfate, dibasic sodium phosphate titrated with monobasic sodium phosphate to give a pH of 7.5 and a phosphate concentration of 125 mM, sodium iodide, sodium nitrate, or sodium chloride. We measured the mating efficiencies of wild-type cells (Fig. 5A) and mprF mutants (Fig. 5B) and then directly compared mprF to wild-type cells (Fig. 5C).
FIG 5.
Some salts enhance conjugation of wild-type but not mprF cells. Filter matings were performed as described in Materials and Methods. Equal numbers of donor and recipient cells were mixed, collected on a filter, and placed on a mating support with the indicated composition. Chemical supplements were added at 125 mM. The samples tested with TSS plus NaHPO4 also contain dibasic sodium phosphate titrated with monobasic sodium phosphate to give a pH of 7.5. The dashed horizontal line in each panel indicates the conjugation frequency on TSS. The mean and standard deviation from ≥3 experiments are shown for each condition. Asterisks indicate that the difference in conjugation frequency on the given support compared to conjugation frequency on TSS is statistically significant (P < 0.05, t test). (A and B) The conjugation frequency (transconjugants per donor) is shown for wild-type donor (CMJ348) and wild-type recipient (CMJ161) (A) and for an mprF null mutant donor (CMJ476) and an mprF null mutant recipient (CMJ162) (B). (C) The conjugation frequencies obtained from panels A and B are directly compared and plotted as the ratio of the conjugation frequencies of the mprF mutant (B) and the wild-type strains (A) under each of the indicated conditions.
We found that addition of sodium fluoride or sodium citrate to TSS caused a decrease in the mating efficiency of wild-type cells (Fig. 5A). There was also a decrease in the mating efficiency of mprF mutants (Fig. 5B). With sodium fluoride, this decrease was somewhat less for the mprF mutants than for wild-type cells (Fig. 5C). With sodium citrate, the decrease was about the same for mprF and wild-type cells (Fig. 5C).
In contrast, we found that addition of magnesium chloride to TSS caused an increase of 16-fold in the mating efficiency of wild-type cells (Fig. 5A). Likewise, there was a similar or somewhat greater increase in the mating efficiency of mprF mutants (33-fold) (Fig. 5B and C). These results indicate that the use of TSS supplemented with magnesium chloride as a solid support for filter matings allows for highly efficient conjugation. We do not know how magnesium chloride stimulates the conjugation efficiency. It might act by affecting the activity of a cell surface component involved in conjugation or stabilizing mating pairs.
Addition of several other salts, including sodium sulfate, sodium phosphate, sodium iodide, sodium nitrate, and sodium chloride, to TSS either stimulated or had relatively little effect on the mating efficiency of wild-type cells (Fig. 5A). The stimulatory effects were less than that of magnesium chloride. The same salts had little or no effect or caused a small increase in the mating efficiency of mprF mutants (Fig. 5B). The stimulatory effects on wild-type cells were larger than the effects on mprF mutants, and this is most easily seen in the ratio of the mating efficiencies of mprF and wild-type cells (Fig. 5C). These ratios are <0.37, the ratio of efficiencies when mating is done on TSS without any modifications.
Together, our results (Fig. 4 and 5) indicate that mating efficiencies are affected by the external ionic environment and that several salts that enhance conjugation of wild-type cells do not have the same stimulatory effect on mprF mutants. Since mprF mutants do not produce lysyl-phosphatidylglycerol, we infer that the different effects of salts are due to the presence or absence of this phospholipid. The presence of mprF and hence lysyl-phosphatidylglycerol enables cells to have efficient conjugation under a variety of different ionic conditions.
DISCUSSION
Our findings indicate that lysyl-phosphatidylglycerol plays a role in stimulating conjugation. Preventing or reducing lysyl-phosphatidylglycerol synthesis in either the donor or the recipient reduces conjugation. Overproduction of lysyl-phosphatidylglycerol in either partner enhances conjugation. Accumulation of lysyl-phosphatidylglycerol was eliminated in mprF null mutants and increased in ugtP mutants or upon overexpression of mprF. Our results also indicate that alterations in phospholipid content that do not detectably affect lysyl-phosphatidylglycerol also alter conjugation efficiencies.
mprF and ugtP.
We found that mprF is epistatic to ugtP for the conjugation phenotype. That is, the mprF ugtP double mutant had the same phenotype as the mprF single mutant. This is consistent with the interpretation that the conjugation phenotypes of ugtP and mprF mutants are due to alterations in lysyl-phosphatidylglycerol and that loss of ugtP causes an increase in phosphatidylglycerol, which then leads to an increase in lysyl-phosphatidylglycerol (Fig. 1). mprF is epistatic because it is needed to make lysyl-phosphatidylglycerol.
Loss of ugtP caused an increase in the amount of lysyl-phosphatidylglycerol, indicating that UgtP normally plays a role limiting the amount of lysyl-phosphatidylglycerol in the cell. ugtP is also known to affect cell division (42), primarily by directly interacting with and inhibiting the cell division protein FtsZ (42, 43). The effects of ugtP on cell division and conjugation are most likely not related. We infer this mainly because the effects on cell division appear to be direct and the effects on conjugation are likely through mprF.
ugtP mutants also appear to have many alterations in gene expression in rich medium (27). The effects of mprF mutations on gene expression are not known, but based on analyses of an mprF pssA ywnE (clsA) triple mutant, there are fewer effects than in a ugtP single mutant (27). It is possible that the effects of mprF and ugtP on conjugation are due to alterations in gene expression. However, the simplest model is that these genes affect conjugation due to alterations in lysyl-phosphatidylglycerol and that the composition of the cell envelope directly affects activity of the conjugation machinery (see below).
mprF and lysyl-phosphatidylglycerol enable efficient conjugation under various ionic conditions.
Our results demonstrate that the effects of lysyl-phosphatidylglycerol on conjugation are dependent on the environmental conditions. That is, the ratio of mating efficiencies of mprF mutants and wild-type cells was affected by the ionic conditions used for mating. For example, the mprF mutants had a much more severe mating defect on SMS agar (∼30-fold) than of TSS agar (∼3-fold). Together with analysis of the differences between SMS and TSS, our results indicate that mprF and lysyl-phosphatidylglycerol normally facilitate efficient mating under a variety of external ionic conditions. We suggest that the presence of lysyl-phosphatidylglycerol buffers conjugation against the some of the otherwise inhibitory effects of different salts and enhances conjugation in the presence of others, allowing the conjugation machinery to function reasonably well under a range of different ionic conditions.
mprF homologs, and by extension lysyl-phosphatidylglycerol, affect cell surface properties of other organisms. For example, mprF in Staphylococcus aureus acts as a virulence factor and potentiates resistance to several cationic antimicrobials, including those produced by potential human hosts (reviewed in reference 22). mprF homologs impact the ability of Enterococcus faecium and Listeria monocytogenes to adapt to different environmental conditions (44, 45). We suggest that in Gram-positive bacteria, mprF and lysyl-phosphatidylglycerol ensure that the cell envelope is buffered from some of the variations in the chemistry of the environment and enable the cell to perform physiological functions in a regular manner under different environmental conditions.
A model for how membrane phospholipids affect conjugation efficiencies.
We suspect that alterations in the phospholipid content of the recipient (and donor) might affect the function of the conjugation machinery. This could be through changes to the physical properties of the membrane (e.g., fluidity) that might affect assembly of the machinery. This could also be through inhibition of a component of the machinery. Transfer of DNA through the ICEBs1-encoded conjugation machinery depends on CwlT (46), a secreted cell wall hydrolase encoded by ICEBs1 (46, 47). Components of the cell envelope, including lipoteichoic acids (35, 36), wall teichoic acids (37, 38), and the phospholipids cardiolipin (36), phosphatidylglycerol (39), and lysyl-phosphatidylglycerol (36, 40), can modulate the function of at least some cell wall hydrolases. Cell wall teichoic acids inhibit hydrolase activity, at least in part, by preventing hydrolase binding to the peptidoglycan of the cell wall (37, 38). Phospholipids can stimulate or inhibit the function of particular hydrolases; for example, phosphatidylglycerol can either enhance or inhibit the N-acetylmuramoyl-l-alanine amidase of E. coli, depending on concentration, but has no effect on the major autolysin of Clostridium acetobutylicum under the conditions tested (39, 40). Altering the phospholipid content of the donor and/or recipient may affect a postulated interaction between the cell wall hydrolase CwlT and the cell envelope, either enhancing or inhibiting the ability of the conjugation machinery to deliver DNA. This interaction could be binding of the conjugation machinery to the recipient cell envelope and/or digestion of the donor and recipient cell wall. If this model is correct, it strongly predicts that the cell wall hydrolase acts on both donor and recipient cells.
Cell wall hydrolases are encoded by many conjugative elements (10, 48–51). Where tested, they have been found to be critical for efficient conjugation. Based on this conservation, it seems likely that the composition of the cell wall affects the efficiencies of many different conjugative elements. Perhaps the cell wall hydrolases have evolved in ways that help determine the host range of the cognate element.
ACKNOWLEDGMENTS
We thank Tony DeBono (Anthony Sinsky lab) for help with thin-layer chromatography, John Helmann for strains, Suzanne Walker, Bernhardt Trout, Barbara Imperialli, and Thomas Bernhardt for helpful conversations, and Laurel Wright and Monika Avello for comments on the manuscript.
Funding Statement
Any opinions, findings, and conclusions or recommendations expressed in this report are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
REFERENCES
- 1.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toleman MA, Walsh TR. 2011. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev 35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 3.Johnson CM, Grossman AD. 2015. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97. doi: 10.1016/S0147-619X(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 6.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 8.Auchtung JM, Lee CA, Garrison KL, Grossman AD. 2007. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CA, Auchtung JM, Monson RE, Grossman AD. 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol Microbiol 66:1356–1369. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez-Mendoza D, de la Cruz F. 2009. Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10:71. doi: 10.1186/1471-2164-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, Arai T, Hattori T. 1970. Effects of cell wall polysaccharide on the mating ability of Salmonella typhimurium. Nature 225:70–71. doi: 10.1038/225070a0. [DOI] [PubMed] [Google Scholar]
- 13.Skurray RA, Hancock RE, Reeves P. 1974. Con− mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J Bacteriol 119:726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havekes L, Tommassen J, Hoekstra W, Lugtenberg B. 1977. Isolation and characterization of Escherichia coli K-12 F− mutants defective in conjugation with an I-type donor. J Bacteriol 129:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanderson KE, Janzer J, Head J. 1981. Influence of lipopolysaccharide and protein in the cell envelope on recipient capacity in conjugation of Salmonella typhimurium. J Bacteriol 148:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiwa A, Komano T. 2004. PilV adhesins of plasmid R64 thin pili specifically bind to the lipopolysaccharides of recipient cells. J Mol Biol 343:615–625. doi: 10.1016/j.jmb.2004.08.059. [DOI] [PubMed] [Google Scholar]
- 17.Trotter KM, Dunny GM. 1990. Mutants of Enterococcus faecalis deficient as recipients in mating with donors carrying pheromone-inducible plasmids. Plasmid 24:57–67. doi: 10.1016/0147-619X(90)90025-8. [DOI] [PubMed] [Google Scholar]
- 18.Bensing BA, Dunny GM. 1993. Cloning and molecular analysis of genes affecting expression of binding substance, the recipient-encoded receptor(s) mediating mating aggregate formation in Enterococcus faecalis. J Bacteriol 175:7421–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrenfeld EE, Kessler RE, Clewell DB. 1986. Identification of pheromone-induced surface proteins in Streptococcus faecalis and evidence of a role for lipoteichoic acid in formation of mating aggregates. J Bacteriol 168:6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. doi: 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst CM, Peschel A. 2011. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol 80:290–299. doi: 10.1111/j.1365-2958.2011.07576.x. [DOI] [PubMed] [Google Scholar]
- 23.Jaacks KJ, Healy J, Losick R, Grossman AD. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol 171:4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 26.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salzberg LI, Helmann JD. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J Bacteriol 190:7797–7807. doi: 10.1128/JB.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 30.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150:45–51. doi: 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto J, Shimizu M, Yamane K. 1991. Molecular cloning and analysis of nucleotide sequence of the Bacillus subtilis lysA gene region using B. subtilis phage vectors and a multi-copy plasmid, pUB110. Agric Biol Chem 55:1615–1626. doi: 10.1271/bbb1961.55.1615. [DOI] [PubMed] [Google Scholar]
- 33.Jorasch P, Wolter FP, Zahringer U, Heinz E. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol 29:419–430. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 34.Wormann ME, Corrigan RM, Simpson PJ, Matthews SJ, Grundling A. 2011. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol Microbiol 79:566–583. doi: 10.1111/j.1365-2958.2010.07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtje JV, Tomasz A. 1975. Lipoteichoic acid: a specific inhibitor of autolysin activity in Pneumococcus. Proc Natl Acad Sci U S A 72:1690–1694. doi: 10.1073/pnas.72.5.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleveland RF, Wicken AJ, Daneo-Moore L, Shockman GD. 1976. Inhibition of wall autolysis in Streptococcus faecalis by lipoteichoic acid and lipids. J Bacteriol 126:192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. 2008. The major and minor wall teichoic acids prevent the sidewall localization of vegetative dl-endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- 38.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol 75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 39.Vanderwinkel E, De Vlieghere M. 1985. Modulation of Escherichia coli N-acetylmuramoyl-l-alanine amidase activity by phosphatidylglycerol. Biochim Biophys Acta 838:54–59. doi: 10.1016/0304-4165(85)90249-1. [DOI] [PubMed] [Google Scholar]
- 40.Croux C, Canard B, Goma G, Soucaille P. 1992. Autolysis of Clostridium acetobutylicum ATCC 824. J Gen Microbiol 138:861–869. doi: 10.1099/00221287-138-5-861. [DOI] [PubMed] [Google Scholar]
- 41.Hutton CA, Perugini MA, Gerrard JA. 2007. Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol Biosyst 3:458–465. doi: 10.1039/b705624a. [DOI] [PubMed] [Google Scholar]
- 42.Weart RB, Lee AH, Chien A-C, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chien AC, Zareh SK, Wang YM, Levin PA. 2012. Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol Microbiol 86:594–610. doi: 10.1111/mmi.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AM, Harrison JS, Sprague KM, Roy H. 2013. A conserved hydrolase responsible for the cleavage of aminoacylphosphatidylglycerol in the membrane of Enterococcus faecium. J Biol Chem 288:22768–22776. doi: 10.1074/jbc.M113.484402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dare K, Shepherd J, Roy H, Seveau S, Ibba M. 2014. LysPGS formation in Listeria monocytogenes has broad roles in maintaining membrane integrity beyond antimicrobial peptide resistance. Virulence 5:534–546. doi: 10.4161/viru.28359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeWitt T, Grossman AD. 2014. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol 196:1588–1596. doi: 10.1128/JB.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, Shida T, Sekiguchi J. 2008. Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting n-acetylmuramidase and dl-endopeptidase activities. J Biol Chem 283:11117–11125. doi: 10.1074/jbc.M706626200. [DOI] [PubMed] [Google Scholar]
- 48.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci 60:2371–2388. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett 318:1–9. doi: 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- 50.Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455–3467. doi: 10.1099/mic.0.28141-0. [DOI] [PubMed] [Google Scholar]
- 51.Abajy MY, Kopec J, Schiwon K, Burzynski M, Doring M, Bohn C, Grohmann E. 2007. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J Bacteriol 189:2487–2496. doi: 10.1128/JB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menard KL, Grossman AD. 2013. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet 9:e1003623. doi: 10.1371/journal.pgen.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]