ABSTRACT
Escherichia coli RtcB is a founding member of a family of manganese-dependent RNA repair enzymes that join RNA 2′,3′-cyclic phosphate (RNA>p) or RNA 3′-phosphate (RNAp) ends to 5′-OH RNA (HORNA) ends in a multistep pathway whereby RtcB (i) hydrolyzes RNA>p to RNAp, (ii) transfers GMP from GTP to RNAp to form to RNAppG, and (iii) directs the attack of 5′-OH on RNAppG to form a 3′-5′ phosphodiester splice junction. The crystal structure of the homologous archaeal RtcB enzyme revealed an active site with two closely spaced manganese ions, Mn1 and Mn2, that interact with the GTP phosphates. By studying the reactions of wild-type E. coli RtcB and RtcB alanine mutants with 3′-phosphate-, 2′,3′-cyclic phosphate-, and 3′-ppG-terminated substrates, we found that enzymic constituents of the two metal coordination complexes (Cys78, His185, and His281 for Mn1 and Asp75, Cys78, and His168 for Mn2 in E. coli RtcB) play distinct catalytic roles. For example, whereas the C78A mutation abolished all steps assayed, the D75A mutation allowed cyclic phosphodiester hydrolysis but crippled 3′-phosphate guanylylation, and the H281A mutant was impaired in overall HORNAp and HORNA>p ligation but was able to seal a preguanylylated substrate. The archaeal counterpart of E. coli RtcB Arg189 coordinates a sulfate anion construed to mimic the position of an RNA phosphate. We propose that Arg189 coordinates a phosphodiester at the 5′-OH end, based on our findings that the R189A mutation slowed the step of RNAppG/HORNA sealing by a factor of 200 compared to that with wild-type RtcB while decreasing the rate of RNAppG formation by only 3-fold.
IMPORTANCE RtcB enzymes comprise a widely distributed family of manganese- and GTP-dependent RNA repair enzymes that ligate 2′,3′-cyclic phosphate ends to 5′-OH ends via RNA 3′-phosphate and RNA(3′)pp(5′)G intermediates. The RtcB active site includes two adjacent manganese ions that engage the GTP phosphates. Alanine scanning of Escherichia coli RtcB reveals distinct contributions of metal-binding residues Cys78, Asp75, and His281 at different steps of the RtcB pathway. The RNA contacts of RtcB are uncharted. Mutagenesis implicates Arg189 in engaging the 5′-OH RNA end.
INTRODUCTION
Escherichia coli RtcB exemplifies a novel family of RNA ligases implicated in tRNA splicing and RNA repair (1–7). Unlike classic RNA and DNA ligases, which join 3′-OH and 5′-phosphate ends, RtcB seals broken RNAs with 5′-OH (HORNA) and either 2′,3′-cyclic phosphate (RNA>p) or 3′-phosphate (RNAp) ends. E. coli RtcB executes a multistep ligation pathway (5–7) entailing (i) reaction of the enzyme with GTP to form a covalent RtcB-(His337-N)-GMP intermediate, (ii) hydrolysis of RNA>p to RNAp, (iii) transfer of guanylate from His337 to the polynucleotide 3′-phosphate to form a polynucleotide-(3′)pp(5′)G intermediate, and (iv) attack of a 5′-OH on the −NppG end to form the splice junction and liberate GMP (Fig. 1). The E. coli RtcB reaction pathway requires manganese as a divalent cation cofactor. The catalytic repertoire of E. coli RtcB is not limited to RNA transactions. RtcB efficiently “caps” DNA 3′-phosphate (DNAp) ends to form a stable DNAppG structure (8). Moreover, RtcB can ligate DNAp and 5′-OH DNA (HODNA) ends within a stem-loop structure (8).
FIG 1.
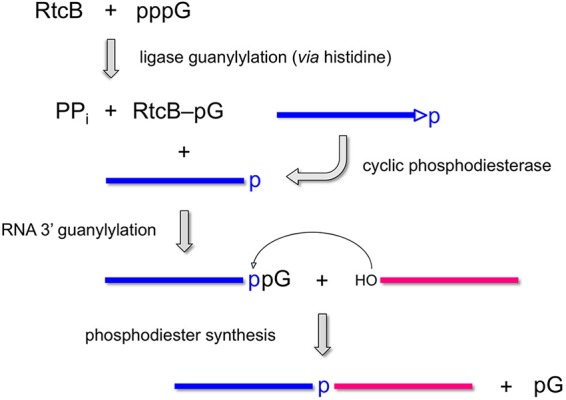
Multistep pathway of RtcB-catalyzed RNA ligation. See the text for details.
The wide phylogenetic distribution of RtcB enzymes (in bacteria, bacteriophages, archaea, and metazoa) and the inauguration by RtcB of a novel enzymology based on 3′-end activation prompt us here to interrogate the enzymic requirements for several of the component steps. We aim to distinguish between models in which (i) all pathway steps are performed by a full ensemble of active-site functional groups and cofactors or (ii) individual steps are driven by a subset of active-site constituents. In principle, this issue can be addressed by identifying RtcB changes that cripple the end-joining pathway and then testing them for separations of function, i.e., whether such mutations selectively affect one or more of the chemical steps while sparing others. The utility of this approach was exemplified by an analysis of the H337A mutant of E. coli RtcB, which eliminates the site of covalent attachment of GMP to the enzyme, rendering the H337A mutant protein inert in RtcB guanylylation and, hence, formation of the polynucleotide-ppG intermediate (6). The instructive findings were that the H337A mutant was adept at joining DNAppG and HODNA ends in a reaction that required Mn2+ but not GTP (8), signifying that GTP and His337 play no essential role in the pathway downstream of the step of 3′-end activation.
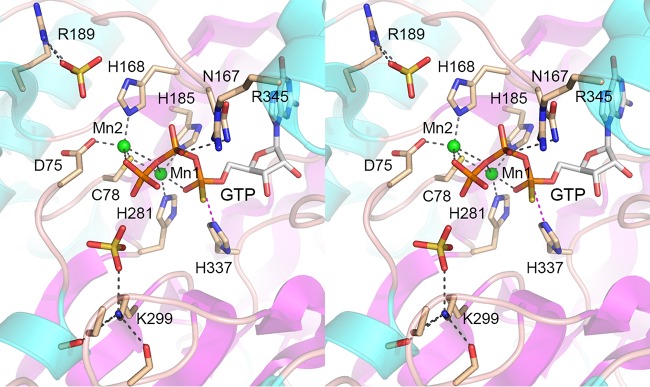
Although the biochemistry of RtcB is best understood for the E. coli enzyme, available structural insights stem from Pyrococcus horikoshii RtcB, which has been crystallized in various functional states: as the RtcB apoenzyme, as RtcB complexes with Mn2+ or Mn2+-GTP(αS), and as the covalent RtcB-pG–Mn2+ intermediate (9–11). A key insight from the crystal structures was that RtcB binds two Mn2+ ions, separated by 3.6 Å, one of which (Mn1) (Fig. 2) coordinates the α phosphate of GTP (and the GMP phosphate in the covalent RtcB-pG complex), while the other (Mn2) (Fig. 2) contacts the GTP γ phosphate (11). Both metal ions also contact the β-γ bridging oxygen of GTP. For convenience, we have labeled the conserved amino acids in the archaeal RtcB structure in Fig. 2 according to their positions in E. coli RtcB. Mn1 is coordinated by a cysteine and two histidines (Cys78, His185, and His281 in E. coli RtcB); Mn2 is coordinated by aspartate, cysteine, and histidine side chains (Asp75, Cys78, and His168 in E. coli RtcB). The cysteine uniquely bridges the two metal ions (Fig. 2). The predominance of “soft” metal ligands (histidine nitrogens and cysteine sulfur) in the active site explains the observed metal activity and inhibition spectrum of E. coli RtcB (1, 5, 8), whereby (i) the overall ligation pathway and the component steps of RtcB guanylylation and phosphodiester synthesis rely on Mn2+, which is favorably coordinated by soft ligands; (ii) “hard” metals Mg2+ and Ca2+, which favor oxygen ligands, neither support RtcB activity themselves nor inhibit RtcB function in the presence of Mn2+; and (iii) alternative soft metals Zn2+, Co2+, Cu2+, and Ni2+ that do not support activity are potent inhibitors of activity in the presence of Mn2+, presumably because they compete with Mn2+ for binding to one or both metal sites, whence bound they are unable to sustain catalysis. It is an open question whether both Mn2+ sites in the binuclear cluster are required for each step in the RtcB pathway or if some of the steps rely on just one of the metal ions.
FIG 2.
Structure of an RtcB-GTP-(Mn2+)2 complex. Shown is a stereo view of the active site of P. horikoshii RtcB (PDB accession number 4ISZ) highlighting atomic interactions of two manganese ions (Mn1 and Mn2) (depicted as green spheres) and GTPαS (stick model with gray carbons). The RtcB fold is shown as a cartoon model with magenta β strands and cyan α helices. Selected RtcB amino acids are depicted as stick models with beige carbons. Amino acids are numbered according to their equivalents in E. coli RtcB. Two sulfate anions (stick models) in the vicinity of GTPαS are suggested to mimic RNA phosphates. Atomic contacts are indicated by black dashed lines. His337-Nε is poised for nucleophilic attack on the GTP α phosphorus, as denoted by the magenta dashed line.
In the Pyrococcus RtcB-Mn2+-GTP(αS) structure, the His404-Nε nucleophile (His337 in E. coli RtcB) is situated 3.3 Å from the GTP α phosphorus in a favorable apical orientation (168°) to the bridging oxygen of the pyrophosphate leaving group, signifying that the structure plausibly mimics the Michaelis complex of the RtcB guanylylation reaction (11). The structure highlights additional active-site amino acids that contact GTP or that contact nearby sulfate anions, which might mimic the phosphates at and flanking the polynucleotide ends that RtcB joins.
In a preliminary study (2) antedating the elucidation of the multistep RtcB pathway, we tested the effects of a set of alanine mutations of E. coli RtcB, at positions chosen based on their phylogenetic conservation and spatial proximity in the then-available apoenzyme structure of Pyrococcus RtcB. The mutated set included the side chains depicted in Fig. 2 (except His281, which was mutated presently) plus two amino acids not shown in Fig. 2 (Arg341, a conserved residue, and His280, a nonconserved residue corresponding to Ala328 in Pyrococcus RtcB). Initial tests of sealing of a broken tRNA-like stem-loop with 2′,3′-cyclic phosphate and 5′-OH RNA ends at a single level of input recombinant enzyme showed that (i) the H280A mutant retained full sealing activity; (ii) the R345A, N167A, H185A, R189A, and K299A mutants had low activity; and (iii) the D75A, C78A, H168A, H337A, and R341A mutants had no activity under the conditions tested (2). Since then, our laboratory has improved the purification of recombinant RtcB protein, such that it is fully dependent on exogenous GTP for sealing activity (5). Moreover, we have developed new substrates to assay the cyclic phosphodiesterase (CPDase), 3′-PO4/5′-OH ligation, 3′-PO4 guanylylation, and 3′-ppG/5′-OH ligation reactions under single-turnover conditions (5–8). Here we applied these interval technical advances to reexamine the effects of active-site alanine mutations on E. coli RtcB function in vitro. Our results identify separation-of-function mutations and step-specific defects. We discuss our findings in light of the available crystal structures.
MATERIALS AND METHODS
RtcB purification.
Wild-type RtcB and RtcB alanine (RtcB-Ala) mutants were produced in E. coli as His10Smt3-RtcB fusions and isolated from soluble lysates by nickel-agarose chromatography as described previously (5). The tags were excised by treatment with the Smt3-specific protease Ulp1. The tag-free RtcB proteins were separated from His10Smt3 by a second round of nickel-agarose chromatography, during which RtcB was recovered in the flowthrough fractions, which were immediately adjusted to 20 mM EDTA and incubated for 1 h before dialysis overnight against a solution containing 10 mM Tris-HCl (pH 8.0), 350 mM NaCl, 1 mM dithiothreitol (DTT), and 1 mM EDTA. The RtcB preparations were stored at −80°C. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin (BSA) as the standard.
RNA and DNA substrates.
The 20-mer HORNAp oligonucleotide labeled with 32P at the penultimate phosphate was prepared by T4 Rnl1-mediated addition of 5′-[32P]pCp to a 19-mer synthetic oligoribonucleotide as described previously (5). The HORNAp was treated with E. coli RNA 3′-terminal phosphate cyclase (RtcA) and ATP to generate a 2′,3′-cyclic phosphate derivative, HORNA>p (5). The HORNAp and HORNA>p substrates were gel purified prior to use in RtcB assays. The 12-mer DNAp oligonucleotide was 5′-phosphorylated by reaction with T4 Pnkp-D167N and [γ32P]ATP to form pDNAp and then gel purified. The preguanylylated pDNAppG strand was prepared by reaction with RtcB, Mn2+, and GTP and then gel purified and annealed to a complementary cold HODNA strand to form a DNA stem-loop as described previously (8).
RtcB activity assays.
Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM radiolabeled nucleic acid substrate, and 1 μM RtcB were incubated at 37°C. The reactions were initiated by adding RtcB to the mixtures and quenched at the times specified by adding an equal volume of 90% formamide–50 mM EDTA to the mixtures. Alternatively, in reaction mixtures with the HORNA>p substrate, the reactions were quenched by adding 0.5 volumes of 40 mM EDTA to the mixture, followed by digestion for 15 min at 37°C with 1,000 U RNase T1 (Fermentas). The RNase T1 digests were then mixed with an equal volume of formamide-EDTA. The reaction products were analyzed by electrophoresis (at a constant power of 50 W) through a 40-cm 20% polyacrylamide gel containing 8 M urea in 45 mM Tris borate–1.2 mM EDTA. The 32P-labeled nucleic acids were visualized by autoradiography of the gel and, where specified, quantified by scanning of the gel with a Fuji Film BAS-2500 imager.
RESULTS
Effects of active-site mutations on HORNAp ligation.
Wild-type RtcB and 11 Ala mutants were produced in E. coli as His10Smt3 fusions and isolated from bacterial soluble extracts by adsorption to Ni-agarose and elution with imidazole. After cleavage of the tag with the Smt3 protease Ulp1, the RtcB proteins were separated from His10Smt3 by a second round of Ni-agarose chromatography. SDS-PAGE of the protein preparations showed that the RtcB polypeptide was the major species in each case (Fig. 3).
FIG 3.
RtcB mutants. Aliquots (5 μg) of the indicated RtcB preparations were analyzed by SDS-PAGE. A Coomassie blue-stained gel is shown. The positions and sizes (kilodaltons) of marker polypeptides are indicated on the left. WT, wild type.
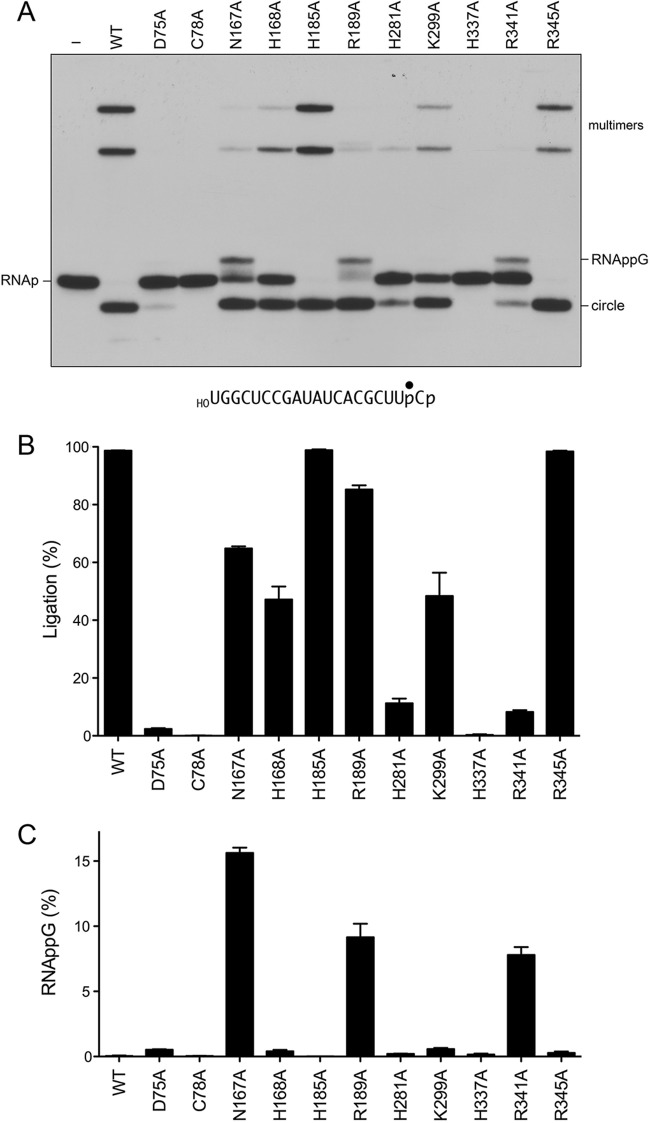
To initially gauge mutational effects on RNA ligation activity, the wild-type RtcB and RtcB-Ala proteins (1 μM) were reacted for 5 min at 37°C with 0.1 μM HORNAp (a 20-mer oligoribonucleotide, labeled with 32P at the penultimate phosphate) in the presence of 2 mM Mn2+ and 100 μM GTP. The products were analyzed by urea-PAGE and visualized by autoradiography (Fig. 4A). Wild-type RtcB converted the HORNAp substrate into a circular intramolecular ligation product that migrated ahead of the substrate strand. RtcB also generated more slowly migrating multimers via intermolecular end joining. The H337A mutant, which lacks the histidine nucleophile for the RtcB reaction with GTP to form RtcB-pG (6), was inert in HORNAp ligation, as was the C78A mutant (Fig. 4A and B), which lacks the cysteine that coordinates both of the manganese ions in the active site (Fig. 2). The D75A mutant, which lacks an aspartate that coordinates Mn2, was crippled in HORNAp ligation, with only 4% of the input substrate being sealed (Fig. 4A and B). In contrast, the H185A (lacking one of the ligands for Mn1) and R345A mutants were similar to the wild type with respect to their high extent of sealing and mixture of circular and multimer products (Fig. 4A and B). The H168A (missing one of the Mn2 ligands) and K299A mutants had diminished ligase activity, whereby half of the input HORNAp substrate was sealed. The H281A mutant was affected more severely, sealing only 13% of the HORNAp substrate.
FIG 4.
Mutational effects on HORNAp ligation. (A) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 20-mer HORNAp (depicted at the bottom, with the radiolabeled phosphate denoted by ●), and either 1 μM RtcB as specified or no RtcB (lane −) were incubated at 37°C for 5 min. The products were analyzed by urea-PAGE. An autoradiograph of the gel is shown. The identities of the radiolabeled RNAs are indicated at the left and right. (B and C) The extents of RNA ligation (circle plus multimers) (B) and RNAppG accumulation (C), expressed as a percentage of the total radiolabeled RNA, are plotted in bar graph format. Each datum is the average of results from three experiments ± the standard error of the mean.
The most instructive findings pertained to the N167A, R189A, and R341A mutants, which, notwithstanding their various extents of RNA sealing (65%, 85%, and 8%, respectively), all accumulated an HORNAppG intermediate that migrated just above the HORNAp substrate (Fig. 4A). The HORNAppG intermediate comprised between 8% and 16% of the labeled RNA in these reactions (Fig. 4C).
The R189A mutant selectively affects the kinetics of phosphodiester synthesis.
Previous kinetic studies of the single-turnover HORNAp ligation reaction of wild-type RtcB detected only trace amounts of HORNAppG (1% of the total labeled RNA) at the earliest times sampled: 15 and 30 s (6). Here we revisited the kinetics of single-turnover HORNAp ligation by sampling the reaction products at shorter intervals. HORNAppG comprised 12% and 9% of the labeled RNA at 1 and 2 s, respectively, and then declined, concomitant with a steady increase in the levels of ligated RNA (Fig. 5). Fitting of the data to a simple two-step kinetic scheme yielded apparent rate constants of 17.7 min−1 for the formation of HORNAppG (kguan) and 137 min−1 for subsequent phosphodiester synthesis (klig).
FIG 5.
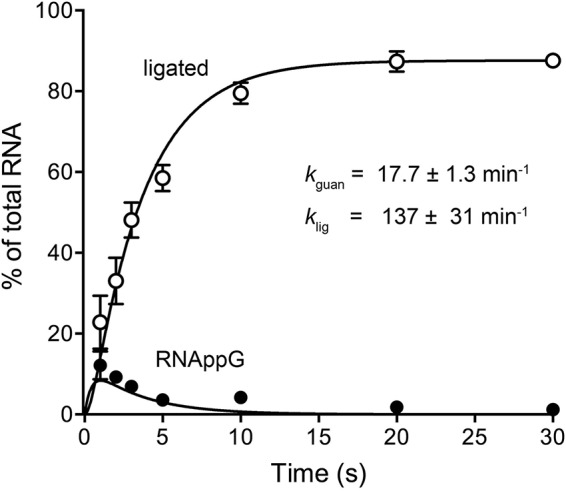
Kinetic profile of HORNAp ligation by wild-type RtcB. Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 1 μM RtcB, and 0.1 μM HORNAp were incubated at 37°C. The reactions were quenched with formamide-EDTA at the times specified. The products were resolved by urea-PAGE and quantified by scanning of the gel. The levels of RNAppG and ligated RNAs are plotted as a function of time. Each datum is the average of results from three experiments ± the standard error of the mean. The data were fit by nonlinear regression in Prism to a two-step kinetic mechanism (HORNAp → HORNAppG → ligated RNA) with rate constants, kguan and klig, as indicated.
The kinetic profile of the HORNAp ligation reaction of the R189A mutant was markedly different with respect to the time scale (endpoint approached in 10 min) and the transient accumulation of very high levels of the HORNAppG intermediate, which comprised 58% of the total RNA at 15 and 30 s (Fig. 6, top). Indeed, the R189A mutant profile verified the precursor-product relationship between HORNAppG and sealed RNA. Fitting of the R189A data to a two-step scheme yielded kguan and klig values of 5.75 ± 0.55 min−1 and 0.66 ± 0.033 min−1, respectively. Thus, whereas the R189A change decreased kguan by a factor of 3 compared to wild-type RtcB, it decreased klig by a factor of 200. These results implicate Arg189 specifically in the catalysis of the 5′-OH attack on RNAppG to form a phosphodiester splice junction.
FIG 6.
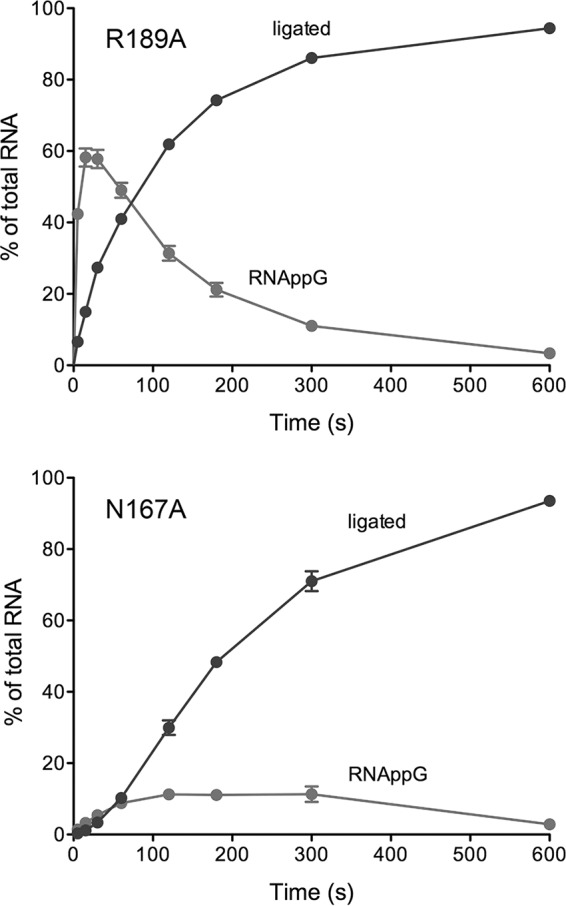
Kinetic profiles of HORNAp ligation by the R189A and N167A mutants. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM HORNAp, and 1 μM the RtcB mutant R189A or N167A were incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified and quenched immediately with formamide-EDTA. The products were resolved by urea-PAGE and quantified by scanning of the gel. The levels of RNAppG and ligated RNAs are plotted as a function of time. Each datum is the average of results from three experiments ± the standard error of the mean.
The N167A mutant displayed slower kinetics of RNA guanylylation (with HORNAppG comprising 11% of the total RNA at 2, 3, and 5 min) and a lag in the accumulation of the ligated product (Fig. 6, bottom). Whereas these data did not fit a simple two-step scheme, we derived a rate constant of 0.25 ± 0.018 min−1 for HORNAppG formation by plotting HORNAppG plus ligated RNA as a function of time and fitting these data to a single exponential curve. In comparison to wild-type RtcB, the N167A change slowed HORNAppG formation by a factor of 70.
Mutational effects on the reaction of RtcB with HORNA>p.
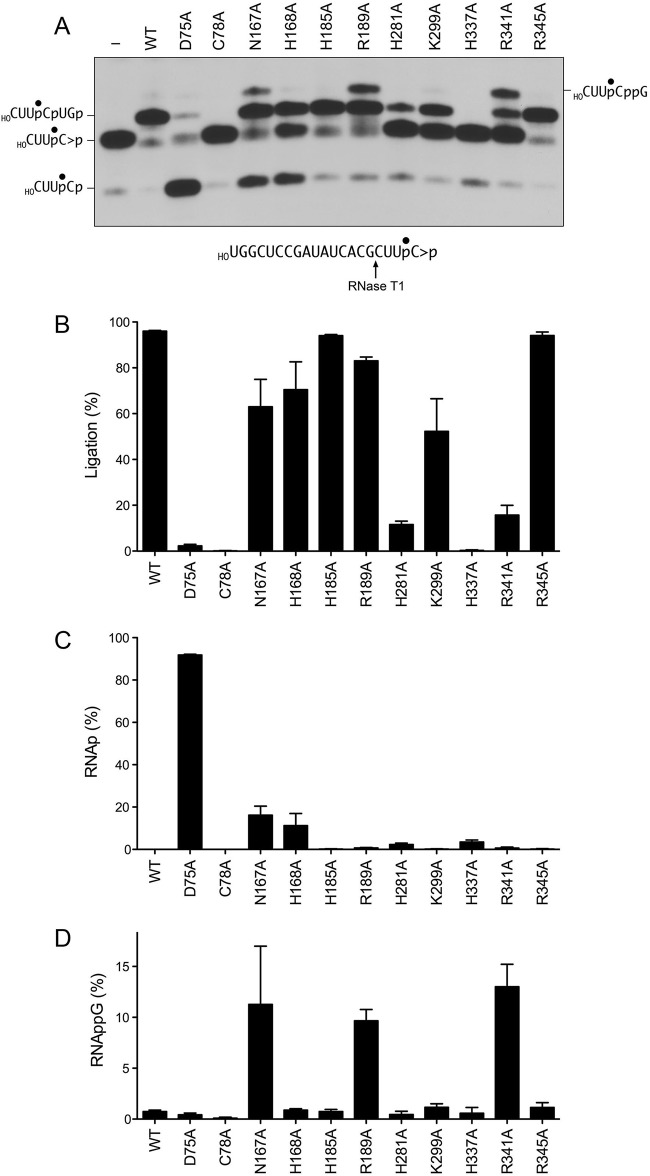
To illuminate mutational effects on the cyclic phosphodiesterase phase of the RNA repair/splicing pathway, wild-type RtcB and RtcB-Ala proteins (1 μM) were reacted for 5 min at 37°C with 0.1 μM HORNA>p (a 20-mer RNA strand with 5′-OH and 2′,3′-cyclic phosphate ends and a single radiolabel between the 3′-terminal and penultimate nucleosides) in the presence of 2 mM Mn2+ and 0.1 mM GTP. The reactions were quenched with EDTA. The mixtures were digested with RNase T1 and then analyzed by denaturing PAGE. RNase T1 incised the substrate 3′ of the most distal guanosine to yield the 32P-labeled tetranucleotide HOCUUpC>p (where p denotes the radiolabeled phosphate) (Fig. 7A, lane −). Wild-type RtcB caused a depletion of the HOCUUpC>p T1 fragment and the appearance of a more slowly migrating T1 fragment that corresponded to a 6-mer oligonucleotide, HOCUUpCpUGp, released by T1 incision at the guanosines flanking the ligation junction (Fig. 7A). As reported previously (7), the H337A mutant did not convert RNA>p to RNAp, signifying that RtcB-pG formation precedes cyclic phosphodiester hydrolysis (Fig. 1). The C78A mutant was also unable to hydrolyze HORNA>p to HORNAp (Fig. 7A).
FIG 7.
Mutational effects on HORNA>p ligation. (A) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 20-mer HORNA>p (depicted at the bottom, with the radiolabeled phosphate denoted by ●), and either 1 μM RtcB as specified or no RtcB (lane −) were incubated at 37°C for 5 min. The mixtures were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiograph of the gel is shown. The identities of the radiolabeled RNase T1 fragments are indicated at the left and right. (B to D) The extents of RNA ligation (HOCUUpCpUGp) (B) and accumulation of RNAp (HOCUUpCp) (C) and RNAppG (HOCUUpCppG) (D), expressed as a percentage of the total radiolabeled RNA, are plotted in bar graph format. Each datum is the average of results from three experiments ± the standard error of the mean.
The H185A and R345A mutants ligated the HORNA>p substrate with high efficiency (94%), with little or no apparent accumulation of the HORNAp or HORNAppG intermediate species (Fig. 7A to C). These data accord with the activity of the H185A and R345A mutants in HORNAp ligation (Fig. 4) and signify that the H185A and R345A mutants are competent for CPDase activity. The H281A and K299A mutants ligated 12% and 52% of the HORNA>p substrate, respectively (consistent with the HORNAp sealing data in Fig. 4), and did not accumulate HORNAp or HORNAppG intermediates (Fig. 7). The R189A and R341A mutants ligated 83% and 16% of the HORNA>p substrate, respectively, and both mutants accumulated HORNAppG (to an extent of 10 to 13% of the total RNA, again in accordance with the HORNAp sealing data shown in Fig. 4), although neither mutant accumulated HORNAp in the single-point assay format.
The key findings from the HORNA>p sealing experiment were that the H168A, N167A, and D75A mutants accumulated HORNAp and did so in a distinct fashion with respect to downstream steps in the RtcB pathway. The reaction of the H168A mutant with HORNA>p yielded ligated RNA and HORNAp, without accumulating HORNAppG. The N167A mutant generated ligated RNA and HORNAp while also accumulating HORNAppG (Fig. 7). The kinetic profile of the single-turnover reaction of the N167A mutant with HORNA>p revealed a steady increase in HORNAp levels up to 1 min, followed by a crest of HORNAp at the 2-min and 3-min times, where it comprised 28% of the total RNA (Fig. 8, green symbols). The HORNAp level declined thereafter, concomitant with the appearance of HORNAppG (Fig. 8, red symbols), which comprised 9% of the total RNA at 2, 3, and 5 min, and the steady conversion of HORNAppG to ligated RNA (Fig. 8, blue symbols) from 1 to 10 min. These data verify that HORNAp is an intermediate in the HORNA>p ligation reaction. We derived a rate constant of 0.53 ± 0.053 min−1 for the N167A CPDase, by plotting HORNAp plus HORNAppG plus ligated RNA as a function of time (Fig. 8, open circles) and fitting these data to a single exponential curve.
FIG 8.
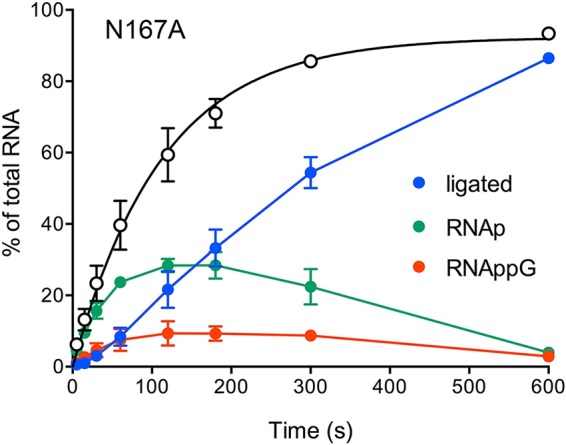
Kinetic profile of HORNA>p ligation by the N167A mutant. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 20-mer HORNA>p, and 1 μM RtcB N167A mutant were incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified, quenched with EDTA, digested with RNase T1, mixed with formamide-EDTA, and then analyzed by urea-PAGE. The extents of RNA ligation and accumulation of RNAp and RNAppG, expressed as a percentage of the total radiolabeled RNA, are plotted as a function of time. Each datum is the average of results from three experiments ± the standard error of the mean. The open circles denote RNAp plus RNAppG plus ligated RNA (as a percentage of the total RNA) with a fit of the data to a single exponential curve.
The most salient observation was that the D75A mutant converted 92% of the HORNA>p substrate to HORNAp but achieved only 4% ligation (Fig. 7). These results, and those shown in Fig. 4, signify that Asp75 is not strictly essential for RtcB CPDase activity but is essential for RNA guanylylation.
Characterization of D75A mutant CPDase activity.
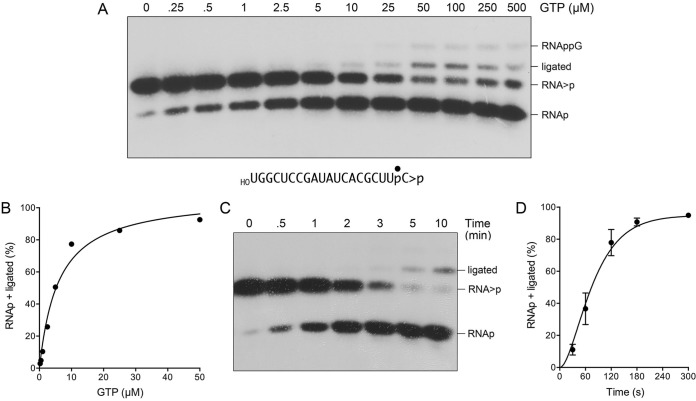
Prior studies established that the CPDase activity of wild-type RtcB is dependent on GTP (7). Here we queried whether this was the case for the D75A mutant by changing the concentration of GTP included for the single-turnover reaction of the D75A mutant with HORNA>p, in the range of 0 to 500 μM GTP. The conversion of HORNA>p to HORNAp by the D75A mutant required GTP, and the extent of conversion increased with the GTP concentration, attaining saturation at 50 μM GTP with 91% conversion to HORNAp and 1.8% ligation (Fig. 9A). The data for the D75A mutant fit a one-site binding model with half-saturation at 6 μM GTP (Fig. 9B). In comparison, assays of the GTP dependence of the RNA 3′-PO4/5′-OH ligation and RNA 2′,3′-cyclic-PO4/5′-OH ligation reactions of wild-type RtcB revealed half-saturation between 0.4 μM and 1 μM GTP (see reference 6, and data not shown). The kinetic profile of the D75A mutant CPDase reaction highlighted a steady conversion of HORNA>p to HORNAp to attain an endpoint after 3 min (Fig. 9C and D). The shape of the reaction curve was notable for a slight initial lag in HORNAp product accumulation by the D75A mutant, suggestive of a partially rate-limiting upstream step not directly detected by the assay format (presumably, the formation of an RtcB-D75A–pG intermediate). Half of the substrate was hydrolyzed to HORNAp in 1.25 min, from which we can estimate a CPDase rate of ∼0.55 min−1.
FIG 9.
CPDase activity of the D75A mutant. (A and B) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 0.1 μM 20-mer HORNA>p (depicted at the bottom, with the radiolabeled phosphate denoted by ●), 1 μM RtcB D75A mutant, and GTP as specified were incubated at 37°C for 5 min. The mixtures were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiograph of the gel is shown. The identities of the radiolabeled RNase T1 fragments are indicated at the right. The extent of 2′,3′-cyclic phosphodiester hydrolysis (RNAp plus ligated RNA) is plotted as a function of the GTP concentration (B). (C) A reaction mixture containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 20-mer HORNA>p, and 1 μM RtcB D75A mutant was incubated at 37°C. Aliquots (10 μl) were withdrawn at the times specified and quenched with EDTA. The RNAs were digested with RNase T1, and the products were analyzed by urea-PAGE. An autoradiograph of the gel is shown. (D) The extent of 2′,3′-cyclic phosphodiester hydrolysis (RNAp plus ligated RNA) is plotted as a function of time. Each datum is the average of results from three experiments ± the standard error of the mean.
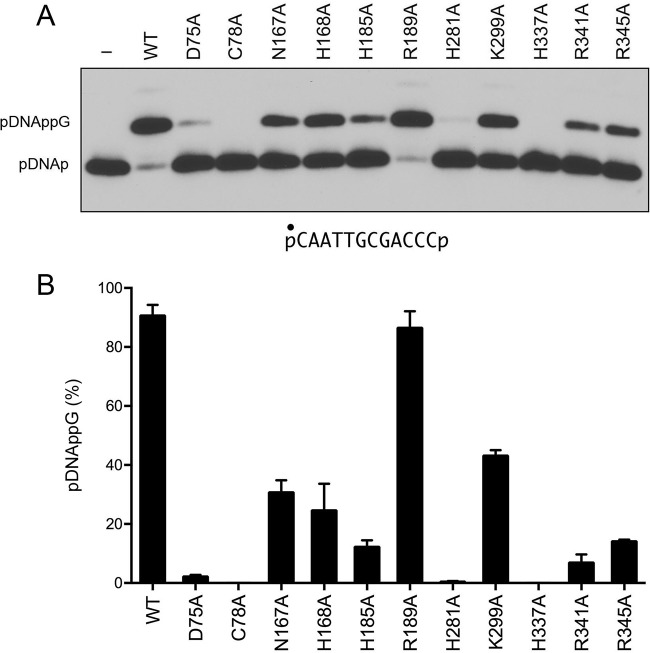
Mutational effects on DNA 3′-phosphate capping.
Bacterial RtcB enzymes transfer GMP from GTP to a DNA 3′-phosphate end (DNAp) to form a DNAppG cap structure (8, 12). To probe the enzymic requirements for DNA capping, we reacted the E. coli RtcB proteins (1 μM) for 5 min at 37°C with 0.1 μM 5′-32P-labeled 12-mer pDNAp substrate (Fig. 10A) in the presence of 2 mM Mn2+ and 100 μM GTP. The products were analyzed by urea-PAGE and visualized by autoradiography. Wild-type RtcB converted 91% of the pDNAp substrate into a slower-migrating pDNAppG capped product. For most of the mutants, the effects on DNA capping (Fig. 10) echoed what was seen for HORNAp ligation (Fig. 4). To wit, (i) the H337A and C78A mutants were inert in DNA capping; (ii) the D75A, H281A, and R341A mutants were severely compromised (2%, 0.5%, and 7% pDNAp capping, respectively); (iii) the N167A (31%), H168A (25%), and K299A (43%) mutants were modestly impaired; and (iv) the R189A mutant retained near-wild-type capping activity in the single-point assay (Fig. 10B). The exceptions were the H185A and R345A mutants, which were feeble at DNA capping (effecting 12% and 14% conversion of pDNAp to DNAppG, respectively) (Fig. 10B) but relatively unaffected in HORNAp ligation (Fig. 4B).
FIG 10.
Mutational effects on DNA 3′-phosphate capping. (A) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 12-mer pDNAp (depicted at the bottom, with the radiolabeled phosphate denoted by ●), and either 1 μM RtcB as specified or no RtcB (lane −) were incubated at 37°C for 5 min. The products were analyzed by urea-PAGE. An autoradiograph of the gel is shown. The pDNAp substrate and capped pDNAppG product are indicated at the left. (B) The extents of DNAppG formation are plotted in bar graph format. Each datum is the average of results from three experiments ± the standard error of the mean.
Effects on DNAppG ligation and deguanylylation.
The GTP-dependent steps of the RtcB ligation pathway can be bypassed by presenting the enzyme with a “broken” stem-loop DNA substrate composed of a preguanylylated 5′-32P-labeled pDNAppG strand annealed to an unlabeled 5′-OH DNA strand (Fig. 11A). As shown previously (8), wild-type RtcB will catalyze manganese-dependent phosphodiester synthesis, in the absence of added GTP, via attack of the DNA 5′-OH on the 3′-phosphate of pDNAppG to yield a sealed DNA stem-loop (ligation) (Fig. 11A). RtcB will also catalyze “backward” GMP transfer from the pDNAppG cap to the enzyme to form RtcB-pG and a 5′-32P-labeled pDNAp strand (deguanylylation) (Fig. 11A). Thus, the fate of the pDNAppG strand of the broken stem-loop reflects partitioning between ligation and deguanylylation. For example, in the experimental setup for Fig. 11, wild-type RtcB ligated 67% of the input substrate (Fig. 11B) and deguanylylated 24% (Fig. 11C). Mutational effects on the RtcB reaction with the preguanylylated DNA stem-loop fell into several instructive categories. The C78A mutant abolished phosphodiester synthesis and deguanylylation, thereby attesting to the central role of the manganese-bridging cysteine at every step of the RtcB pathway. As noted previously (8), mutation of the His337 nucleophile to alanine abolished deguanylylation (Fig. 11C) but preserved phosphodiester synthesis, such that 89% of the input pDNAppG substrate was sealed (Fig. 11B). Mutations H281A and K299A elicited similar effects on the pDNAppG reaction outcome, i.e., suppressing deguanylylation (1% and 6%, respectively) and enhancing ligation (86% and 88%, respectively), relative to that of wild-type RtcB (Fig. 11B and C). These results establish that the H281A mutation is a separation-of-function mutation that strongly impaired the GTP-dependent ligations of HORNAp (Fig. 4) and HORNA>p (Fig. 7) and the GTP-dependent capping of pDNAp (Fig. 10) but had comparatively little effect on GTP-independent sealing of pDNAppG. We surmise that the contact of His281 with Mn1 (Fig. 2) is critical for the various GTP-dependent steps but not for the step of phosphodiester synthesis at a preguanylylated end.
FIG 11.
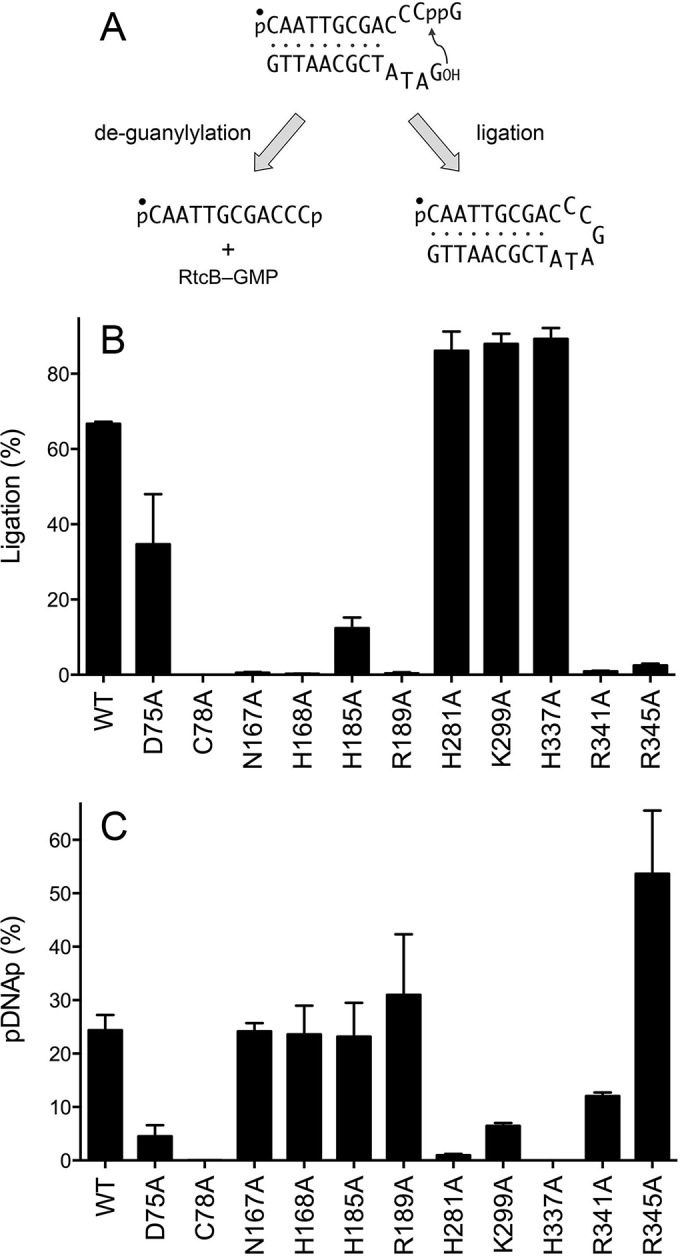
Mutational effects of DNAppG ligation and deguanylylation. (A) Reactions of RtcB with a preguanylylated pDNAppG/HODNA stem-loop substrate. The 5′ 32P label on the pDNAppG strand is denoted by ●. (B and C) Reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, 0.1 μM 12-mer pDNAppG stem-loop, and 1 μM RtcB as specified were incubated at 37°C for 5 min. The products were resolved by urea-PAGE and quantified by scanning of the gel. The extents of DNAppG ligation (B) and deguanylylation to pDNAp (C) are plotted in bar graph format. Each datum is the average of results from three experiments ± the standard error of the mean.
Other mutations exerted the opposite effect on the reaction of RtcB with the pDNAppG stem-loop; i.e., they suppressed ligation and favored deguanylylation. This class included the R189A mutant, which ligated 0.4% of the pDNAppG strand yet deguanylylated 31% (Fig. 11B and C). This result accords with the R189A mutant accumulating HORNAppG during the HORNAp and HORNA>p sealing reactions and fortifies the conclusion that Arg189 is especially important for the step of phosphodiester synthesis. Other mutants with outcomes heavily skewed toward deguanylylation of the pDNAppG stem-loop were the N167A (0.5% ligation and 24% deguanylylation), H168A (0.2% ligation and 24% deguanylylation), R341A (0.9% ligation and 12% deguanylylation), and R345A (2.5% ligation and 54% deguanylylation) mutants (Fig. 11B and C). The H185A mutant displayed a more modest preference in this direction (12% ligation and 23% deguanylylation).
The D75A mutant, although less active than wild-type RtcB with the pDNAppG stem-loop, nonetheless retained a bias toward sealing as the reaction outcome (35% ligation and 5% deguanylylation) (Fig. 11B and C). Thus, the D75A mutant is revealed as a separation-of-function mutant that is (relatively) competent in 2′,3′-cyclic phosphodiester hydrolysis and phosphodiester synthesis but severely defective in RNAp and DNAp guanylylation.
DISCUSSION
By studying the reactions of wild-type E. coli RtcB and RtcB-Ala mutants with 3′-phosphate, 2′,3′-cyclic phosphate, and 3′-ppG terminated substrates, we provide new evidence in support of the multistep ligation pathway in Fig. 1, and we illuminate the distinctive contributions of certain active-site amino acids to individual steps of the ligation pathway. The mutational data, when interpreted in light of archaeal RtcB structures (11), suggest that (i) the enzymic constituents of the two manganese coordination complexes play distinct catalytic roles and (ii) Arg189 is a plausible candidate to position the 5′-OH polynucleotide end during the step of phosphodiester synthesis.
Initial characterization of E. coli RtcB established a requirement for manganese as the cofactor for ligation (1) by virtue of its essentiality for both RtcB-pG formation (5) and sealing of a preguanylylated stem-loop (8). Pyrococcus RtcB structures highlighted RtcB as a binuclear metalloenzyme with closely spaced Mn2+ ions bridged by an invariant cysteine (10, 11). Mutations of this cysteine abolished the overall ligase activity of E. coli, Pyrococcus, and mammalian RtcBs (2, 4, 10). In the present study, we showed that Cys78 of E. coli RtcB was essential for every reaction step tested. However, subtractions of other enzymic side-chain metal ligands elicited defects of various magnitudes at different component pathway steps.
His281 and His185 are the unique amino acid components of the Mn1 coordination complex, but their mutations had disparate effects. The loss of His281 was quite deleterious to overall HORNAp and HORNA>p ligation and to pDNAp capping but was well tolerated with respect to sealing of a preguanylylated stem-loop, implying that His281 and its contact with Mn1 are critical for RtcB's reaction with GTP. In contrast, the loss of His185 was relatively well tolerated with respect to HORNAp and HORNA>p ligation in the presence of 2 mM manganese, suggesting that the H185A mutant's reactivity with GTP was preserved. In this vein, it is worth noting that the reaction of Pyrococcus RtcB with GTP was preserved when the equivalent His234 Mn1 ligand was mutated to alanine (10). (The H185A mutant had a detrimental effect on pDNAp capping and pDNAppG ligation by E. coli RtcB. The selective impact on DNA substrates is a feature shared with the R345A mutant. The basis for this effect is unclear, absent a structure of RtcB in complex with nucleic acid).
His168 and Asp75 are the unique constituents of the Mn2 coordination complex that includes the GTP γ phosphate, and their mutations also had disparate effects. Subtraction of Asp75 virtually effaced overall HORNAp and HORNA>p ligation and DNA 3′-phosphate capping while permitting the hydrolysis, in high yield, of an RNA 2′,3′-cyclic phosphate in a manner that requires GTP, just as the CPDase activity of wild-type RtcB is GTP dependent (7). No other mutants in our collection displayed this biochemical phenotype. One early model for the RtcB mechanism invoked a concerted single-step reaction of RtcB-pG with RNA>p to form RNAppG (10), but the concerted model does not stand up to the findings that (i) wild-type RtcB is fully capable of joining RNAp ends via an RNAppG intermediate and (ii) the D75A mutation separates the CPDase activity from downstream steps. Our findings implicate Mn2 ligand Asp75 as being critical for the step of polynucleotide-3′-phosphate guanylylation, and they are in accordance with the observation that the equivalent D95A change in Pyrococcus RtcB preserved enzyme guanylylation but abolished RNA ligation (10).
In contrast, the loss of Mn2 ligand His168 had a modest effect on HORNAp and HORNA>p ligation and DNAp capping (e.g., compared to that of the D75A mutant). The detection of RNAp (but not RNAppG) among the products of the reaction of the H168A mutant with HORNA>p (Fig. 7) suggests that RNA 3′-phosphate guanylylation is most sensitive to this mutation.
The counterpart of E. coli RtcB Arg189 (Arg238 in Pyrococcus RtcB) coordinates a sulfate anion in the RtcB-GTP-(Mn2+)2 structure (Fig. 2) and the RtcB-pG–(Mn2+)2 structure (11) and may be construed to mimic the position of a phosphate in one of the RNA substrate strands. The sulfate sulfur atom is located 11 Å away from the GMP phosphorus atom of the RtcB-pG–(Mn2+)2 covalent intermediate (11). Based on our findings that a mutation of Arg189 to alanine selectively affected phosphodiester synthesis while sparing RNAp guanylylation, we speculate that Arg189 promotes the catalysis of phosphodiester synthesis by coordinating the 5′-terminal phosphodiester of the HORNA strand (HON1pN2pN3−) and thereby positioning the 5′-OH for its attack on the 3′-phosphate of RNAppG. This is analogous to the established role of an essential arginine in polynucleotide kinase enzymes, whereby the arginine coordinates the 5′-terminal phosphodiester of the HORNA or HODNA strand to poise the 5′-OH for its attack on the γ phosphate of the nucleotide triphosphate (NTP) phosphate donor (13, 14). Needless to say, the proof of this speculation for the RtcB arginine residue will hinge on capturing crystal structures of RtcB-pG in complex with RNAp and/or HORNA strands.
The N167A mutation affected the CPDase, RNA guanylylation, and phosphodiester synthesis steps of the RtcB pathway. The involvement of Asn167 in multiple reaction steps is in keeping with the atomic interactions of the equivalent Asn202 side chain in Pyrococcus RtcB. The Asn contacts a GTP β phosphate nonbridging oxygen in the RtcB-GTP-(Mn2+)2 structure (Fig. 2); the Asn undergoes a rotamer shift in the RtcB-pG–(Mn2+)2 structure so that it contacts one nonbridging oxygen and the 5′-bridging oxygen of the covalently bound GMP (11).
Finally, we suspect that the functional consequences of the K299A, R341A, and R345A mutations reflect the “structural” roles of these functional groups. The Pyrococcus RtcB equivalent of Lys299 (Lys351) donates hydrogen bonds to three main-chain carbonyls that tether multiple secondary structure elements of the RtcB fold (Fig. 2). The counterpart of Arg345 (Arg412) makes a salt bridge to a conserved glutamate. The equivalent of Arg341 (Arg408) makes a salt bridge to the same glutamate and also donates hydrogen bonds to a main-chain carbonyl (11).
ACKNOWLEDGMENTS
This research was supported by NIH grant GM46330 (S.S.) and by MSKCC core grant P30-CA008748.
REFERENCES
- 1.Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka N, Meineke B, Shuman S. 2011. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem 286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. 2011. Archaeal 3′-phosphate RNA splicing ligase characterization identified the missing component in tRNA maturation. Proc Natl Acad Sci U S A 108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lürhmann R, Söll D, Martinez J. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci U S A 109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarty AK, Shuman S. 2012. The sequential 2′,3′ cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res 40:8558–8567. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das U, Chakravarty AK, Remus BS, Shuman S. 2013. Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc Natl Acad Sci U S A 110:20437–20442. doi: 10.1073/pnas.1314289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada C, Maegawa Y, Yao M, Tanaka I. 2006. Crystal structure of an RtcB homolog protein (PH1502-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins 63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 10.Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yoa M, Eom SH, Konigsberg WH, Söll D, Wang J. 2012. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining between 3′-terminal phosphate and 5′-OH. Proc Natl Acad Sci U S A 109:15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai KK, Bingman CA, Phillips GN, Raines RT. 2013. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry 52:2518–2525. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maughan WP, Shuman S. 2015. Characterization of 3′-phosphate RNA ligase paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus highlights DNA and RNA 5′-phosphate capping activity of RtcB3. J Bacteriol 197:3616–3624. doi: 10.1128/JB.00631-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eastberg JH, Pelletier J, Stoddard BL. 2004. Recognition of DNA substrates by bacteriophage T4 polynucleotide kinase. Nucleic Acids Res 32:653–660. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das U, Wang LK, Smith P, Jacewicz A, Shuman S. 2014. Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP·Mg2+ and 5′-OH oligonucleotide and a product complex with GDP·Mg2+ and 5′-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition. Nucleic Acids Res 42:1152–1161. doi: 10.1093/nar/gkt936. [DOI] [PMC free article] [PubMed] [Google Scholar]