Abstract
Cryo-electron tomography (cryo-ET) has emerged as a leading technique for three-dimensional visualization of large macromolecular complexes and their conformational changes in their native cellular environment. However, the resolution and potential applications of cryo-ET are fundamentally limited by specimen thickness, preventing high-resolution in situ visualization of macromolecular structures in many bacteria (such as Escherichia coli and Salmonella enterica). Minicells, which were discovered nearly 50 years ago, have recently been exploited as model systems to visualize molecular machines in situ, due to their smaller size and other unique properties. In this review, we discuss strategies for producing minicells and highlight their use in the study of chemotactic signaling, protein secretion, and DNA translocation. In combination with powerful genetic tools and advanced imaging techniques, minicells provide a springboard for in-depth structural studies of bacterial macromolecular complexes in situ and therefore offer a unique approach for gaining novel structural insights into many important processes in microbiology.
INTRODUCTION
A living cell can be viewed as a miniature factory of a large collection of dedicated molecular machines (1). These machines, optimized by billions of years of evolution, orchestrate nearly every major biochemical process in the cell. A grand challenge for cell biology is to study the assembly and function of the machines at the molecular level, particularly within their cellular environment. Advances in imaging techniques and novel sample preparation methods are providing unparalleled opportunities to visualize the molecular machines and subcellular structures in cells.
Electron microscopy (EM) is one imaging technique and has been instrumental to cell biology through seminal discoveries on cellular organization and ultrastructure (2). Traditional EM, however, has been limited to the production of images of subcellular structures lacking molecular details. Moreover, traditional sample preparations comprising chemical fixation, staining, dehydration, and sectioning unavoidably disturb the native state of cells and introduce artifacts that complicate our understanding of cellular organization. The development of an easily applicable method for sample vitrification made it possible to preserve the native structures of cells, organelles, and biomolecules (3). Together with recent breakthroughs in electron microscopic hardware and image processing software, cryo-EM is now capable of determining the atomic structures of biochemically purified biomolecules (4). On a broader spectrum, cryo-electron tomography (cryo-ET) can provide three-dimensional (3D) structural information ranging from the cellular to the molecular level, enabling a better understanding of fundamental processes in eukaryotic and prokaryotic cells (2, 5–9).
Cryo-ET is particularly powerful for studying bacterial cells because of their relatively small size. It has extensively been used for investigating bacterial chemotaxis systems (10, 11), cytoskeletal filaments (12–14), motility machineries (15–20), cell division (21, 22), and phage assembly and infection (23–25). However, most bacterial cells are still too large for high-resolution in situ characterizations of cellular structures.
Many techniques have been used to produce frozen-hydrated bacterial samples that are thin enough for cryo-ET. Bacteria of <300 nm in diameter, such as spirochetes, can be used directly for preparing thin frozen-hydrated specimens (16, 17, 26–28). Larger bacteria can be treated with antibiotics or lysozyme to flatten them (29). Alternatively, the expression of a phage lysis gene (30) has been used to produce partially lysed flatter cells that contain native membranes and membrane-associated protein complexes. Vitreous sectioning (31) and focused ion beam milling (FIB) (32, 33) have been developed to produce thin and vitreous sections, opening a new window to view the ultrastructure of larger cells and tissues in their native state. Nevertheless, both techniques require expensive instrumentation and remain technically challenging (34–36).
Recently, bacterial minicells have rapidly emerged as a valuable system for studying molecular machines in situ by cryo-ET, as minicells are considerably smaller than normal bacterial cells, and their preparation does not require specialized equipment. Here, we review various methods for producing, isolating, and purifying minicells and highlight many recent applications.
A MINIHISTORY OF MINICELLS
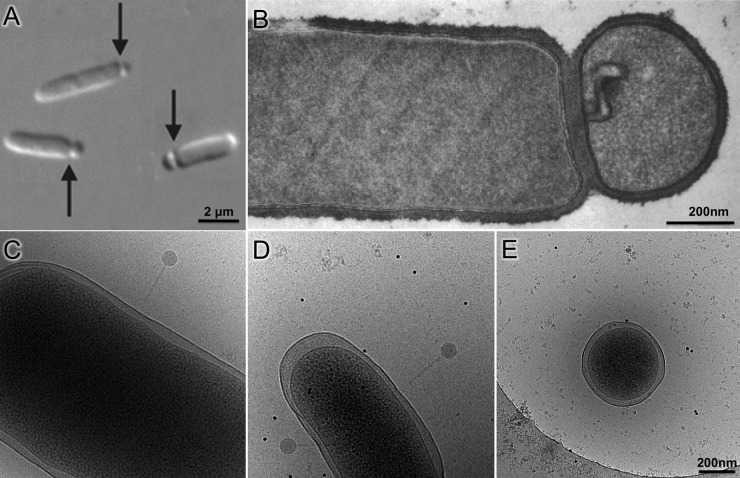
Although the first report of minicell-producing bacteria dates back to 1930 (37), the term minicell was coined in 1967 by Howard Adler's group after discovering miniature cells in a mutant strain of Escherichia coli (38). Minicell production has since been documented in a variety of bacterial species, both Gram negative and Gram positive (Fig. 1A and B) (37). Minicells are generally produced by aberrant cell divisions at chromosome-free polar ends of rod-shaped bacteria. Like their parent cells, minicells contain membranes, peptidoglycan, ribosomes, RNA, protein, and often plasmids but no chromosome (37). As a result, minicells cannot divide or grow, but they can continue other cellular processes, such as ATP synthesis, replication and transcription of plasmid DNA, and translation of mRNA. The minicell system was widely exploited in the late 1960s through the 1970s to study a variety of processes, including cell division, molecular transport, bacteriophage infection, and polypeptide synthesis (37). Now, there is renewed interest in developing minicells into drug delivery systems (39, 40) and vaccines that activate an immune response (41), because minicells often retain the virulence properties of the parent cells and yet cannot proliferate (42). Importantly, their unique properties emphasize the potential of using minicells and minicell-producing mutants as a toolbox for in situ structural biology.
FIG 1.
Bacterial minicell production and comparison of minicells to parent cells. (A) Composite differential interference contrast image of minicell formation through polar division in E. coli. (B) Thin-sectioned negative-stain micrograph of B. subtilis minicell (adapted from reference 55 with permission of the publisher). (C to E) Micrographs of cryopreserved wild-type E. coli (C), skinny E. coli (D), and a minicell (E) generated from the skinny E. coli (republished from reference 61 with permission of the publisher). Phage P1 is visible attached to cells in panels C and D. The scale of panels C and D is the same as that in panel E.
MINICELL-PRODUCING MUTANTS
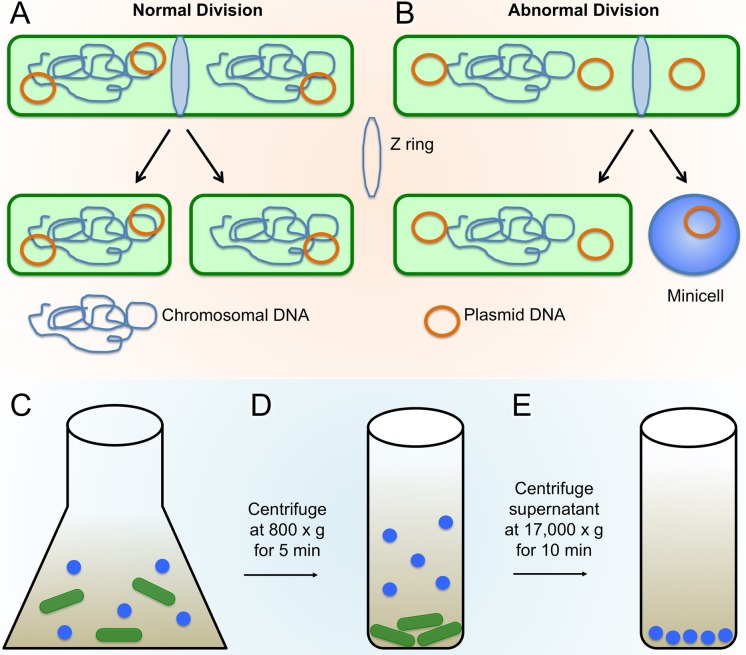
Genetic and biochemical characterization of E. coli minicell-producing mutants revealed that most are defective in a multiprotein system known as the Min system, which mediates the proper midcell placement of the cell division septum (43, 44). The Min system is best characterized in E. coli and Bacillus subtilis but is present in diverse bacterial species and in chloroplasts (45). In E. coli, three Min proteins, MinC, MinD, and MinE, synergistically mediate the proper placement of the cell division machinery, or the divisome, by inhibiting its development at sites other than midcell (46). FtsZ, a bacterial homolog of tubulin, is an essential component of this complex, in that it initiates the development of the divisome by forming a ring-like structure, termed the Z-ring, at the potential site of division, ultimately recruiting additional divisome proteins to form the complete divisome (47, 48). In E. coli, the Min proteins rapidly transit from one cell pole to the opposite cell pole, forming a bipolar inhibitory gradient that blocks assembly of the Z-ring near the poles but allows it near midcell (Fig. 2A) (49–51).
FIG 2.
Minicell production and enrichment. (A and B) Normal bacterial cell division (A) compared with abnormal division (B), resulting in minicell generation due to polar Z-ring placement (adapted from reference 41 with permission of the publisher). (C to E) Minicells are separated from typical rod-shaped cells through centrifugation; first, the larger rod-shaped cells are pelleted through a short low-speed spin, and then minicells are pelleted from the resulting supernatant fraction through a longer and higher-speed centrifugation.
Mutant strains of E. coli deficient in the Min system are not able to spatially restrict the Z-ring to midcell. Another negative spatial regulator of Z-rings called “nucleoid occlusion,” which inhibits Z-ring assembly in the space occupied by the chromosomes, is still intact in Δmin cells (52). The result is that Z-rings in Δmin mutants assemble at either midcell between separated chromosomes or at the chromosome-free cell poles (Fig. 2B). The polar division from a polar Z-ring leaves behind a viable mother cell containing chromosomal DNA and a chromosome-less minicell; the production of mother cells with chromosomes along with normal cells from divisions at midcell maintains the overall viability of min mutants of E. coli similar to that of wild-type cells. B. subtilis has an analogous system, although some of the protein components are different (53), and they do not oscillate. The result is a similar bipolar gradient that prevents inappropriate Z-rings from assembling near cell poles (54). Inactivation of the Min system in B. subtilis, as with E. coli, results in polar septation and minicell formation (Fig. 1B) (55, 56). Growth in rich medium significantly increases the percentage of B. subtilis minicells (55), although it is not clear if this is generally applicable to other species.
Inactivation of Min or Min-like systems has proven successful in generating minicells from many species, including Salmonella enterica (41), Pseudomonas aeruginosa (39), and Corynebacterium glutamicum (40). However, inactivation of the Min system in some bacterial species may not be sufficient for generating high minicell yields. For example, inactivation of the Listeria monocytogenes Min system produces minicells at very low frequency (57), perhaps because of the action of additional Z-ring spatial regulators in this species. Alternatively, the overproduction of FtsZ can overwhelm the Min system and produce minicells in an otherwise wild-type strain of E. coli (58). A similar strategy was recently used to overproduce FtsZ and resulted in a 2-fold increase in minicell production of L. monocytogenes (57).
Many rod-shaped bacteria lack a recognizable Min system, but alternative mechanisms for mediating divisome placement can be exploited to generate minicells in those species (59). For example, Myxococcus xanthus uses a novel protein called PomZ to positively regulate the assembly of the divisome by recruiting FtsZ to midcell (60). Interestingly, cells deficient in PomZ readily produce minicells, suggesting that PomZ acts as a spatial regulator of Z-ring placement (60). Inactivation of these spatial regulatory systems, as they become known, should be considered when attempting to generate minicells from species that lack typical Min systems.
Although minicells are much shorter than the rod-shaped parent cells, they may not always be small enough for optimal cryo-ET studies. One solution has been to obtain thinner E. coli cells. The actin-like MreB protein is required for the normal rod shape and width of E. coli cells (Fig. 1C), but a mutant mreB gene encoding an A125V change results in cells with significantly smaller diameters (Fig. 1D) than those of wild-type cells (61). Inactivation of the min locus in this skinny E. coli strain results in minicells with a reduced diameter (Fig. 1E), which is better for cryo-ET analysis (61). Furthermore, for reasons that are yet unclear, minicells obtained from the above-mentioned Δmin mreB-A125V double mutant are even smaller when the strain harbors a plasmid expressing the flagellar flhDC genes. An alternative approach for obtaining minicells with smaller diameters is to use a multistep minicell isolation procedure that enriches for smaller minicells (see “Isolating Minicells”).
ISOLATING MINICELLS
To prepare frozen-hydrated samples, it is ideal to isolate the minicells from the nucleated nonminicells so they may be loaded onto EM grids and vitrified. Several methods have been developed for the purification of minicells, such as differential centrifugation (Fig. 2C to E), multiple density gradient centrifugation, and multistep filtration (38, 62) (see below). For specific methodologies, see the references listed in Table 1. Each method presents certain drawbacks, including limited sample size, low recovery from density gradients, and increased cost of production.
TABLE 1.
Minicell-producing bacterial strains
| Species | Strain description and/or source | Application(s) | Reference(s) |
|---|---|---|---|
| E. coli | P678-54 treated with triethylenemelamine (0.5 mg/ml) | Basic properties of E. coli minicells | 38 |
| BS100(λDB173), a min mutant | Z-ring formation is associated with minicell formation | 106 | |
| Minicell mutant X-1488 (F− str hst hsm+ minA minB purE pdxC his ile met ade ura [r− mK+]) | Identification of membrane proteins enriched in minicells | 107 | |
| WM3433 (ΔminCDE::kan introduced into DS612 [mreB-A125V linked to yhdE::cat]) | In situ structural studies of phage P1 infection | 61 | |
| WM4011 (pBAD30-flhDC in WM3433 with ΔclpX::kan fadR::Tn10) | In situ structural studies of receptor arrays, phage adsorption, and DNA translocation | 66, 88, 90, 92 | |
| B. subtilis | CV403 + divIVBI mutant | Gram-positive minicell production and properties | 55 |
| Molecular transport | 108 | ||
| Bacteriophage-directed polypeptide synthesis | 109 | ||
| C. glutamicum | parA ΔNCgl1366 | Drug delivery system | 40 |
| Haemophilus influenzae | LB11 mutagenesis with N-methyl-N′-nitro-N-nitrosoguanidine | Phage induction | 110 |
| L. monocytogenes | ΔminCDE mutant | Drug delivery system | 39 |
| P. aeruginosa | ΔminCDE mutant | Drug delivery system | 39 |
| S. enterica | TH17261 ParaBAD1091::ftsZ+ | In situ structure of chemoreceptor array | 65 |
| SJW1103/pBAD24-ftsZ | In situ structure of injectisome and flagellum | 75 | |
| SL1344 minD::cat | Minicell virulence | 41 | |
| S. flexneri | M90T-Sm/pB558 (E. coli ftsQAZ genes) | In situ structure of injectisome | 78 |
| Y. enterocolitica | Y. enterocolitica E40 × E. coli Sm10λpir+ (BW19610) | In situ structure of injectisome | 77 |
To prepare minicells that are uniformly very small, a more complex isolation method can be used (39). In this approach, cultures of minicell-producing strains are grown under high-stress conditions (5% NaCl) known to cause bacterial cells to become filamentous, which increases the size differential between the minicells and remaining cells. Although this strategy is not always necessary with Δmin mutants of E. coli, which are already a mixture of filaments and minicells, it should be particularly useful for cells that modestly overproduce FtsZ and are generally short. Once they reach adequate cell density, the bacterial cultures are then centrifuged at low speed to remove the majority of the large cells. Supernatant fractions are collected, and minicells are purified away from larger cells through three sequential density gradient centrifugation steps. The centrifugation speed and time are specific to each particular strain and should be modified depending on the type of minicell-producing bacteria by comparing the distribution of minicells to that of nonminicells in each fraction. Enriched minicell suspensions are then passed through a 0.45-μm-pore-size cross-flow filter to homogenize minicell size. The filtrate is then finally passed through a 0.20-μm-pore-size cross-flow filter to filter out very small contaminants, such as membrane vesicles and cellular debris, producing a high minicell yield in the retentate with uniform size and minimal contamination (39).
It should be emphasized that minicells are not simply small cells that lack a chromosome. Because polar Z-rings assemble at various distances from the cell pole, probably as a result of the variability of nucleoid positioning, minicells of different sizes are generated. In particular, tiny minicells most likely arise from Z-rings that form very close to cell poles. Minicells generated from Δmin mutants may have a different physiology than that of minicells generated by the overexpression of ftsZ. Once formed, minicells may also have the potential to reorganize their structure, both internal and external, as they remain metabolically active. We are unaware of any studies directly addressing this possibility, but it does provide a reasonable explanation for the observed variation in minicell sizes derived from a single bacterial culture. We have also noted that the cytoplasm in a fraction of minicells is smaller than expected and that the width of the minicell periplasm is more varied than that in intact cells. These are substantial concerns that need to be considered before conclusions are drawn. In part, these concerns are ameliorated by analyzing many individual tomograms and by use of classification and subtomogram-averaging procedures, which are routinely used in generating the high-resolution structures. However, minicells may not be appropriate for studies in which the focus is on the kinetics of a biological process, specific requirements for membrane curvature, or the colocalization of independent macromolecular structures. Further potential challenges in interpreting data from phage infection of minicells are discussed below.
CRYO-ET APPLICATIONS OF BACTERIAL MINICELLS
Architecture of chemoreceptor arrays.
Chemotaxis allows bacteria to sense the chemical signals of their environment and respond to changes by either moving away from unfavorable conditions or moving toward favorable ones (63, 64). This process is mediated by arrays of chemotaxis signaling proteins that sense chemical signals and transduce the signal to motility organelles to modulate the direction of bacterial motility. The chemotaxis signaling complex is composed of three proteins: a methyl-accepting chemotaxis protein (MCP), CheA (histidine kinase), and CheW (complex coupling protein). The chemotaxis signaling complexes are assembled into large arrays, which are essential for signal amplification and remarkable sensitivity of the system. Extensive biochemical and structural analyses have provided atomic structures of individual components and their interactions (63, 64). However, it was not clear how these components assemble to form arrays until recent in situ structural studies.
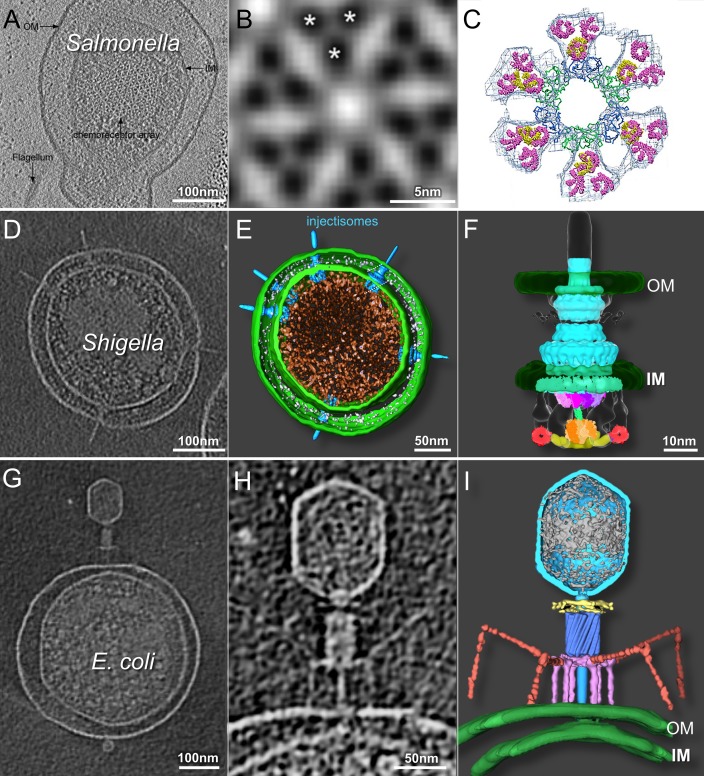
The spatial organization of the functional arrays was initially visualized by cryo-ET studies of Caulobacter crescentus (10) and E. coli cells (11). These studies produced low-resolution structures of the chemotaxis signaling complex, revealing that MCPs are anchored to the cytoplasmic side of the inner membrane, where they form pillar-like structures, and that CheA/W interacts with the cytoplasmic tip of MCPs (10, 11). To determine higher-resolution structures that could distinguish individual MCP dimers, S. enterica minicells were used for cryo-ET studies of chemoreceptor arrays (65). These minicells (Fig. 3A) were produced through the introduction of a second copy of the ftsZ gene. The small S. enterica minicells allowed for higher-resolution structural determination of the receptor arrays (Fig. 3B). Moreover, the in situ structure revealed individual MCP dimer densities (Fig. 3B) for the first time, which allowed better docking of the crystal structures (Fig. 3C) (65).
FIG 3.
Cryo-ET of minicells provides 3D structures of chemoreceptor arrays, injectisomes, and phage-host interactions. (A) Slice-through tomographic reconstruction of an S. enterica minicell showcasing the top-down view of the native chemoreceptor array. (B) Subtomographic-average structure of the receptor array resolving the organization of the trimer of dimers, as indicated by asterisks. (C) EM density map fit with the crystal structures of the individual components (republished from reference 65 with permission of the publisher). (D to F) Central tomographic slice of a Shigella minicell with multiple injectisomes (D), corresponding 3D surface rendering (E), and surface rendering of the injectisome basal body (republished from reference 78 with permission of the publisher) fit with structures of the isolated components (F) (republished from reference 105 with permission of the publisher). (G to I) Tomographic slice through the center of an E. coli minicell infected with phage T4 (G), magnified view of the same adsorbed phage particle penetrating and transferring DNA into the cell (H), and the corresponding 3D rendering (republished from reference 92 with permission of the publisher) (I). OM, outer membrane; IM, inner membrane.
Similarly, tiny E. coli minicells produced from a skinny mreB (A125V) mutant strain were used to visualize receptor arrays in detail (66). Larger arrays were specifically generated by the overproduction of FlhDC from a plasmid copy, which upregulates the expression of flagellar and chemotaxis genes. After the images from hundreds of minicells were averaged, EM structures of the intact arrays in their native cell envelope were achieved at 3.2-nm resolution. Importantly, this level of resolution is sufficient to build a model of the chemotaxis signaling complex in the context of the receptor array. Together, these studies provide a structural basis for array formation, which is required to understand the high sensitivity and cooperativity of chemotaxis signaling in bacteria (65, 66).
Minicells help elucidate type III secretion machine structures.
Type III secretion systems (T3SSs) are widely used by Gram-negative bacteria to assemble flagella for locomotion and to transport virulence effectors into eukaryotic cells. The flagellar T3SS (the flagellum) and the nonflagellar T3SS (the injectisome) diverge greatly in terms of function. However, they share an evolutionarily related core structure consisting of a multiring basal structure that traverses the periplasmic space and is embedded in the inner and outer membranes (67, 68). The injectisome, consisting of approximately 20 proteins, directly contacts a eukaryotic host cell and serves as a conduit for the delivery of bacterial effector proteins into the host (67, 69). The flagellum is more complex. Approximately 40 proteins are involved in mediation of the assembly and rotation of the large complex (6, 70–72).
EM studies of purified basal body complexes have revealed considerable structural information for both the flagellum and injectisome (73, 74). However, published structures of these organelles lack structural detail for many critical components, because they are often lost after detergent extraction during basal body purification. To determine in situ structures of the intact flagellar motor and injectisome, Kawamoto et al. (75) employed S. enterica minicells to visualize both motors and injectisomes embedded in a common cell envelope. Similar to the S. enterica chemoreceptor study (65), minicells were induced by increasing the cellular levels of FtsZ. Although both in situ structures have relatively low resolution (75), particularly compared to the cryo-EM structures from purified basal complexes (73, 74), this study demonstrates the potential of minicell systems for investigating flagella and injectisomes in situ.
Minicells were also used to visualize the injectisome in Yersinia enterocolitica. In this case, a mutant strain forming minicells was generated by two-step allelic exchange (76). The combined use of Yersinia minicells and subtomogram averaging resulted in an ∼4-nm-resolution in situ structure of the injectisome (77). Notably, the injectisome structure from Y. enterocolitica minicells is considerably more informative than the ∼7-nm-resolution injectisome structure derived from whole cells of Shigella flexneri, partly because of reduced cell thickness (77).
To obtain higher-resolution structures of an intact injectisome and its cytoplasmic sorting platform, an S. flexneri minicell-producing strain was recently constructed by overproducing FtsZ from a plasmid (along with two other cell division proteins, FtsA and FtsQ, encoded by adjacent genes) (78). It was previously reported that S. flexneri minicells are capable of invading host cells (42) and that S. enterica minicell injectisomes can effectively deliver proteins into eukaryotic cells (41). Cryo-ET reconstructions of S. flexneri minicells interacting with red blood cells represent the first use of minicells to visualize the interaction between bacteria and a eukaryotic host (78). Tiny S. flexneri minicells (Fig. 3D and E) were selected to generate thousands of 3D tomograms (78). The small diameter of these minicells made it possible to resolve the injectisome and its cytoplasmic sorting platform (Fig. 3F), which had not been visualized in earlier injectisome structures (75, 77). This platform, consisting of a central hub and six spokes, with a pod-like structure at the terminus of each spoke, functions to sort effector proteins for the delivery and recognition of specific effectors, and it is required for T3SS assembly and function (79). By docking known atomic structures onto the resulting density maps, this study determined that the hub consists mainly of a hexamer of the ATPase Spa47, while the MxiN protein and Spa33 protein comprise the spokes and pods, respectively (78). The structure not only provides the basis for further dissection of the secretion mechanisms of the injectisome but also underscores the major structural distinctions between flagella and injectisomes.
Minicells as model hosts for bacteriophage adsorption and DNA ejection.
Phages are the most abundant biological entity in the biosphere and are responsible for much of bacterial evolution. Most phages utilize elaborate tail machines to eject their genome into a host cell. Despite major efforts and astonishing progress in achieving atomic structures of individual components and even entire virions (80, 81), only limited progress has been made in understanding the dynamic process of phage infection since the 1967 landmark papers showing T4 infecting cells by conventional EM (82, 83). The development of cryo-ET has helped redress this gap in our knowledge. For example, cryo-ET of E. coli cells infected by the tailless lipid-containing phage PRD1 revealed that a proteolipid tube forms after infection and crosses the cell envelope, presumably providing a conduit for DNA transport into the cell (84). Cryo-ET studies on phage infection of the naturally thin cells of Prochlorococcus spp. (23), Synechococcus spp. (24), and C. crescentus (25) have yielded exciting and significant information on phage development. However, a detailed molecular understanding of the infection process has been hampered by the lack of genetic and physiological information about these host-parasite interactions relative to E. coli and its phages. A similar drawback is associated with interpreting ε15 structural intermediates during infection by cryo-ET (85). This work was made even more difficult by the size of the host bacterium, S. enterica serovar Anatum, which limited the resolution of the structures that were obtained. The development of E. coli minicells as a host should allow the generation of higher-resolution structures. In turn, higher resolution can lead to a deeper understanding and new insights into the mechanisms of phage infection initiation.
E. coli minicells were initially used to structurally characterize interactions between the host cell and P1, a contractile-tailed phage most commonly used for generalized transduction (86). Cryo-ET reconstructions of minicells infected with P1 revealed three stages of phage infection (61). First, phages interact with the cell surface via their tail fibers, initially with one tail fiber contacting the outer membrane and then with multiple fibers, positioning the tail baseplate perpendicular to the cell surface. The tail sheath subsequently contracts, moving away from the cell surface and exposing the inner tail tube while penetrating the cell envelope. Tomographic images of phage-host interactions suggest that contraction of the phage tail and penetration of the outer membrane do not necessarily lead to the injection of genetic material, implicating additional mechanisms for phage DNA to penetrate the inner membrane and enter the cell.
A subsequent study pushed the limits of resolution and provided an example of the extraordinary potential of minicells for understanding the molecular mechanism of infection by T7, a short-tailed phage (87). Cryo-ET of infected minicells revealed structures of the phage during three sequential stages of infection: transient interaction with the cell surface, stable attachment to the surface, and formation of an extended tail that penetrates both outer and inner membranes for DNA translocation (88). Importantly, although a high-resolution structure of the T7 phage was published previously (89), only the cryo-ET study was able to resolve individual phage tail fibers by subtomogram averaging and classification. In contrast to the conventional depiction of a phage particle, the cryo-ET-derived structure showed that the majority of tail fibers were bound to the capsid, with only one or two fibers being unbound, presumably aiding the tail in locating its cell surface receptors. Once stably adsorbed, all tail fibers contact the outer membrane. In addition, this study provided the first structural evidence that the internal core proteins of T7 are ejected from the capsid and that they assemble inside the cell to form a transenvelope channel that allows DNA translocation from the phage to the cytoplasm.
Similar E. coli minicells were employed to study infection by a ϕX174-like coliphage, ST-1 (90). The icosahedral ST-1 particle lacks an external tail. The pilot protein H is required for DNA translocation (91). Indeed, cryo-ET of ST-1-infected minicells showed that a fraction of the phages contacting cells forms a novel tail tube, which was often observed to penetrate the inner membrane. The tail tubes were absent after genome delivery, suggesting that they disassemble after DNA ejection.
Recently, E. coli minicells were used to resolve the infection mechanism of phage T4 (92), a classic model system for studying DNA ejection (93). Phage T4 has a contractile tail with a baseplate and both short tail fibers (STFs) and long tail fibers (LTFs). There is high structural similarity between the contractile tail of the phage and type VI bacterial secretion systems (81). The application of cryo-ET and subtomogram averaging to the study of T4 infection of skinny E. coli minicells provided novel insights into T4 infection initiation (Fig. 3G to I) and confirmed the phage structure and individual components previously resolved by cryo-EM and X-ray crystallography (81). This study also provided direct structural evidence that prior to adsorption, the majority of LTFs are bound to the sheath, allowing two or three extended LTFs to transiently interact with the cell surface. Once several LTFs have bound to the surface, the baseplate undergoes a conformational change from a hexagonal to a star conformation, releasing all STFs that bind irreversibly to the cell surface. Following sheath contraction, the tail tube penetrates the outer membrane. The tail-associated lysozyme degrades the cell wall, and the tail tube then traverses the host cell periplasm. Further penetration into the host cytoplasm is accompanied by a dramatic local outward curvature of the cytoplasmic membrane as it fuses with the phage tail tip. This study not only provided new molecular insights into the mechanistic pathway of T4 phage infection but also demonstrated the potential of the combination of thin E. coli minicells and cryo-ET to provide high-resolution 3D structures of phage infection intermediates in near-native states.
By definition, minicells represent only a portion of the cell, and the cell envelope is not of uniform composition. Differences in lipid, cell wall, and protein composition at the polar and septal regions are perhaps best studied (94–96). For example, relative to their intact E. coli parent cells, minicell cytoplasmic membranes are enriched for cardiolipin and specific polar membrane proteins, and their cell walls are enriched for inert peptidoglycan (97–99). Although these properties may raise concerns about the physiological relevance of minicells as model systems compared with parental cells, many structures derived from minicells, such as chemoreceptors, naturally occur at cell poles. Furthermore, several phages exhibit a distinct preference for adsorbing to cell poles or a future pole at midcell (100), which makes minicells eminently suitable for studying structural changes occurring during the initiation of infection. We are not aware of any compelling data showing that the fundamental mechanism of phage infection initiation, distinct from any kinetics of the process, is different following polar or sidewall adsorption. As long as conditions can be found under which phage adsorption results in a complex that is sufficiently stable for cryo-ET analysis, it seems likely that comparable structural changes during the initial stages of infection will be similar to those of infection of intact cells. Some phages have been shown to produce progeny after infection of minicells (88, 101, 102), but the kinetics of phage development in minicells is likely aberrant and cannot be compared to that of infections of exponentially growing parent cells.
OUTLOOK AND POTENTIAL APPLICATIONS
Although bacterial minicells have been known to exist for nearly 50 years, their use in cryo-ET is relatively new and opens tremendous opportunities for many novel and exciting applications. Specifically, the decreased thickness of bacterial minicells allows direct visualization of molecular machines within intact cells without stains, fixatives, or additional steps for thinning or sectioning. Given that E. coli and many other bacteria are model organisms widely used as protein expression systems, bacterial minicells can be considered a miniature living toolbox for the expression and assembly of a variety of molecular machines, including cell surface structures that do not require the chromosome or midcell components. This toolbox is particularly valuable for structural characterization of macromolecular complexes in situ, as highlighted by the many examples in this review. Moreover, as minicells retain functional and often infectious properties of the parent bacteria (41, 42), they promise to be useful model systems for understanding the molecular basis of bacterium-host interactions, a central process during bacterial infection. Given the increase in antibiotic-resistant bacteria that could alternatively be targeted by phage treatment (103, 104), minicells present ideal host systems for understanding the molecular basis of host specificity during adsorption and infection. Most studies highlighted in this review have focused on applications of minicells from a variety of Gram-negative bacteria, but minicells from Gram-positive bacteria promise to be equally fruitful.
ACKNOWLEDGMENT
We thank Ian Molineux for his comments on the manuscript.
REFERENCES
- 1.Alberts B. 1998. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92:291–294. doi: 10.1016/S0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Lučič V, Rigort A, Baumeister W. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202:407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. 1988. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21:129–228. doi: 10.1017/S0033583500004297. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y. 2015. Single-particle cryo-EM at crystallographic resolution. Cell 161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoenger A. 2014. High-resolution cryo-electron microscopy on macromolecular complexes and cell organelles. Protoplasma 251:417–427. doi: 10.1007/s00709-013-0600-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne JL, Subramaniam S. 2009. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat Rev Microbiol 7:666–675. doi: 10.1038/nrmicro2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Jensen GJ. 2009. Electron cryotomography: a new view into microbial ultrastructure. Curr Opin Microbiol 12:333–340. doi: 10.1016/j.mib.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridman K, Mader A, Zwerger M, Elia N, Medalia O. 2012. Advances in tomography: probing the molecular architecture of cells. Nat Rev Mol Cell Biol 13:736–742. doi: 10.1038/nrm3453. [DOI] [PubMed] [Google Scholar]
- 10.Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. 2008. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol 69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A 104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kürner J, Frangakis AS, Baumeister W. 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436–438. doi: 10.1126/science.1104031. [DOI] [PubMed] [Google Scholar]
- 13.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 14.Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, McBride MJ, Subramaniam S. 2007. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. 2010. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, Chiu W. 2010. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol 17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, Nagayama K, Schmid MF, King JA, Chiu W. 2013. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 502:707–710. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci U S A 108:9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. 2009. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N. 2009. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol 191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. 2009. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briegel A, Ladinsky MS, Oikonomou C, Jones CW, Harris MJ, Fowler DJ, Chang YW, Thompson LK, Armitage JP, Jensen GJ. 2014. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife 3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X, Himes BA, Ke D, Rice WJ, Ning J, Zhang P. 2014. Controlled bacterial lysis for electron tomography of native cell membranes. Structure 22:1875–1882. doi: 10.1016/j.str.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Amoudi A, Norlen LP, Dubochet J. 2004. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J Struct Biol 148:131–135. doi: 10.1016/j.jsb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Rigort A, Bäuerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM. 2012. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A 109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. 2007. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods 4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 34.Rigort A, Plitzko JM. 2015. Cryo-focused-ion-beam applications in structural biology. Arch Biochem Biophys 581:122–130. doi: 10.1016/j.abb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Villa E, Schaffer M, Plitzko JM, Baumeister W. 2013. Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr Opin Struct Biol 23:771–777. doi: 10.1016/j.sbi.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Al-Amoudi A, Studer D, Dubochet J. 2005. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J Struct Biol 150:109–121. doi: 10.1016/j.jsb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Frazer AC, Curtiss R III. 1975. Production, properties and utility of bacterial minicells. Curr Top Microbiol Immunol 69:1–84. [DOI] [PubMed] [Google Scholar]
- 38.Adler HI, Fisher WD, Cohen A, Hardigree AA. 1967. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci U S A 57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP, Haasdyk JE, Dickson KA, Brahmbhatt VN, Pattison ST, James AC, Al Bakri G, Straw RC, Stillman B, Graham RM, Brahmbhatt H. 2007. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell 11:431–445. doi: 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, Choy HE, Lee JH, Kim GJ. 2015. Generation of minicells from an endotoxin-free Gram-positive strain Corynebacterium glutamicum. J Microbiol Biotechnol 25:554–558. doi: 10.4014/jmb.1408.08037. [DOI] [PubMed] [Google Scholar]
- 41.Carleton HA, Lara-Tejero M, Liu X, Galán JE. 2013. Engineering the type III secretion system in non-replicating bacterial minicells for antigen delivery. Nat Commun 4:1590. doi: 10.1038/ncomms2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hale TL, Sansonetti PJ, Schad PA, Austin S, Formal SB. 1983. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect Immun 40:340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 44.Rowlett VW, Margolin W. 2013. The bacterial Min system. Curr Biol 23:R553-556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 45.Rothfield L, Taghbalout A, Shih YL. 2005. Spatial control of bacterial division-site placement. Nat Rev Microbiol 3:959–968. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- 46.Rowlett VW, Margolin W. 2015. The Min system and other nucleoid-independent regulators of Z ring positioning. Front Microbiol 6:478. doi: 10.3389/fmicb.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busiek KK, Margolin W. 2015. Bacterial actin and tubulin homologs in cell growth and division. Curr Biol 25:R243–R254. doi: 10.1016/j.cub.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raskin DM, de Boer PA. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181:6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A 96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meinhardt H, de Boer PA. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci U S A 98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu XC, Margolin W. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 53.Edwards DH, Errington J. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol 24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 54.Bramkamp M, van Baarle S. 2009. Division site selection in rod-shaped bacteria. Curr Opin Microbiol 12:683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Reeve JN, Mendelson NH, Coyne SI, Hallock LL, Cole RM. 1973. Minicells of Bacillus subtilis. J Bacteriol 114:860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levin PA, Margolis PS, Setlow P, Losick R, Sun D. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol 174:6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaval KG, Rismondo J, Halbedel S. 2014. A function of DivIVA in Listeria monocytogenes division site selection. Mol Microbiol 94:637–654. doi: 10.1111/mmi.12784. [DOI] [PubMed] [Google Scholar]
- 58.Ward JE Jr, Lutkenhaus J. 1985. Overproduction of FtsZ induces minicell formation in E. coli. Cell 42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 59.Monahan LG, Harry EJ. 2013. Identifying how bacterial cells find their middle: a new perspective. Mol Microbiol 87:231–234. doi: 10.1111/mmi.12114. [DOI] [PubMed] [Google Scholar]
- 60.Treuner-Lange A, Aguiluz K, van der Does C, Gomez-Santos N, Harms A, Schumacher D, Lenz P, Hoppert M, Kahnt J, Muñoz-Dorado J, Søgaard-Andersen L. 2013. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol 87:235–253. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. 2011. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology 417:304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shepherd N, Dennis P, Bremer H. 2001. Cytoplasmic RNA polymerase in Escherichia coli. J Bacteriol 183:2527–2534. doi: 10.1128/JB.183.8.2527-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones CW, Armitage JP. 2015. Positioning of bacterial chemoreceptors. Trends Microbiol 23:247–256. doi: 10.1016/j.tim.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. 2012. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A 109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. 2012. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A 109:E1481-1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diepold A, Armitage JP. 2015. Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci 370:20150020. doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2:a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 72.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 73.Schraidt O, Marlovits TC. 2011. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 74.Thomas DR, Francis NR, Xu C, DeRosier DJ. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawamoto A, Morimoto YV, Miyata T, Minamino T, Hughes KT, Kato T, Namba K. 2013. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep 3:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 77.Kudryashev M, Stenta M, Schmelz S, Amstutz M, Wiesand U, Castano-Diez D, Degiacomi MT, Munnich S, Bleck CK, Kowal J, Diepold A, Heinz DW, Dal Peraro M, Cornelis GR, Stahlberg H. 2013. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife 2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A 112:1047–1052. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson JE, Chiu W. 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr Opin Struct Biol 17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 81.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. 2010. Morphogenesis of the T4 tail and tail fibers. Virol J 7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon LD, Anderson TF. 1967. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology 32:279–297. [DOI] [PubMed] [Google Scholar]
- 83.Simon LD, Anderson TF. 1967. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. II. Structure and function of the baseplate. Virology 32:298–305. [DOI] [PubMed] [Google Scholar]
- 84.Peralta B, Gil-Carton D, Castaño-Diez D, Bertin A, Boulogne C, Oksanen HM, Bamford DH, Abrescia NG. 2013. Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS Biol 11:e1001667. doi: 10.1371/journal.pbio.1001667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang JT, Schmid MF, Haase-Pettingell C, Weigele PR, King JA, Chiu W. 2010. Visualizing the structural changes of bacteriophage Epsilon15 and its Salmonella host during infection. J Mol Biol 402:731–740. doi: 10.1016/j.jmb.2010.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yarmolinsky MS N. 1988. Bacteriophage P1. In Calendar R. (ed), The bacteriophages, vol 1, 2nd ed Plenum Press, New York, NY. [Google Scholar]
- 87.Casjens SR, Molineux IJ. 2012. Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv Exp Med Biol 726:143–179. doi: 10.1007/978-1-4614-0980-9_7. [DOI] [PubMed] [Google Scholar]
- 88.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. 2005. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J 24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. 2014. Icosahedral bacteriophage PhiX174 forms a tail for DNA transport during infection. Nature 505:432–435. [DOI] [PubMed] [Google Scholar]
- 91.Jazwinski SM, Lindberg AA, Kornberg A. 1975. The gene H spike protein of bacteriophages phiX174 and S13. I. Functions in phage-receptor recognition and in transfection. Virology 66:283–293. [DOI] [PubMed] [Google Scholar]
- 92.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goldberg E, Grinius L, Letellier L. 1994. Recognition, attachment, and injection. In Karam JD, Kreuzer KN, Hall DH (ed), Molecular biology of bacteriophage T4. ASM Press, Washington, DC. [Google Scholar]
- 94.Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laloux G, Jacobs-Wagner C. 2014. How do bacteria localize proteins to the cell pole? J Cell Sci 127:11–19. doi: 10.1242/jcs.138628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsumoto K, Hara H, Fishov I, Mileykovskaya E, Norris V. 2015. The membrane: transertion as an organizing principle in membrane heterogeneity. Front Microbiol 6:572. doi: 10.3389/fmicb.2015.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. 1997. Murein segregation in Escherichia coli. J Bacteriol 179:2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li G, Young KD. 2012. Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol Microbiol 84:276–295. doi: 10.1111/j.1365-2958.2012.08021.x. [DOI] [PubMed] [Google Scholar]
- 99.Renner LD, Weibel DB. 2011. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci U S A 108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Edgar R, Rokney A, Feeney M, Semsey S, Kessel M, Goldberg MB, Adhya S, Oppenheim AB. 2008. Bacteriophage infection is targeted to cellular poles. Mol Microbiol 68:1107–1116. doi: 10.1111/j.1365-2958.2008.06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roozen KJ, Fenwick RG Jr, Curtiss R III. 1971. Synthesis of ribonucleic acid and protein in plasmid-containing minicells of Escherichia coli K-12. J Bacteriol 107:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reeve JN. 1977. Bacteriophage infection of minicells: a general method for identification of “in vivo” bacteriophage directed polypeptide biosynthesis. Mol Gen Genet 158:73–79. doi: 10.1007/BF00455121. [DOI] [PubMed] [Google Scholar]
- 103.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M, Wakiguchi H, Imai S. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother 11:211–219. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 104.Lu TK, Koeris MS. 2011. The next generation of bacteriophage therapy. Curr Opin Microbiol 14:524–531. doi: 10.1016/j.mib.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 105.Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da Fonseca PC, Morris EP, Wall JS, Lea SM, Blocker AJ. 2009. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol 16:477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bi E, Lutkenhaus J. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol 175:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lai EM, Nair U, Phadke ND, Maddock JR. 2004. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol Microbiol 52:1029–1044. doi: 10.1111/j.1365-2958.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 108.Reeve JN, Mendelson NH. 1974. Minicells of Bacillus subtilis. A unique system for transport studies. Biochim Biophys Acta 352:298–306. doi: 10.1016/0005-2736(74)90221-1. [DOI] [PubMed] [Google Scholar]
- 109.Reeve J. 1979. Use of minicells for bacteriophage-directed polypeptide synthesis. Methods Enzymol 68:493–503. doi: 10.1016/0076-6879(79)68038-2. [DOI] [PubMed] [Google Scholar]
- 110.Setlow JK, Boling ME, Allison DP, Beattie KL. 1973. Relationship between prophage induction and transformation in Haemophilus influenzae. J Bacteriol 115:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]