ABSTRACT
It is well established that the RpoN-RpoS sigma factor (σ54-σS) cascade plays an essential role in differential gene expression during the enzootic cycle of Borrelia burgdorferi, the causative agent of Lyme disease. The RpoN-RpoS pathway is activated by the response regulator/σ54-dependent activator (also called bacterial enhancer-binding protein [bEBP]) Rrp2. One unique feature of Rrp2 is that this activator is essential for cell replication, whereas RpoN-RpoS is dispensable for bacterial growth. How Rrp2 controls cell replication, a function that is independent of RpoN-RpoS, remains to be elucidated. In this study, by generating a series of conditional rrp2 mutant strains, we demonstrated that the N-terminal receiver domain of Rrp2 is required for spirochetal growth. Furthermore, a D52A point mutation at the phosphorylation site within the N terminus of Rrp2 abolished cell replication. Mutation of the ATPase motif within the central domain of Rrp2 did not affect spirochetal replication, indicating that phosphorylation-dependent ATPase activity of Rrp2 for σ54 activation is not required for cell growth. However, deletion of the C-terminal domain or a 16-amino-acid truncation of the helix-turn-helix (HTH) DNA-binding motif within the C-terminal domain of Rrp2 abolished spirochetal replication. It was shown that constitutive expression of rpoS is deleterious to borrelial growth. We showed that the essential nature of Rrp2 is not due to an effect on rpoS. These data suggest that phosphorylation-dependent oligomerization and DNA binding of Rrp2 likely function as a repressor, independently of the activation of σ54, controlling an essential step of cell replication in B. burgdorferi.
IMPORTANCE Bacterial enhancer-binding proteins (bEBPs) are a unique group of transcriptional activators specifically required for σ54-dependent gene transcription. This work demonstrates that the B. burgdorferi bEBP, Rrp2, has an additional function that is independent of σ54, that of its essentiality for spirochetal growth, and such a function is dependent on its N-terminal signal domain and C-terminal DNA-binding domain. These findings expand our knowledge on bEBP and provide a foundation to further study the underlying mechanism of this new function of bEBP.
INTRODUCTION
Borrelia burgdorferi, the Lyme disease spirochetal pathogen, survives in nature in a complex enzootic cycle involved in two markedly different hosts, an arthropod vector and a mammalian host (1–3). During the process of tick feeding, B. burgdorferi dramatically regulates its gene expression in order to transmit and adapt to the diverse host environments, and the RpoN-RpoS sigma factor (σ54-σS) cascade plays an essential role in this process (4–6). In this pathway, a bacterial enhancer-binding protein (bEBP)/σ54-dependent activator Rrp2, along with RpoN (σ54), directly controls transcription of rpoS from a −24/−12 RpoN-type promoter (4, 7, 8). RpoS, functioning as a global regulator, further controls the expression of numerous genes important for the enzootic cycle of B. burgdorferi (for a review, see references 2, 3, and 9). Since the first discovery of this pathway, additional factors, including BosR (10–12), BadR (13, 14), DsrA (15), Rrp1 (16–18), and the posttranscriptional regulation of rpoS (19), have been identified.
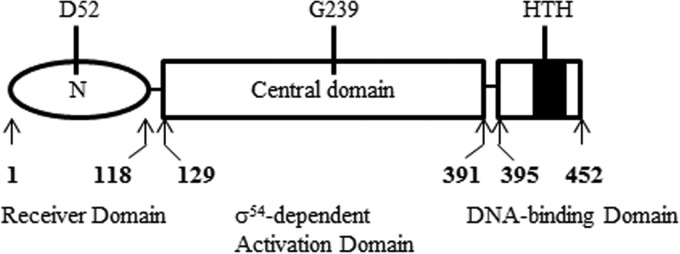
Rrp2 is the only bEBP encoded in the B. burgdorferi genome (7). Rrp2 is an NtrC-type bEBP that contains three putative functional domains (7, 20, 21) (Fig. 1): the N-terminal receiver domain conserved among two-component response regulators, the central domain conserved among bEBPs for ATPase activity and RpoN activation, and the C-terminal domain involved in oligomerization and DNA binding. A G239C mutation in an ATP-binding motif in the central domain of Rrp2 abolished rpoS expression in B. burgdorferi (7). Similar to other NtrC proteins, phosphorylation at the conserved D52 residue in the N-terminal domain of Rrp2 activates the central-domain ATPase activity and subsequently is required for transcriptional activation of rpoS via RpoN (22–24). The C terminus of Rrp2 has DNA-binding capability (22). Interestingly, Rrp2 does not appear to require a specific enhancer-binding sequence to activate rpoS (22); therefore, the function of the C-terminal DNA-binding domain currently remains unclear.
FIG 1.
Domain structure of B. burgdorferi Rrp2. Rrp2 is an NtrC-type bEBP that contains three putative functional domains: the N-terminal receiver domain, the central domain for ATPase activity and σ54 activation, and the C-terminal domain involved in oligomerization and DNA binding. The numbers denote amino acid residues. D52 is a phosphorylation site. G239 is a conserved residue in the Walker B motif for ATP hydrolysis. HTH, helix-turn-helix motif.
One unexpected feature of Rrp2 is that, in addition to being the activator for RpoN-dependent rpoS activation, it is essential for B. burgdorferi growth (25). Unlike rpoN or rpoS, which can readily be inactivated in B. burgdorferi, attempts by multiple groups to generate an rrp2 deletion mutant have failed (7, 22, 23). Recently, a conditional rrp2 mutant was constructed in the presence of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible copy of rrp2 (25). The conditional rrp2 mutant was unable to replicate in the absence of IPTG, demonstrating that Rrp2 is indispensable for borrelial growth. To the best of our knowledge, Rrp2 is the first bEBP that was shown to play two distinct roles: activating the RpoN (σ54) regulon and controlling cell growth. Questions still exist. First, how does Rrp2 perform these two different functions? Second, what is the molecular mechanism for the essential nature of Rrp2? In this study, we attempted to determine the structural basis for the essentiality of Rrp2 by dissecting the various domains of Rrp2. These results provide insight into the dual functions of Rrp2 and set the foundation for further elucidation of mechanisms underlying the essential nature of Rrp2.
MATERIALS AND METHODS
Bacterial strains.
All strains and plasmids used in this study are described in Table 1. Low-passage-number, virulent B. burgdorferi strain B31-A3 was kindly provided by P. Rosa (Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD) (26). B. burgdorferi was cultivated in Barbour-Stoenner-Kelly II (BSK-II) medium supplemented with 6% normal rabbit serum (Pel-Freez Biologicals, Rogers, AR) at 37°C with 5% CO2. Relevant antibiotics were added to the cultures at the following final concentrations: 300 μg/ml for kanamycin (Kan), 100 μg/ml for streptomycin (Strep), and 50 μg/ml for gentamicin (Gent). The constructed suicide vectors and shuttle vector were maintained in Escherichia coli strain DH5α.
TABLE 1.
Strains, plasmids, and primers used
| Strain, plasmid, or primer | Description or sequencea | Source |
|---|---|---|
| B. burgdorferi strains | ||
| BbYY31 | B31-A3 transformed with pRrp2-HA(pYY14) | This study |
| BbYP17 | Conditional rrp2 deletion mutant; BbYY31 transformed with pYP24 | This study |
| BbYP18 | Conditional rrp2 strain with a C-terminal deletion in Rrp2; BbYP17 transformed with pYP40 | This study |
| BbYP22 | Conditional rrp2-D52A point mutant; BbYP17 transformed with pYP38 | This study |
| BbYP24 | A3-83 transformed with pYY14 | This study |
| BbYP25 | Conditional rrp2 mutant with a C-terminal HTH deletion in Rrp2; BbYP17 transformed with pYP48 | This study |
| BbYP26 | Conditional rrp2 mutant with a C-terminal deletion in Rrp2; BbYP17 transformed with pYP47 | This study |
| BbYP27 | Conditional rrp2 strain with the central domain and C terminus of Rrp2; BbYP17 transformed with pYP39 | This study |
| BbYP28 | Conditional rrp2 rpoS double-knockout strain; BbYP24 transformed with pYP24 | This study |
| A3-83 | rpoS mutant of B31-A3 | 5 |
| Plasmids | ||
| pXY206A | Suicide vector containing a wild-type rrp2 gene linked to an aadA marker; Strr | 7 |
| pYY14 | pRrp2-HA, shuttle vector carrying IPTG-inducible, HA-tagged Rrp2; Strr | This study |
| pYP24 | Suicide vector for constructing rrp2 deletion; Genr | This study |
| pYP38 | Suicide vector for constructing Rrp2-D52A; Kanr | This study |
| pYP39 | Suicide vector for constructing Rrp2 central and C-terminal domains; Kanr | This study |
| pYP40 | Suicide vector for constructing Rrp2 C-terminal domain; Kanr | This study |
| pYP47 | Suicide vector for constructing Rrp2 C-terminal HTH deletion; Kanr | This study |
| pYP48 | Suicide vector for constructing Rrp2 C-terminal deletion; Kanr | This study |
| Primers | ||
| PRYP24 | TTGCAGTACTCTTAAAAGCTTGACTTGAATTTCA | |
| PRYP25 | AAGAACTAGTTAGAACAGAAGATAAAATAATAGG | |
| PRYP26 | CGCGGATCCTCAATACCCTTCAATTAAAATATT | |
| PRYP27 | GAGGGATCCTTAGGTGGCGGTACTTGGGT | |
| PRYP28 | CGCGGATCCTCATTAGAAAAACTCATCGAGCATC | |
| PRYY55 | GATCGGATCCCAGCTTTTTTTTGA | |
| PRYY59 | CTCAGAGCGCTTATTATTACATCAAGATTTTCA | |
| PRYY60 | TAATAAGCGCTCTGAGAATGCCTCAGATATCTGG | |
| P1 (aacC1-ID-F) | TTCGGAGACGTAGCCACCTA | |
| P2 (PRYP48) | TGGAAGAGCCTCCCTCAAAT | |
| P3 (PRYY34) | GCAGCGGATCCTTAATCAATATTATATTCGA | |
| Del rrp2-N-F | TTTAATTGAAGGGTATTGAAAATGAGCAATAAAAAAGAAAATAA CGATGATGAAAATCC | |
| Del rrp2-N-R | TTCATCATCGTTATTTTCTTTTTTATTGCTCATTTTCAATACCCTTC AATTAAAATA | |
| rrp2-C-F | TTTAATTGAAGGGTATTGAAAATGAGCAAACAAATCACTAAAGA AGATTTACCAGC | |
| rrp2-C-R | TGGTAAATCTTCTTTAGTGATTTGTTTGCTCATTTTCAATACCCTT CAATTAAAATA |
Restriction sites are indicated by underlining.
Construction of the conditional rrp2 mutant.
A conditional rrp2 mutation was previously constructed in strain 297 (25). In this study, we generated a conditional rrp2 mutation in wild-type B. burgdorferi strain B31-A3. To do so, we first constructed a shuttle vector, pYY14, that harbors an IPTG-inducible, wild-type rrp2 gene (with a human influenza virus hemagglutinin [HA] tag at the 3′ end of the coding region). pYY14 was then transformed into B31-A3. The resulting Strep-resistant strain, BbYY31 (B31-A3/pRrp2-HA), was confirmed by PCR and immunoblot analyses. Next, pYP24, the suicide vector containing a Gent resistance marker flanked by the upstream and downstream sequences of rrp2, was constructed. To construct pYP24, we used the previously generated pXY206A, which harbors a wild-type copy of rrp2 linked to an aadA marker (which confers Strep resistance) (27). First, pXY206A was restriction enzyme digested with BamHI and SpeI, resulting in the excision of the aadA marker, the entire rrp2 gene, and a portion of the hk2 gene. Next, the aacC1 marker (which confers Gent resistance [28]) and the missing portion of hk2 were PCR amplified with the primer pairs PRYP27/PRYY55 and PRYP25/PRYP26 (Table 1), respectively, restriction enzyme digested, and ligated back into the digested pXY206A fragment. pYP24 was confirmed by sequencing. To generate the conditional rrp2 deletion mutant BbYP17, the suicide vector pYP24 was transformed into BbYY31. Transformants were selected with Gent and Strep, and deletion of the chromosomal rrp2 gene was verified by PCR and immunoblotting analyses.
Construction of an rrp2 partial deletion mutant.
To construct the conditional Rrp2 N-terminal deletion mutant BbYP27, we constructed a suicide vector (pYP39) that contained an rrp2 gene encoding only the central and C-terminal domains linked to a Kan resistance marker, along with the upstream and downstream sequences of rrp2. Briefly, the DNA fragment encoding the Rrp2 central and C-terminal regions was PCR amplified with the primer pair Del rrp2-N-F and Del rrp2-N-R (Table 1), which introduced NdeI and ClaI restriction sites at the 5′ and 3′ ends of the sequence, respectively. The digested fragment was then cloned into pXY206A to replace the full-length rrp2 gene fragment, resulting in pHX207. Then the Strep resistance marker in pHX207 was replaced with a Kan resistance marker (PCR amplified with primer pair PRYY55 and PRYP28 and digested with BamHI). The resulting suicide plasmid, pYP39, was confirmed by restriction enzyme digestion and by DNA sequencing. pYP39 was then transformed into BbYP17, the conditional full-length rrp2 deletion strain. BbYP17 was used for generating all of the partial rrp2 deletion strains used in this study. BbYP17 is both Gent and Strep resistant. Upon transformation of BbYP17 with pYP39, Kan- and Strep-resistant clones were selected. Clones were analyzed by PCR and immunoblotting to confirm successful replacement of the chromosomal copy of full-length rrp2 with the rrp2 mutant encoding Rrp2 with an N-terminal deletion. The strain was designated BbYP27 (expressing Rrp2 with the central and C-terminal domains [Rrp2-Cen-Cter] and pRrp2-HA).
To generate a conditional Rrp2-D52A mutant, the suicide vector pYP38 was constructed. pYP38 contains an rrp2 gene encoding Rrp2-D52A linked to a Kan resistance marker, along with the upstream and downstream sequences of rrp2. The rrp2-D52A fragment was PCR amplified by a two-step PCR method. The first step was undertaken with two primer pairs, PRYP24/PRYY59 and PRYY60/PRYP25, resulting in two DNA fragments, namely, F1, upstream of D52A, and F2, downstream of D52A, which also introduced an AfeI restriction site at D52A. The second step of PCR used F1 and F2 as the DNA template with the primer pair PRYP24/PRYP25 to generate an rrp2-D52A fragment, which also introduced ScaI and SpeI restriction sites into the 5′ and 3′ ends of the fragment. This DNA fragment was cloned into pHX207 at the ScaI and SpeI restriction sites to generate plasmid pYP23. Finally, the Strep resistance marker in pYP23 was replaced with a Kan resistance marker. The resulting suicide plasmid, pYP38, was confirmed by restriction digestion and by DNA sequencing. pYP38 was then transformed into the conditional full-length-rrp2 deletion strain BbYP17 to generate the conditional Rrp2-D52A mutant. Positive Kan- and Strep-resistant clones were selected and analyzed by PCR and immunoblotting to confirm successful replacement of the chromosomal copy of full-length rrp2 with the rrp2 mutant encoding Rrp2-D52A. The strain was designated BbYP22 (Rrp2-D52A mutant/pRrp2-HA).
To construct a conditional Rrp2 C-terminal deletion mutant, the suicide vector pYP47 was generated. pYP47 contains an rrp2 gene encoding Rrp2 with its C terminus deleted (without the C-terminal amino acid residues 395 to 452) linked to a Kan resistance marker, along with the upstream and downstream sequences of rrp2. First, a stop codon mutation was introduced into pXY206A at the nucleotide position corresponding to amino acid residue 394 of Rrp2, along with a BglII restriction site, using the QuikChange kit (Stratagene, CA). An NdeI/ClaI fragment from the mutated plasmid was then ligated into pXY206A at the same restriction sites, resulting in plasmid pJSB332. pJSB332 was verified by restriction digestion and by DNA sequencing. Next, the Strep resistance marker in pJSB332 was replaced with a Kan resistance marker, resulting in pYP47. pYP47 was then transformed into the conditional full-length rrp2 deletion strain BbYP17 to generate the conditional Rrp2 C-terminal deletion mutant. Positive Kan- and Strep-resistant clones were selected and subjected to PCR and immunoblot analyses to confirm successful replacement of the chromosomal copy of full-length rrp2 with the rrp2 mutant encoding Rrp2 with only the N-terminal and the central domains (Rrp2-Nter-Cen). The strain was designated BbYP26 (Rrp2-Nter-Cen mutant/pRrp2-HA).
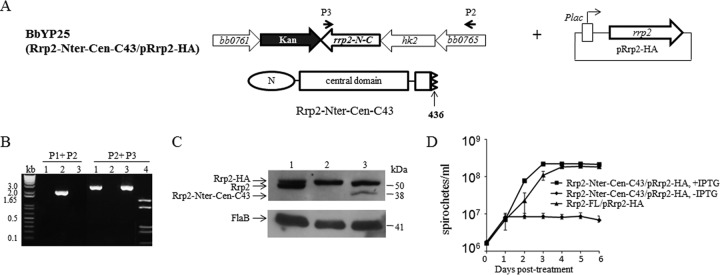
To construct a conditional Rrp2 mutant that lacks the helix-turn-helix (HTH) motif, the pYP48 suicide vector containing an rrp2 gene encoding Rrp2 without the HTH motif (lacking the last 16 amino acid residues) linked to a Kan resistance marker, along with the upstream and downstream sequences of rrp2, was constructed. First, a stop codon mutation was introduced into pXY206A at the nucleotide position corresponding to amino acid residue 437 of Rrp2, along with a HindIII restriction site, in a manner similar to that described above, resulting in plasmid pJSB333. pJSB333 was verified by restriction digestion and by DNA sequencing. Then the Strep resistance marker in pJSB332 was replaced with a kanamycin resistance marker, resulting in pYP48. pYP48 was then transformed into the conditional full-length-rrp2 deletion strain BbYP17 to generate the conditional Rrp2 mutant lacking the HTH motif. Positive Kan- and Strep-resistant clones were selected and analyzed by PCR and immunoblotting to confirm successful replacement of the chromosomal copy of full-length rrp2 with the rrp2 mutant encoding Rrp2 lacking the HTH motif. The strain was designated BbYP25 (Rrp2-Nter-Cen-C43 mutant/pRrp2-HA).
To construct a conditional Rrp2 mutant that contains only the C terminus, the suicide vector pYP40 was constructed. pYP40 contains an rrp2 gene encoding the C terminus of Rrp2 linked to a Kan resistance marker, along with the upstream and downstream sequences of rrp2. First, the section of rrp2 encoding the C-terminal fragment was PCR amplified with the primer pair rrp2-C-F and rrp2-C-R (Table 1), which also introduced NdeI and ClaI restriction sites into the 5′ and 3′ ends of the fragment, respectively. The digested fragment was then cloned into pXY206A to replace the full-length rrp2 gene fragment, resulting in pHX208. Next, the Strep resistance gene in pHX208 was replaced with a Kan resistance marker (PCR amplified with the primer pair PRYY55 and PRYP28 and digested with BamHI). The resulting suicide plasmid, pYP40, was confirmed by restriction digestion and DNA sequencing. pYP40 was then transformed into the conditional full-length-rrp2 deletion strain BbYP17. Kan- and Strep-resistant clones were analyzed by PCR and immunoblotting to confirm successful replacement of the chromosomal copy of full-length rrp2, with the rrp2 mutant gene encoding only the C terminus of Rrp2. The strain was designated BbYP18 (Rrp2-Cter mutant/pRrp2-HA).
Construction of the conditional rrp2 deletion in the B. burgdorferi strain lacking rpoS.
We used a strategy similar to that described above for construction of the conditional rrp2 deletion in wild-type strain BbYP17, except the parental strain used in this case was the B31 rpoS mutant A3-83 (5). We first transformed the shuttle vector pYY14 into A3-83, resulting in strain BbYP24 (ΔrpoS mutant/pRrp2-HA, which confers Kan and Strep resistance). Then the suicide vector pYP24, containing the Gent resistance marker flanked by the upstream and downstream sequences of rrp2, was transformed into BbYP24 to generate the conditional rrp2 deletion mutation in the rpoS mutant (designated BbYP28, with Gent resistance [Genr], Kan resistance [Kanr], and Strep resistance [Strr]). Positive clones were verified by PCR and immunoblot analyses (data not shown).
Growth curve analyses.
Growth curve analyses were performed as previously described (25). Briefly, various conditional rrp2 mutant strains were inoculated to an initial density of 1 × 104 spirochetes/ml in the presence of 50 μM IPTG. When the culture density reached to the early exponential phase of growth (∼1 × 107 spirochetes/ml), spirochetes were collected by centrifugation and washed twice with BSK-II medium. The washed spirochetes were then resuspended in fresh BSK-II to a final density of 1 × 106 spirochetes/ml and divided into two cultures; one was supplemented with 50 μM IPTG, and the other remained untreated. Cell numbers were counted daily for 7 days using dark-field microscopy.
SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting were performed as previously described (29). The images were developed using both the colorimetric (4-chloro-1-naphthol) and the chemiluminescent method (Pierce ECL Western blotting substrate; Thermo Scientific, IL). All proteins were separated by 15% SDS-PAGE, except for that shown in Fig. 7B, for which a 12.5% gel was used in order to separate full-length Rrp2 from the mutated Rrp2 lacking the HTH motif. Spirochetes were collected at 48 to 72 h postdepletion for immunoblotting. Antibodies directed against Rrp2 and FlaB were described previously (24).
FIG 7.
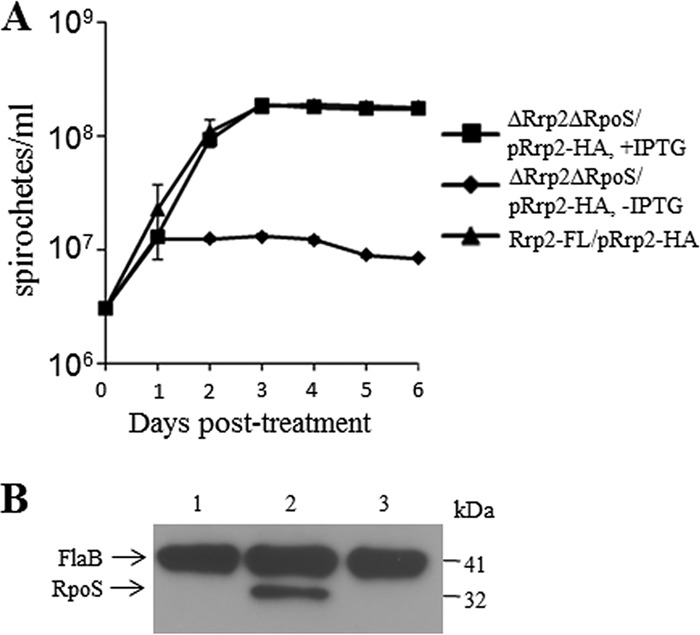
Growth curve analysis of the conditional rrp2 mutant in the rpoS mutant background. (A) The Rrp2-FL mutant/pRrp2-HA and ΔRrp2 ΔRpoS mutant/pRrp2-HA strains were grown for 6 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures. (B) Expression of RpoS. Lane 1, ΔRrp2 mutant/pRrp2-HA without IPTG (day 2); lane 2, Rrp2-FL mutant/pRrp2-HA without IPTG; lane 3, ΔRrp2 ΔRpoS mutant/pRrp2-HA without IPTG (day 2). Bands corresponding to each protein are labeled on the left. Molecular masses are indicated on the right.
RESULTS
Rrp2 is required for spirochetal growth.
Rrp2 is a bacterial enhancer-binding protein that is essential for RpoN-dependent rpoS activation. Groshong et al. reported that Rrp2 is also essential for B. burgdorferi growth in infectious wild-type B. burgdorferi strain 297 (25). We first thought to confirm this finding in another strain, B. burgdorferi B31-A3. To do so, B31-A3 was transformed with a shuttle vector carrying an IPTG-inducible, HA-tagged, wild-type rrp2 gene (B31-A3/pRrp2-HA, pYY14). The resulting strain, BbYY31, was then electroporated with the suicide vector pYP24 to inactivate the chromosomal copy of rrp2 in the presence of IPTG (Fig. 2A). PCR (Fig. 2B) and the immunoblotting (Fig. 2C) experiments were performed to demonstrate that the native rrp2 gene on the chromosome was disrupted in the resulting strain, BbYP17 (ΔRrp2 mutant/pRrp2-HA), which produces Rrp2-HA in the presence of IPTG.
FIG 2.
Construction of the conditional rrp2 deletion mutation in strain B31. (A) Diagram showing the generation of the conditional rrp2 deletion mutant. Wild-type B31-A3 was first transformed with the shuttle vector pRrp2-HA carrying an IPTG-inducible, HA-tagged, wild-type rrp2 gene, resulting in strain BbYY31 (full-length Rrp2 [Rrp2-FL] Rrp2-HA). Then the suicide plasmid pYP24 was transformed into BbYY31 to generate the conditional rrp2 deletion strain BbYP17 (ΔRrp2 Rrp2-HA). X's denote the approximate positions of recombination (double-crossover) events. P1, P2, and P3, primers used to identify the rrp2 conditional knockout strain BbYP17. (B) Characterization of BbYP17 (ΔRrp2 Rrp2-HA) by PCR analysis. Lane 1, strain BbYY31 (Rrp2-FL Rrp2-HA); lane 2, strain BbYP17 (ΔRrp2 Rrp2-HA). The P1, P2, and P3 (Table 1) combinations denote primer pairs used for PCR. (C) Immunoblot analysis of BbYP17. The lanes correspond to those in panel B. Bands corresponding to each protein form are indicated on the left. Molecular masses are indicated on the right. (D) Growth curves of the ΔRrp2 mutant/pRrp2-HA and Rrp2-FL mutant/pRrp2-HA strains. Strains were grown for 7 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± standard deviations (SD) from three independent cultures. The immunoblot on the right shows that Rrp2 (day 2) was induced in the ΔRrp2 mutant/pRrp2-HA only in the presence of IPTG, not in the absence of IPTG.
In the presence of 50 μM IPTG, strain BbYP17 (ΔRrp2 mutant/pRrp2-HA) grew similarly to wild-type B. burgdorferi strain B31-A3. In the absence of IPTG, the ΔRrp2 mutant/pRrp2-HA could replicate only for 1 to 2 generations and then ceased (Fig. 2D, left). As expected, no Rrp2-HA was detected in these cells in the absence of IPTG (Fig. 2D, right). These results reinforce the notion that the bacterial enhancer-binding protein Rrp2 has two distinct functions, one required for σ54 activation and the other essential for spirochetal growth.
The inability to grow in the absence of IPTG of the conditional rrp2 mutant suggests that Rrp2 is either required for cell replication or essential for cell viability. To examine whether the conditional rrp2 mutant in the absence of IPTG remained viable, we took cells from this culture at different days starting from day 2 (note that partial growth in day 1 was due to the presence of a trace amount of intracellular IPTG in the parental cells used for inoculation) and inoculated these cells in fresh medium containing IPTG. Virtually no growth was observed for all cultures, suggesting that these cells were no longer viable, and their growth could not be rescued by adding IPTG (data not shown). Thus, Rrp2 is essential for spirochetal viability, not just for active replication.
The N-terminal domain of Rrp2 is required for spirochetal growth.
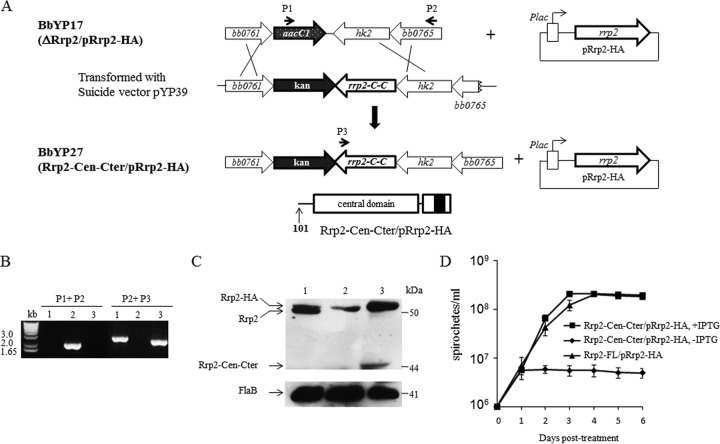
As a two-component response regulator, Rrp2 contains a conserved receiver domain (N terminus) in which phosphorylation of the conserved Asp within the receiver domain controls the activation of the response regulator. In the case of Rrp2, this phosphorylation controls the ATPase activity of Rrp2 that is essential for its activation of RpoN (σ54) (22). To determine whether the essential nature of Rrp2 is also dependent on its N-terminal domain, we constructed a conditional N-terminal Rrp2 deletion strain by transforming strain BbYP17 with a suicide vector carrying an rrp2 variant (Rrp2-Cen-Cter) encoding only the central and C-terminal domains of Rrp2 (pYP39) (Fig. 3A). The resulting strain, BbYP27 (Rrp2-Cen-Cter mutant/pRrp2-HA), was confirmed by PCR analysis (Fig. 3B). Immunoblots also showed that BbYP27 produced two versions of Rrp2, Rrp2-HA and truncated Rrp2 lacking its N terminus (Rrp2-Cen-Cter) (Fig. 3C).
FIG 3.
The Rrp2 N terminus is required for borrelial growth. (A) Diagram showing the generation of the conditional rrp2 mutant encoding Rrp2 lacking the N terminus. BbYP17 (ΔRrp2 Rrp2-HA) was transformed with the suicide vector pYP39 to generate BbYP27 (Rrp2-Cen-Cter mutant/pRrp2-HA) by homologous recombination. P1, P2, and P3 (Table 1), primers used to identify strain BbYP27. rrp2-C-C encodes Rrp2 containing the central domain (Cen) and the C-terminal domain (Cter) but not the N terminus. The domain structure for Rrp2-Cen-Cter in the chromosome is shown at the bottom. (B) PCR analysis of strain BbYP27. Lane 1, Rrp2-FL mutant/pRrp2-HA; lane 2, ΔRrp2 mutant/pRrp2-HA; lane 3, Rrp2-Cen-Cter mutant/pRrp2-HA. (C) Immunoblot analysis of BbYP27. The lanes correspond to those in panel B. Bands corresponding to each protein are indicated on the left. Molecular masses are indicated on the right. (D) Growth curves of the Rrp2-FL mutant/pRrp2-HA and Rrp2-Cen-Cter mutant/pRrp2-HA strains grown for 6 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures.
As shown in Fig. 3D, BbYP27 (Rrp2-Cen-Cter) grew similarly to wild-type B. burgdorferi strain B31-A3 in the presence of IPTG. However, in the absence of IPTG, BbYP27 was no longer able to replicate, a phenotype similar to that of the rrp2 conditional deletion mutant (ΔRrp2 mutant/pRrp2-HA). These results indicate that the N-terminal receiver domain of Rrp2 is essential for borrelial growth.
Phosphorylation of Rrp2 is required for growth.
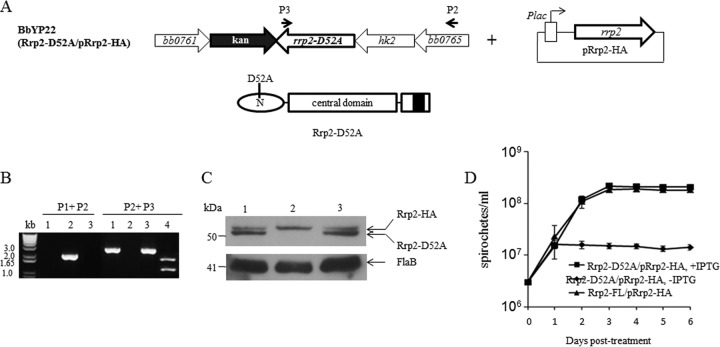
Rrp2 is activated upon phosphorylation of the conserved D52 residue in the N-terminal receiver domain (Fig. 1) (7, 23, 24). To determine whether phosphorylation of D52 is required for spirochetal growth, the pYP38 suicide vector carrying a mutated rrp2 allele encoding Rrp2-D52A was transformed into the rrp2 conditional deletion mutant strain BbYP17. The resulting strain, BbYP22 (Rrp2-D52A mutant/pRrp2-HA) (Fig. 4A), was confirmed by PCR analysis and restriction digestion (Fig. 4B). Immunoblots showed that BbYP22 now produced two versions of Rrp2, Rrp2-HA and non-HA-tagged Rrp2 bearing a D52A mutation (Fig. 4C), suggesting that the chromosomal copy of rrp2-D52A was restored into the conditional rrp2 deletion mutant strain. Similar to the conditional N-terminal Rrp2 deletion strain, the conditional Rrp2-D52A mutant was able to grow only in the presence of 50 μM IPTG (Fig. 4D). This result suggests that phosphorylation of Rrp2 at the N terminus is required for the growth of B. burgdorferi.
FIG 4.
Phosphorylation of Rrp2 at residue D52 is required for cell growth. (A) Diagram showing the generation of the conditional rrp2 mutant strain BbYP22, expressing Rrp2-D52A. The domain structure of Rrp2-D52A is shown below. (B) PCR analysis of BbYP22 (Rrp2-D52A mutant/pRrp2-HA). P1, P2, and P3 (Table 1), primers used to identify the strain BbYP22. P1 was at the same location shown in Fig. 2A. Lane 1, Rrp2-FL mutant/pRrp2-HA; lane 2, ΔRrp2 mutant/pRrp2-HA; lane 3, Rrp2-D52A mutant/pRrp2-HA; lane 4, PCR fragment 3 using P2 plus P3 digested with AfeI. (C) Immunoblot analysis of BbYP22. The lanes correspond to those in panel B. Bands corresponding to each protein are labeled on the right. Molecular masses are indicated on the left. (D) Growth curves of the Rrp2-FL mutant/pRrp2-HA and Rrp2-D52A mutant/pRrp2-HA strains grown for 6 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures.
The C-terminal domain of Rrp2 is required for growth.
We previously showed that phosphorylation controls the central-domain ATPase activity of Rrp2 required for σ54 activation (22), and phosphorylation-controlled ATPase activity of the central domain of Rrp2 is dispensable for spirochetal growth (7). It is well known that in other NtrC family proteins, phosphorylation of the N-terminal domain also enhances oligomerization and DNA-binding activity. Thus, we sought to investigate whether the phosphorylation-associated DNA-binding activity of the C-terminal domain of Rrp2 is involved in regulating cell replication.
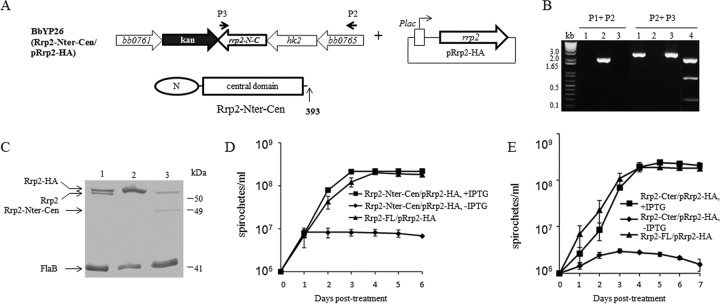
To determine the role of the C-terminal domain of Rrp2 in borrelial replication, we constructed a conditional C-terminal deletion mutant, BbYP26 (Rrp2-Nter-Cen mutant/pRrp2-HA) (Fig. 5A), using a strategy similar to that used with the conditional N-terminal domain deletion mutant. The BbYP26 mutant was confirmed by PCR (Fig. 5B) and immunoblotting (Fig. 5C). The result showed that the conditional Rrp2 C-terminal domain deletion mutant was able to grow only in the presence of 50 μM IPTG, indicating that the C-terminal domain of Rrp2 is also required for spirochetal replication (Fig. 5D).
FIG 5.
The C-terminal domain of Rrp2 is required for borrelial growth. (A) Diagram of the generation of the conditional rrp2 mutant encoding Rrp2 lacking the C-terminal domain. (B) PCR analysis of BbYP26 (Rrp2-Nter-Cen mutant/pRrp2-HA). P1, P2, and P3 (Table 1), primers used to identify strain BbYP26; P1 was at the same location shown in Fig. 2A. Lane 1, Rrp2-FL mutant/pRrp2-HA; lane 2, ΔRrp2 mutant/pRrp2-HA; lane 3, Rrp2-Nter-Cen mutant/pRrp2-HA; lane 4, PCR fragment 3 using P2 plus P3 digested with BglII. (C) Immunoblot analysis of BbYP26. The lanes correspond to those in panel B. Bands corresponding to each protein are labeled on the left. Molecular masses are indicated on the right. (D) Growth curves of the Rrp2-FL mutant/pRrp2-HA and Rrp2-Nter-Cen mutant/pRrp2-HA strains grown for 6 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures. (E) Growth curve analyses of the Rrp2-FL mutant/pRrp2-HA and Rrp2-Cter mutant/pRrp2-HA strains grown for 7 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures.
The C-terminal domain of Rrp2 contains an HTH DNA-binding motif (7, 22). We further investigated whether the HTH motif is required for B. burgdorferi growth. Accordingly, we constructed a conditional rrp2 mutant expressing Rrp2 with a deletion of the second helix of the HTH motif (the last 16 residues), designated BbYP25 (Rrp2-Nter-Cen-C43 mutant/pRrp2-HA) (Fig. 6A). The positive clone was confirmed by both PCR analysis (Fig. 6B) and immunoblotting (Fig. 6C). As shown in Fig. 6D, BbYP25 was not able to replicate in the absence of IPTG, suggesting that the DNA-binding motif within the C terminus of Rrp2 is essential for spirochetal growth. On the other hand, the C terminus alone of Rrp2 is not sufficient for B. burgdorferi growth, since a conditional rrp2 mutant encoding only the C-terminal domain of Rrp2 could not grow in the absence of IPTG (Fig. 5E), further suggesting that DNA binding of Rrp2 is vital for B. burgdorferi growth.
FIG 6.
The HTH motif of Rrp2 is required for borrelial growth. (A) Diagram of the generation of the conditional rrp2 mutant (BbYP25) encoding an Rrp2 variant lacking the HTH motif (Rrp2-Nter-Cen-C43). The domain structure of Rrp2-Nter-Cen-C43 is shown below. (B) PCR analysis of BbYP25 (Rrp2-Cter mutant/pRrp2-HA). P1, P2, and P3 (Table 1), primers used to identify the strain BbYP26. P1 is at the same location shown in Fig. 2A. Lane 1, Rrp2-FL mutant/pRrp2-HA; lane 2, ΔRrp2 mutant/pRrp2-HA; lane 3, Rrp2-Nter-Cen-C43 mutant/pRrp2-HA; lane 4, PCR fragment 3 using P2 plus P3 digested with HindIII. (C) Immunoblot analysis of BbYP25. The lanes correspond to those in panel B. Bands corresponding to each protein are labeled on the left. Molecular masses are indicated on the right. (D) Growth curve analyses of the Rrp2-FL mutant/pRrp2-HA and Rrp2-Nter-Cen-C43 mutant/pRrp2-HA strains grown for 6 days in the presence (50 μM) or absence (0 μM) of IPTG. The values in the growth curves represent the mean cell counts ± SD from three independent cultures.
The essentiality of Rrp2 is not due to an overproduction of RpoS.
The results described above suggest that the phosphorylation and DNA binding of Rrp2 are required for borrelial replication. Since the first function of Rrp2, i.e., transcriptional activation of RpoN-dependent genes, is not required for spirochetal growth, we postulate that the second function of Rrp2, phosphorylation-dependent DNA binding, may be responsible for the essential nature of Rrp2. If so, this function likely represses genes that are deleterious to cell replication. In this regard, it is reported that overproduction of RpoS is lethal to B. burgdorferi growth (25, 30). Lybecker and Samuels showed that in addition to RpoN (sigma54) being necessary for the transcription of rpoS, a sigma70-type promoter is necessary for the transcription of a longer form of rpoS mRNA (15). One possibility is that phosphorylated Rrp2 activates RpoN-dependent rpoS transcription while repressing transcription of the longer form of rpoS mRNA. If so, rrp2 should be readily inactivated in the rpoS mutant.
To test this possibility, we generated a conditional rrp2 deletion mutant in the rpoS mutant using a strategy similar to that used with the construction of BbYP17 (ΔRrp2 mutant/pRrp2-HA), as shown in Fig. 2A. As shown in Fig. 7A, the conditional rrp2 rpoS double mutant (ΔRrp2 ΔRpoS mutant/pRrp2-HA) was not able to replicate in the absence of IPTG. This result indicates that the essentiality of Rrp2 is not due to an overproduction of RpoS. Further immunoblot analysis showed that deletion of Rrp2 did not result in an increase of RpoS (Fig. 7B). Thus, phosphorylated Rrp2 regulates yet-to-be-identified genes essential for borrelial growth.
DISCUSSION
The alternative sigma factor σ54 (RpoN) is a unique sigma factor that is phylogenetically different from other sigma factors (31). It not only recognizes a unique −24/−12 promoter sequence (instead of the −35/−10 sequence for σ70), it also always requires an activator, bEBP (32). All bEBPs reported thus far have a single function, that is, activating σ54-dependent transcription. In this regard, Rrp2 is a unique bEBP in B. burgdorferi. Not only is it required for σ54 activation, but it is also essential for cell replication, independently of σ54. In this study, we provided data showing that phosphorylation of Rrp2, a function that is required for ATPase activity and for σ54 activation, is also indispensable for the essential function of Rrp2. We further showed that regulation of borrelial replication by Rrp2 phosphorylation is likely mediated through the DNA-binding activity of the C-terminal domain of Rrp2.
In this study, we first confirmed the essential nature of Rrp2 in B. burgdorferi strain B31. We then took advantage of the constructed conditional rrp2 deletion mutant by constructing a series of truncated/mutated rrp2 strains in the presence of an inducible wild-type copy of rrp2. Much to our surprise, a single D52A mutation within Rrp2 abolished the growth of B. burgdorferi (Fig. 4). We previously showed that Rrp2 with a D52A mutation could not be phosphorylated in vitro (24). We also demonstrated that as with other NtrC proteins, phosphorylation of recombinant Rrp2 is required for its central-domain ATPase activity (22), a function that is essential for σ54-dependent transcriptional activation. Thus, our data suggest that phosphorylation of Rrp2 controls both functions of Rrp2: triggering the activity of σ54 and promoting B. burgdorferi replication.
The current dogma for the activation of the RpoN-RpoS pathway of B. burgdorferi is that the upstream signals (temperature, cell density, pH, or other host molecules) activate the pathway through the phosphorylation of Rrp2 (2, 3, 7, 23). However, our finding that phosphorylation of Rrp2 is required for cell growth implies that Rrp2 has to be constantly phosphorylated, regardless of the “on” (elevated temperature and pH or low cell density) or “off” (lowered temperature and pH or high cell density) growth conditions for RpoN-RpoS. Given this, how do the environmental signals regulate the RpoN-RpoS pathway? In this regard, one unique feature for RpoN (σ54)-dependent activation in B. burgdorferi is that in addition to phosphorylated Rrp2, another transcription activator, BosR, is required (10–12). We postulate that under the RpoN-RpoS on conditions, both Rrp2-P and BosR are present, whereas under the RpoN-RpoS off conditions, Rrp2-P is present, but BosR is not. Indeed, the level of BosR was shown to be consistent with the production of RpoS under in vitro cultivation conditions (10, 14, 33).
What is the source for phosphorylation of Rrp2? We previously reported that abrogation of the cognate histidine kinase of Rrp2, Hk2, or any combination with other histidine kinases present in the B. burgdorferi genome did not affect either cell growth or RpoN-RpoS activation, suggesting that Hk2 and other histidine kinases are not essential for Rrp2 phosphorylation (23, 24). Subsequently, we showed that the low-molecular-weight high-energy phosphate donor acetyl phosphate (acetyl-P), a metabolic intermediate of the AckA-Pta pathway, can phosphorylate Rrp2-P in vitro (24). We also showed that overexpression of pta inhibited the activation of RpoS, suggesting that pta somehow affects the RpoN-RpoS pathway, either through influencing the level of Rrp2-P or by another unknown mechanism (24). More recently, our collaborators, Richards et al., reported the successful generation of an ackA (does not produce acetyl-P or acetyl coenzyme A [Ac-CoA]) and a pta mutant (overproduces acetyl-P) by supplementing the growth medium with mevalonolactone to bypass the need for mevalonate to grow (34). RpoN-RpoS activation was not affected in these mutants, and cells grew when mevalonolactone was added, thus suggesting that acetyl-P is not essential for Rrp2 phosphorylation and that something else must also contribute to Rrp2 phosphorylation. The B. burgdorferi genome encodes the ArcA-ArcB pathway (arginine deiminase system) (35), in which an intermediate of the pathway is the small high-energy phosphate carbamoyl phosphate (carbamoyl-P). In this regard, we previously showed that the arcA mutant did not affect cell growth or RpoN-RpoS activation (24). It is possible that both acetyl-P and carbamyl-P contribute to Rrp2 phosphorylation and compensate for the loss of each other.
In many bacteria, σ54 is not essential for cell survival. This is also the case for B. burgdorferi, for which the rpoN mutant remains viable during in vitro growth (5, 6). Consistent with this observation, an rrp2 mutant encoding Rrp2-G239C was also viable in vitro (7). Thus, the control of borrelial growth by phosphorylation is unlikely through the central domain of Rrp2. For this reason, we did not generate a central-domain truncation rrp2 mutant in this study. The results from the C-terminal domain deletion and HTH motif deletion strains indicated that the DNA-binding function of Rrp2 is also required for spirochetal growth (Fig. 6). Previously, we reported that Rrp2 does not bind specifically at the rpoS promoter but is capable of binding to DNA nonspecifically (22). Given that both the phosphorylation and DNA binding of Rrp2 are required for cell growth and that both the phosphorylation and DNA binding of E. coli NtrC facilitate the formation of the hexamer, further enhancing binding of Rrp2 to DNA and activating transcription (21, 36), we postulate that phosphorylation-dependent and DNA-binding-enhanced oligomerization plays a key role in the modulation of borrelial growth. Note that although we did not examine whether a D52A or HTH mutation resulted in a defect in phosphorylation or DNA binding in this study, we previously constructed various recombinant Rrp2 proteins and demonstrated that the N-terminal domain of Rrp2 is indeed a receiver domain involved in phosphorylation, that the D52 residue is the phosphorylation site, and that the D52A mutation abolished Rrp2 phosphorylation (24). We also showed previously that the recombinant C-terminal domain of Rrp2 is capable of binding to DNA (22). Although we did not test whether the HTH motif is directly involved in DNA binding in vitro, it is likely so, given that the region encoding the C terminus contains only four helices and that the last helices form an HTH motif that has widely been accepted to be involved in DNA binding in NtrC as well as in other proteins. One concern was that the mutated variants might be misfolded in the cell. This is unlikely, given that all constructed Rrp2 variants were readily detected in the cell by the immunoblots shown in this study. Another observation in this report is that in the presence of IPTG, both HA-tagged wild-type Rrp2 and mutant Rrp2 were produced, and a mix of an Rrp2 dimer or oligomer may be present in the cell. However, we did not observe any defect in growth in the presence of IPTG, suggesting that the amount of the wild-type Rrp2 form of the oligomer is sufficient for its function. The mixed dimer or oligomer does not affect this conclusion because it was based on the result obtained under the condition when IPTG was not added, i.e., no HA-tagged wild-type Rrp2 was produced in the cell and only the mutant version of Rrp2 was present.
How phosphorylation-dependent and DNA-binding-enhanced oligomerization contributes to spirochetal growth remains to be elucidated. One possibility is that there is a specific Rrp2 binding site at a locus that is deleterious to cell growth, and Rrp2 functions as a repressor of such a locus. We tested one such gene, rpoS, whose uncontrolled expression is lethal for B. burgdorferi (25, 30). rpoS has two transcript forms, a short form that is transcribed from the −24/−12 RpoN-type promoter by Rrp2 and RpoN and a long form that is transcribed likely from a sigma70 promoter, which is not activated by Rrp2 and RpoN but posttranscriptionally regulated by the small RNAs DsrA and Hfq of B. burgdorferi (37). In this study, we tested whether Rrp2 represses the expression of the long form of the rpoS transcript and whether deletion of Rrp2 results in overproduction of RpoS, in turn causing cell death. Our data using the rrp2 rpoS double-deletion strain showed that this is not the case. Further work will focus on the identification of other genes regulated by Rrp2.
To the best of knowledge, no bEBP has been reported to be essential in other bacteria. However, there are precedents for two-component systems that are essential for cell growth. Besides the classic example of the essential role of the response regulator CtrA for cell cycle control in Caulobacter (38), the WalRK (YycGF) two-component system in several Gram-positive bacteria has been shown to be essential for cell growth (39, 40). WalRK functions as a regulator that controls peptidoglycan biosynthesis and cell division. It is possible that Rrp2 also regulates peptidoglycan biosynthesis of B. burgdorferi.
In summary, this study demonstrates that phosphorylation of the receiver domain is essential for both functions of Rrp2, the function as a bEBP to activate σ54-dependent gene transcription and the function required for cell growth. Phosphorylation-dependent control of cell growth is likely mediated through the C-terminal DNA-binding activity of Rrp2. Future work will focus on identifying the downstream target(s) of Rrp2 that controls the growth of B. burgdorferi.
ACKNOWLEDGMENT
We thank Patricia Rosa for the rpoS mutant strain.
REFERENCES
- 1.Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J Clin Invest 113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 3.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caimano M, Iyer R, Eggers C, Gonzalez C, Morton E, Gilbert M, Schwartz I, Radolf J. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A 102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XF, Alani SM, Norgard MV. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A 100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. 2008. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun 76:3844–3853. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang Z, Zhou J, Norgard MV. 2014. Synthesis of RpoS is dependent on a putative enhancer binding protein Rrp2 in Borrelia burgdorferi. PLoS One 9:e96917. doi: 10.1371/journal.pone.0096917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyde JA, Shaw DK, Smith Iii R, Trzeciakowski JP, Skare JT. 2009. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol Microbiol 74:1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol 74:1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang Z, Deka RK, Norgard MV. 2011. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog 7:e1001272. doi: 10.1371/journal.ppat.1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller CL, Karna SLR, Seshu J. 2013. Borrelia host adaptation regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol Microbiol 88:105–124. doi: 10.1111/mmi.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang Z, Zhou J. 2015. BadR (BB0693) controls growth phase-dependent induction of rpoS and bosR in Borrelia burgdorferi via recognizing TAAAATAT motifs. Mol Microbiol 98:1147–1167. doi: 10.1111/mmi.13206. [DOI] [PubMed] [Google Scholar]
- 15.Lybecker MC, Samuels DS. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol 64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 16.Rogers EA, Terekhova D, Zhang H, Hovis KM, Schwartz I, Marconi RT. 2009. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol 71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sze CW, Smith A, Choi YH, Yang X, Pal U, Yu A, Li C. 2013. Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect Immun 81:1775–1787. doi: 10.1128/IAI.00050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He M, Zhang J-J, Ye M, Lou Y, Yang XF. 2014. Cyclic di-GMP receptor PlzA controls virulence gene expression through RpoS in Borrelia burgdorferi. Infect Immun 82:445–452. doi: 10.1128/IAI.01238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dulebohn DP, Hayes BM, Rosa PA. 2014. Global repression of host-associated genes of the Lyme disease spirochete through posttranscriptional modulation of the alternative sigma factor RpoS. PLoS One 9:e93141. doi: 10.1371/journal.pone.0093141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wedel A, Kustu S. 1995. The bacterial enhancer-binding protein NtrC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev 9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 21.Bush M, Dixon R. 2012. The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol Mol Biol Rev 76:497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blevins JS, Xu H, He M, Norgard MV, Reitzer L, Yang XF. 2009. Rrp2, a sigma54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J Bacteriol 191:2902–2905. doi: 10.1128/JB.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol 65:277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Caimano MJ, Lin T, He M, Radolf JD, Norris SJ, Gheradini F, Wolfe AJ, Yang XF. 2010. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog 6:e1001104. doi: 10.1371/journal.ppat.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groshong AM, Gibbons NE, Yang XF, Blevins JS. 2012. Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J Bacteriol 194:3336–3342. doi: 10.1128/JB.00253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank K, Bundle S, Kresge M, Eggers C, Samuels D. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol 185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias AF, Bono JL, Kupko JJ III, Stewart PE, Krum JG, Rosa PA. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- 29.Troxell B, Ye M, Yang Y, Carrasco SE, Lou Y, Yang XF. 2013. Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect Immun 81:2743–2752. doi: 10.1128/IAI.00507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Xu Q, Tu J, Ge Y, Liu J, Liang FT. 2013. Increasing RpoS expression causes cell death in Borrelia burgdorferi. PLoS One 8:e83276. doi: 10.1371/journal.pone.0083276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M. 2008. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol Microbiol 68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 32.Reitzer L, Schneider BL. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde JA, Trzeciakowski JP, Skare JT. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J Bacteriol 189:437–445. doi: 10.1128/JB.01109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards CL, Lawrence KA, Su H, Yang Y, Yang XF, Dulebohn DP, Gherardini FC. 2016. Acetyl-phosphate is not a global regulatory bridge between virulence and central metabolism in Borrelia burgdorferi. PLoS One 10:e0144472. doi: 10.1371/journal.pone.0144472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 36.De Carlo S, Chen B, Hoover TR, Kondrashkina E, Nogales E, Nixon BT. 2006. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev 20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lybecker MC, Abel CA, Feig AL, Samuels DS. 2010. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 39.Fritz G, Mascher T. 2014. A balancing act times two: sensing and regulating cell envelope homeostasis in Bacillus subtilis. Mol Microbiol 94:1201–1207. doi: 10.1111/mmi.12848. [DOI] [PubMed] [Google Scholar]
- 40.Fabret C, Hoch JA. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol 180:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]