ABSTRACT
Edwardsiella tarda is an important pathogenic bacterium that can replicate in macrophages. However, how the intramacrophage infection process affects the virulence of this bacterium is essentially unknown. Here, we show that E. tarda replicates and induces a caspase-1-dependent cell pyroptosis in a murine macrophage model. Via pyroptosis, intracellular E. tarda escapes to the extracellular milieu, forming a unique bacterial population. Being different from the bacteria cultured alone, this unique population possesses a reprogrammed transcriptional profile, particularly with upregulated type III secretion system (T3SS)/T6SS cluster genes. Subsequent studies revealed that the macrophage-released population gains enhanced infectivity for host epithelial cells and increases resistance to multiple host defenses and hence displays significantly promoted virulence in vivo. Further studies indicated that T3SS is essentially required for the macrophage infection process, while T6SS contributes to infection-induced bacterial virulence. Altogether, this work demonstrates that E. tarda can utilize macrophages as a niche for virulence priming and for spreading infection, suggesting a positive role for intramacrophage infection in bacterial pathogenesis.
IMPORTANCE Many pathogens can replicate in macrophages, which is crucial for their pathogenesis. To survive in the macrophage cell, pathogens are likely to require fitness genes to counteract multiple host-killing mechanisms. Here, Edwardsiella tarda is proved to exit from macrophages during infection. This macrophage-released population displays a reprogrammed transcriptional profile with significantly upregulated type III secretion system (T3SS)/T6SS-related genes. Furthermore, both enhanced infectivity in epithelial cells and activated resistance to complex host defenses were conferred on this macrophage-primed population, which consequently promoted the full virulence of E. tarda in vivo. Our work provides evidence that E. tarda can utilize macrophages as a niche for virulence priming and for spreading infection, highlighting the importance of the intramacrophage infection cycle for the pathogenesis of E. tarda.
INTRODUCTION
Many bacterial pathogens can survive and multiply within a variety of eukaryotic cells, including macrophages, which is believed to be important for their infection process (1). To promptly adapt to the hostile host environment, many pathogens have been found to adopt precise regulation strategies and generate a global intracellular virulence gene expression profile (2). To date, several high-throughput screening methods (3, 4) have been applied to identify infection-induced virulence genes. For example, a technique named in vivo expression technology (IVET) was used to identify genes that were essential or necessary for bacterial colonization in many bacterial species, such as Salmonella enterica serovar Typhimurium (5), Yersinia enterocolitica (6), Mycobacterium avium (7), and Bordetella bronchiseptica (8). Another method, high-throughput bioluminescence mutant screening (BLMS), was reported to identify virulence genes in Edwardsiella ictaluri (9). Undoubtedly, the infection-responsive virulence profile greatly contributes to the survival and intracellular replication of invading pathogens.
Macrophage death is always observed during the infection of many bacterial pathogens. However, its effect on bacterial infection is rather complicated and controversial. On one hand, the death of macrophages is oftentimes concomitant with the death of the infecting organisms and can promote efficient pathogen clearance. On the other hand, killing phagocytes is also a strategy adopted by invading pathogens for evading immune cells and spreading to the neighborhood. Bacterial pathogens have developed different strategies for “manipulating” host cell death for a successful infection. For instance, Legionella pneumophila delivers a subset of effectors, such as SdhA, to block or delay host cell death and promote its own survival and intracellular replication (10). In contrast, Mycobacterium tuberculosis induces cell death in the later stage of infection to promote bacterial egress from infected macrophages (11), and Yersinia pseudotuberculosis induces apoptosis in vivo to aid in the establishment of a systemic infection of mice (12). Since the pros and cons of host cell death upon infection vary from hosts to pathogens, understanding the underlying mechanisms is vitally necessary.
Edwardsiella tarda is a pathogenic bacterium that infects a wide range of hosts, from fish, birds, and reptiles to human beings (13, 14). As a facultative intracellular bacterial pathogen, E. tarda is capable of surviving within infected host cells, including phagocytes, which is also crucial for its pathogenesis. Once E. tarda has invaded macrophage cells, it resides and replicates in a special vacuole that is largely dependent on its type III secretion system (T3SS) (15). Very recently, E. tarda was reported to induce caspase-1-dependent cell death in macrophages during infection (16). However, the influences of intramacrophage infection on bacterial virulence are largely unknown. Here, we demonstrate that intracellularly replicating E. tarda can exit from macrophages by inducing cell pyroptosis and can form a unique macrophage-released bacterial population. By comparing the gene transcriptional profiles in macrophage-released E. tarda and Dulbecco's minimal Eagle's medium (DMEM)-cultured E. tarda, we identified a reprogrammed gene expression pattern with significantly induced T3SS/T6SS virulence properties. Further studies correlated the infection-induced gene expression with enhanced epithelial invasiveness, resistance to multiple host bactericidal effectors, and in vivo virulence for E. tarda. This work revealed that E. tarda utilizes macrophages as a niche for virulence priming and then induces macrophage death to escape for further dissemination.
MATERIALS AND METHODS
Bacterial strains and cell culture.
Wild-type (WT) Edwardsiella tarda EIB202, the T3SS mutant (ΔT3SS), and the T6SS mutant (ΔT6SS) were grown as described previously (17). Mouse primary lung and kidney cells were prepared as described previously (18). HeLa cells (ATCC CCL-2) and J774A.1 cells (ATCC TIB-67) were cultured at 37°C in a 5% CO2 atmosphere in DMEM supplemented with 10% fetal bovine serum (FBS), which was called growth medium (GM).
Macrophage infection and enumeration of intracellular and extracellular bacteria.
J774A.1 cells were infected as described previously with slight adjustments (17). Briefly, E. tarda was grown overnight in tryptic soy broth (TSB) at 30°C with shaking and then diluted into fresh TSB with shaking at 30°C until the optical density at 600 nm reached 0.8. Harvested bacteria in phosphate-buffered saline (PBS) suspensions were added to macrophage cells at a multiplicity of infection (MOI) of 10:1. Plates were then centrifuged at 600 × g for 10 min. Two hours after incubation, the cells were washed three times with PBS and then incubated with growth medium containing 1,000 μg/ml gentamicin for 30 min to kill the extracellular bacteria, after which the gentamicin concentration was decreased to 10 μg/ml for the remainder of the experiment.
For enumeration of viable intracellular bacteria, Triton X-100 was added at 1% (vol/vol) to the cell culture, a dilution series was plated onto TSB agar plates, and the colonies were enumerated after overnight culture. For enumeration of viable extracellular bacteria, cell supernatant was harvested and viable bacteria (CFU) were counted.
Immunofluorescence and confocal microscopy.
For constitutive expression of green fluorescent protein (GFP) or mCherry, WT E. tarda EIB202 was electroporated with pUTt0456GFP or pUTt0456mCherry, respectively. J774A.1 cells were seeded onto 24-well plates containing sterile coverslips. Following infection with strain EIB202 (GFP) and incubation with gentamicin, the cells were washed with PBS and then fixed in 4% (wt/vol) paraformaldehyde for 10 min at room temperature. Fixed cells were washed in PBS and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After being washed with PBS, actin cytoskeleton was stained with rhodamine-phalloidin (Molecular Probes) for 30 min, and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) for 10 min at room temperature. Fixed samples were viewed on a Nikon A1R confocal microscope. The images were analyzed using ImageJ (NIH).
Cell death assay.
Cells were infected as described above, and supernatants were harvested. Lactate dehydrogenase (LDH) release was quantified using the Cytotox96 assay kit (Promega) according to the manufacturer's instructions. Cytotoxicity was normalized to Triton X-100 treatment (100% of control), and LDH release from uninfected/untreated cells was used for background subtraction. When mentioned, cells were pretreated with the selective caspase-1-specific inhibitor Ac-YVAD-CHO or dimethyl sulfoxide (DMSO) as a control for 60 min before infection (40 μM) and maintained throughout the course of infection.
Preparation of macrophage-released E. tarda.
The procedure to prepare macrophage-released E. tarda (released E. tarda) is described in Fig. S1 in the supplemental material. Briefly, macrophage cells were infected as described above, and the supernatant containing released bacteria was harvested 8 h postinfection. Then, the supernatant was centrifuged at 600 × g for 5 min to discard the cellular debris, and the harvested supernatant was further centrifuged at 13,000 × g for 10 min to collect the macrophage-released bacteria. DMEM-cultured E. tarda (cultured E. tarda) was prepared in the meantime. Briefly, E. tarda was cultured in cell-free growth medium in parallel and harvested by centrifugation.
Transcriptome sequencing (RNA-seq) and RT-qPCR.
The macrophage-released and DMEM-cultured bacteria were prepared as described above. RNA of both samples was extracted by using an RNA isolation kit (Tiangen, Beijing, China). The RNA samples in three biological replicates were sequenced by an Illumina HiSeq 2000 genome sequencer at the Chinese National Human Genome Center (Shanghai, China). One microgram of each RNA sample was used for cDNA synthesis with the Moloney murine leukemia virus (MMLV) reverse transcriptase (ToYoBo, Tsuruga, Japan). Quantitative real-time PCR (RT-qPCR) was performed on an FTC-200 detector (Funglyn Biotech, Shanghai, China) by using the SYBR green real-time PCR kit (ToYoBo).
Epithelial cell infection.
For secondary infection, macrophage-released E. tarda or DMEM-cultured E. tarda was incubated with HeLa cells, primary lung cells, or primary kidney cells at an MOI of 50 for the indicated time. Cell death was assessed as described above. For determination of the internalization ratio, media were washed with PBS and then replaced with GM containing 1,000 μg/ml gentamicin for 30 min to kill the extracellular bacteria, and the intracellular bacteria were counted. The internalization ratio is expressed as the number of CFU of bacteria divided by the number of cells.
Host-killing resistance assay.
Every 2 × 105 CFU of macrophage-released or DMEM-cultured E. tarda was incubated in 1 ml of TSB with the addition of 0.75 mM H2O2 for oxidant stress, incubated in 1 ml of TSB with medium pH of 5.5 for acidic stress, or incubated with 100 μl of mouse serum for serum-killing stress. The surviving bacteria were enumerated by colony counting at several time intervals. Simultaneously, macrophage-released or DMEM-cultured E. tarda was incubated with polymorphonuclear leukocytes (PMNs) prepared as described previously (19) at an MOI of 1. The surviving bacteria were enumerated as described above.
Mouse infection.
Six- to 8-week-old wild-type C57BL/6 mice were obtained from Shanghai SLAC Laboratory Animal Co. Animal experiments were approved by the Experimental Animal Management and Ethics Committees of Shanghai SLAC Laboratory Animal Co. For E. tarda challenges, each mouse was infected intraperitoneally with E. tarda at 1.5 × 106 CFU/g. For competitive index (CI) assays (20), the inoculum was composed equally of macrophage-released E. tarda (labeled with red fluorescence protein [RFP]) and DMEM-cultured E. tarda (labeled with GFP) in doses of 3 × 105 CFU/g. The bacterial burden in the liver of each mouse was determined based on red or green color colony on diluted TSB plates at 6 and 12 h postinfection. CI results are presented as log10 (RFP-expressing bacteria/GFP-expressing bacteria).
Statistical analysis.
Each experiment was performed three times (as indicated in the figure legends). Except for the mortality assay, in which a log-rank test was used to compare the survival distributions of the mice, all other statistical analyses were performed by using the Student t test in the SPSS software (version 11.5; SPSS Inc.). In all cases, the significance level was defined at a P value of ≤0.01, ≤0.05, or ≤0.001.
RESULTS
Intracellular replicating E. tarda exits from macrophages via caspase-1-dependent cell death.
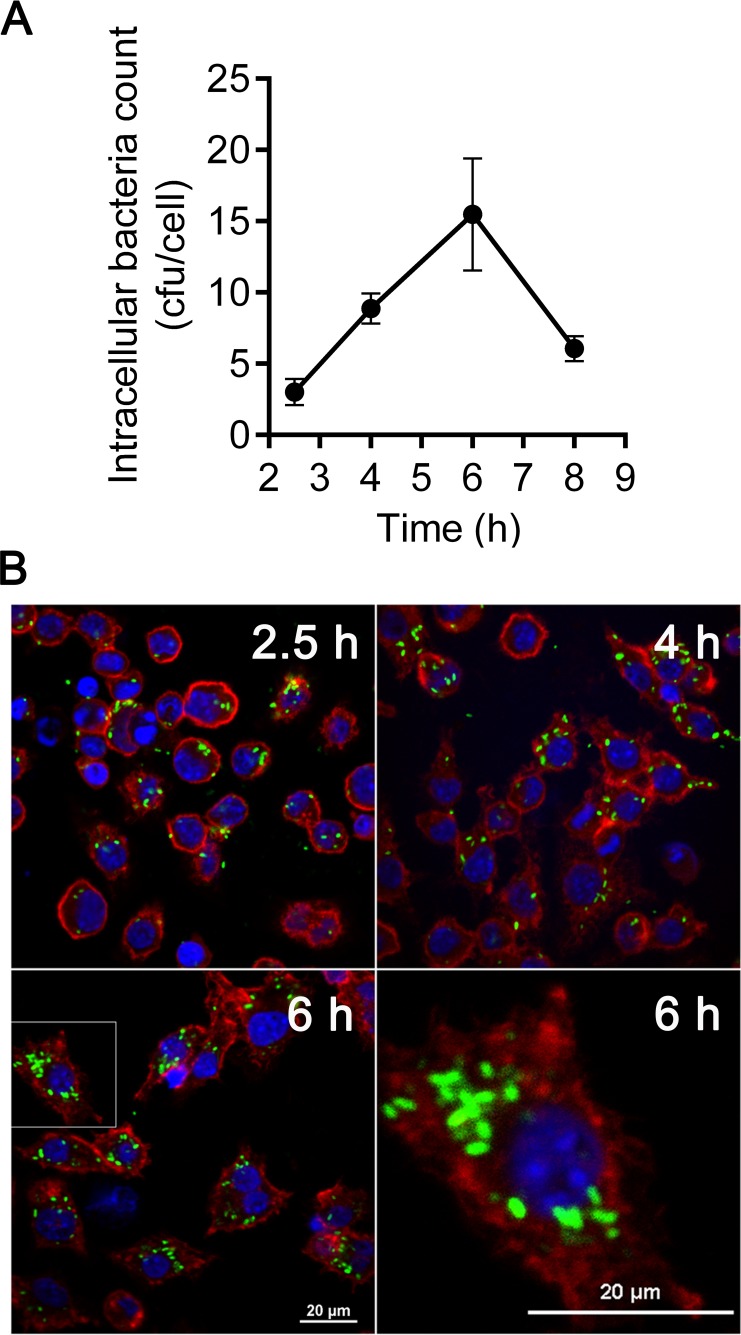
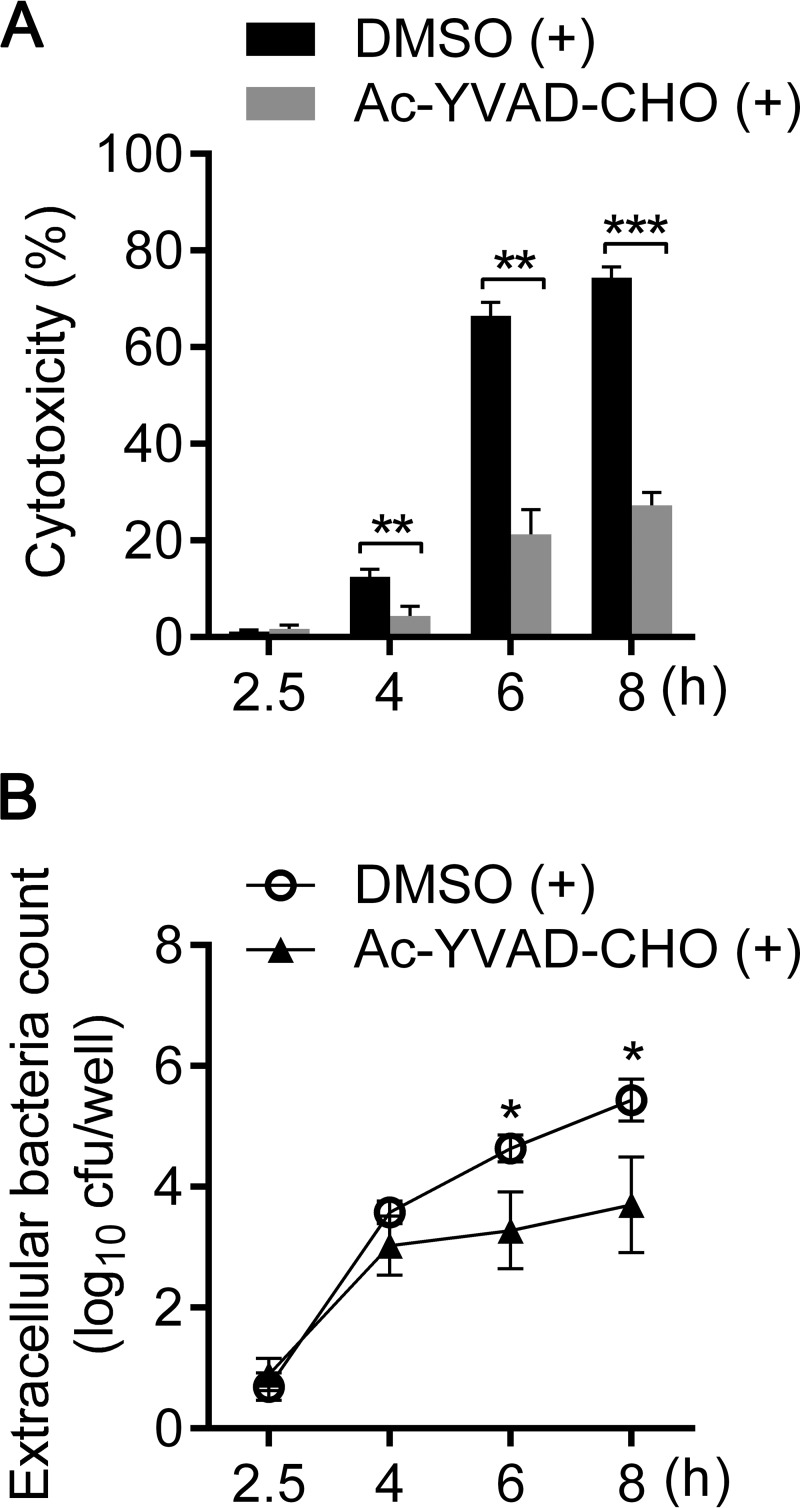
E. tarda prefers an intracellular lifestyle during infection in either epithelial (21, 22) or phagocytic (15, 23, 24) cells, which provide an intracellular niche for this pathogen. In this study, we established an E. tarda infection model in murine macrophage-like cell line J774A.1. In this model, during incubation with J774A.1 at an MOI of 10, E. tarda was efficiently internalized into macrophages within the initial 2 h (Fig. 1A). After treatment with gentamicin to kill the extracellular bacteria, internalized E. tarda replicated inside cells over time and reached a maximal propagation level at approximately 15 bacteria per cell at 6 h (Fig. 1A). Observation of the infected cells under confocal microscopy revealed similar intracellular replicating kinetics (Fig. 1B). Meanwhile, robust cytotoxicity was induced in E. tarda-infected macrophages and was significantly reduced in the presence of the caspase-1-specific inhibitor Ac-YVAD-CHO (Fig. 2A). Simultaneously, caspase-1 cleavage, interleukin 1β (IL-1β) secretion, and LDH release were also detected in bone marrow-derived macrophages (BMDMs) after infection with E. tarda EIB202, indicating the activation of caspase-1-dependent inflammasomes (see Fig. S2A to C in the supplemental material). These results demonstrate that E. tarda EIB202 can replicate in macrophages and induce caspase-1-dependent inflammatory cell death in macrophage cells. Furthermore, the bacterial count in the supernatant significantly increased after 4 h postinfection in accordance with the rising level of LDH release, and Ac-YVAD-CHO treatment notably reduced the viable bacterial count in the supernatant (Fig. 2B). These data suggest that intracellular-replicating E. tarda can escape to the extracellular milieu via caspase-1-dependent macrophage pyroptosis.
FIG 1.
Replication of E. tarda in macrophages. (A) The murine macrophage-like cell line J774A.1 was infected with E. tarda at an MOI of 10 for 2 h, followed by treatment with 1,000 μg/ml gentamicin for 30 min to kill extracellular bacteria. After being washed with PBS, J774A.1 cells were incubated in the growth medium containing 10 μg/ml gentamicin for the time intervals indicated. Triton X-100 was added to the cell culture at 1% (vol/vol) at 2.5, 4, 6, and 8 h postinfection (p.i.). The lysate produced was serially diluted, and the CFU were counted. Graphs show the mean ± SEM of results for triplicate cultures, and data are representative of at least three experiments. (B) Confocal microscopy of macrophages infected with GFP-labeled E. tarda at 2.5, 4, and 6 h p.i. Data are representative of at least three experiments, and representative microscopic images are shown. Filamentous actin was stained by rhodamine-phalloidin (red), and DNA was stained by DAPI (blue).
FIG 2.
Release of E. tarda from macrophages is dependent on activation of caspase-1. (A) Cells were pretreated with either Ac-YVAD-CHO (caspase-1-specific inhibitor; 40 μM) or DMSO as a control and maintained throughout the course of infection. The cytotoxicity of E. tarda-infected macrophages was assessed as the release of lactate dehydrogenase at the indicated times p.i. (B) Cells were pretreated as described for panel A, and viable extracellular bacteria were counted by coating TSB plates with diluted supernatant from E. tarda-infected macrophages. Graphs show the means ± SEM of results of 3 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Released E. tarda exhibits a T3SS/T6SS-responsive transcriptional profile.
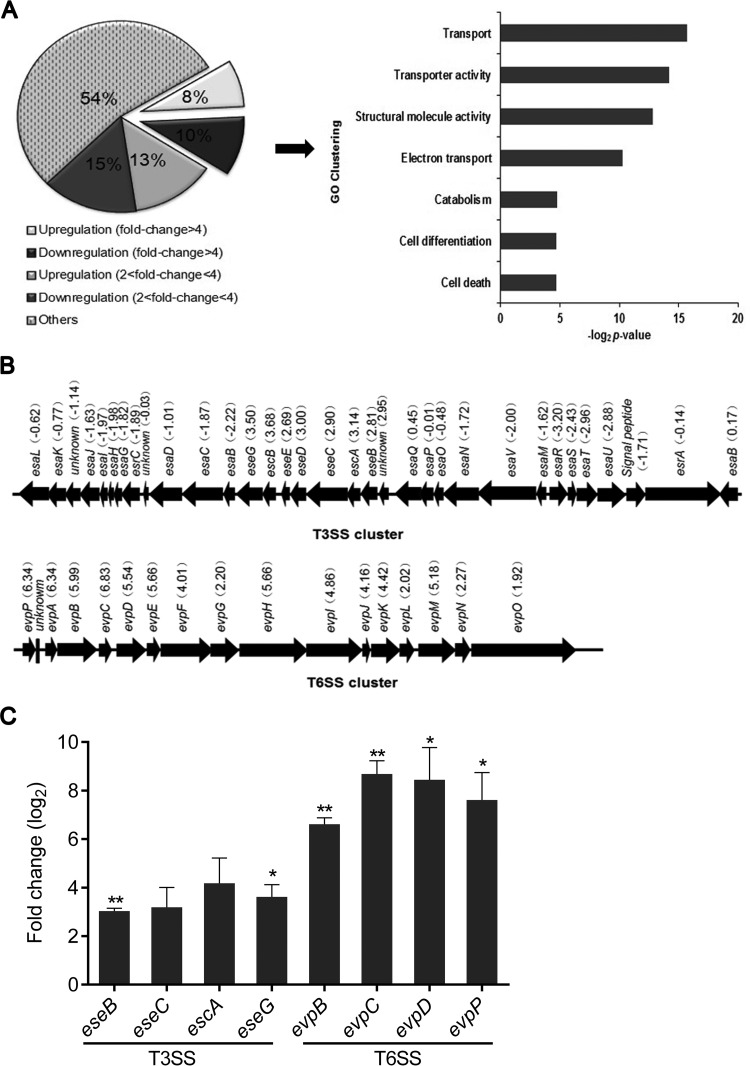
Pathogens, including bacteria, fungi, and viruses, have evolved a multitude of strategies for modulating macrophage defenses to promote their survival. To investigate whether intramacrophage infection induced an altered virulence profile in E. tarda, we compared the global gene transcriptional profiles of macrophage-released E. tarda and DMEM-cultured E. tarda. RNA-seq analysis showed that almost half of the total genes were either downregulated or upregulated by at least 2-fold after infection (Fig. 3A, left). Gene ontology analysis of the upregulated genes with a fold change greater than 4 (q < 0.001) revealed transcriptional activation of genes involved in transport and transporter activity during infection (Fig. 3A, right). Since previous studies have shown that T3SS and T6SS are the most important virulence factors in E. tarda (24–27), we further focused on the different transcriptional profiles of T3SS and T6SS cluster genes in E. tarda. As shown in Fig. 3B, 25 of the 33 T3SS cluster genes and 16 of the 16 T6SS cluster genes were either downregulated or upregulated by >2-fold after infection. Among these genes, those coding for the known T3SS effector EseG (28) and the possible T6SS effector EvpP (27) were greatly upregulated by 11- and 81-fold, respectively. Subsequently, we verified the upregulation of some T3SS/T6SS cluster genes by RT-qPCR (Fig. 3C). Meanwhile, significant upregulation of T3SS/T6SS cluster genes was also found in E. tarda released from BMDMs (see Fig. S3 in the supplemental material). Collectively, these data strongly support the idea that intramacrophage infection induces a T3SS/T6SS-responsive transcriptional profile in E. tarda.
FIG 3.
Macrophage-released E. tarda displays a T3SS/T6SS-responsive transcript profile. (A) Deep RNA-seq was executed to compose the transcript profiles of macrophage-released and DMEM-cultured E. tarda. Genes with different fold changes in transcription were classified. The genes upregulated in at least three biological replicates with >4-fold change (q < 0.001) were assigned to Gene Ontology (GO) categories. (B) Transcriptional alteration of the T3SS and T6SS gene clusters of E. tarda was quantified by RNA-seq. (C) Real-time PCR for the indicated genes of T3SS and T6SS in macrophage-released and DMEM-cultured E. tarda. *, P < 0.05; ***, P < 0.001.
Macrophage infection increases the infectivity of E. tarda in epithelial cells.
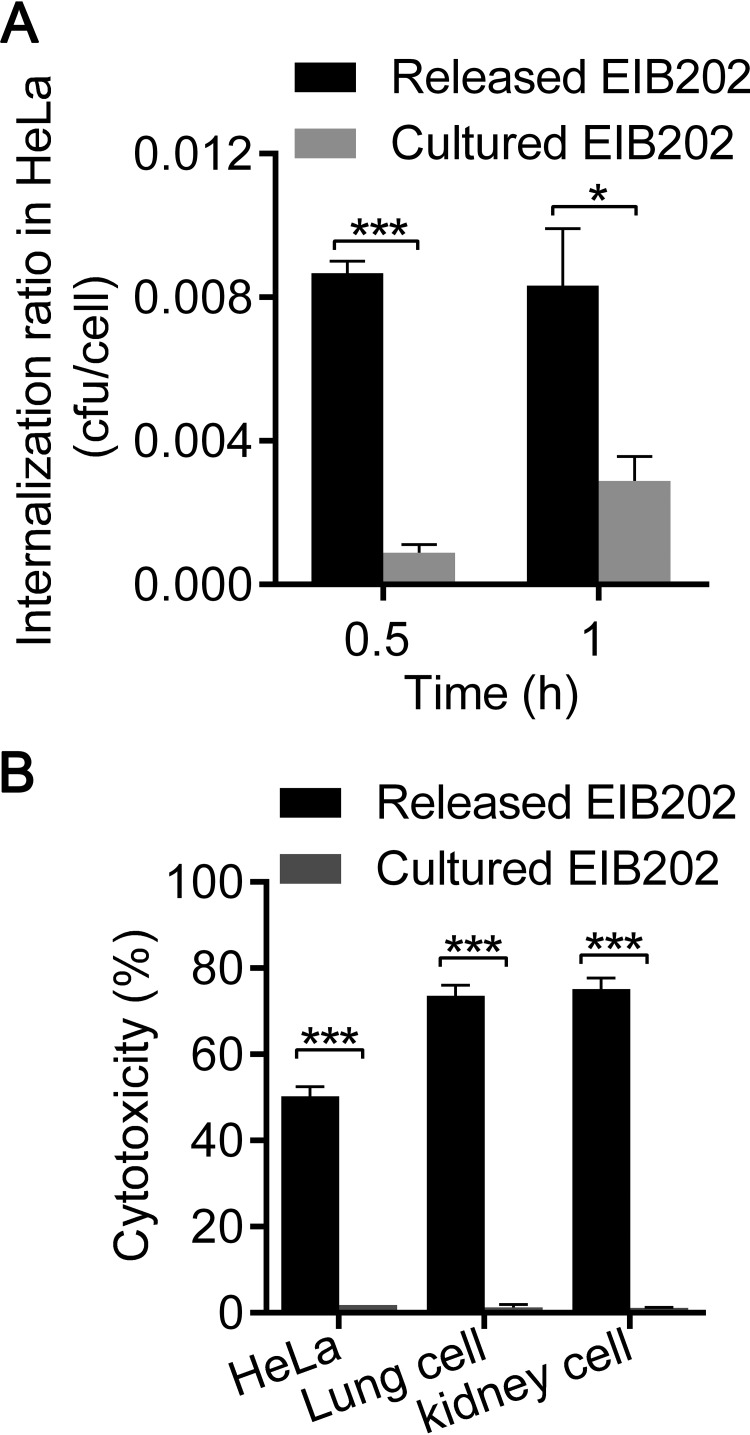
Given the evidence that macrophage infection induces a variety of virulence-related genes in E. tarda, it is interesting to determine whether macrophage-released E. tarda possesses greater virulence than DMEM-cultured E. tarda. Here, we compared the invasion abilities of macrophage-released E. tarda and DMEM-cultured strains in vitro. Epithelial cells were incubated with released E. tarda or cultured E. tarda at an MOI of 50, and both the internalization abilities and the cytotoxicities of the two bacterial populations were analyzed. In contrast to cultured E. tarda, released E. tarda showed a significant increase in the ratio of internalization into HeLa cells within the initial 1 h (Fig. 4A) and further induced dramatic cell death at 2 h postinfection (Fig. 4B). Similarly, significant cell death was also triggered by released E. tarda but not by cultured E. tarda in two primary epithelial cell cultures from mice (Fig. 4B). These data clearly demonstrate that the infectivity of macrophage-released E. tarda is increased in epithelial cells in vitro.
FIG 4.
Macrophage-released E. tarda displays enhanced infectivity in epithelial cells. (A) HeLa cells, incubated with released or cultured E. tarda at an MOI of 50, were washed with PBS 2 h later and subsequently cultured in medium harboring 100 μg/ml gentamicin for 2 h to kill the extracellular bacteria. The intracellular bacteria were counted. Ratios are expressed as the number of bacterial CFU divided by the number of cells. (B) HeLa cells and primary lung and kidney cells from mice were incubated separately with released or cultured E. tarda at an MOI of 50 for 2 h. The death of infected cells was assessed as the release of lactate dehydrogenase. Graphs show the means ± SD of results of triplicate cultures, and data are representative of at least three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Macrophage infection enhances the resistance of E. tarda to multiple host defenses.
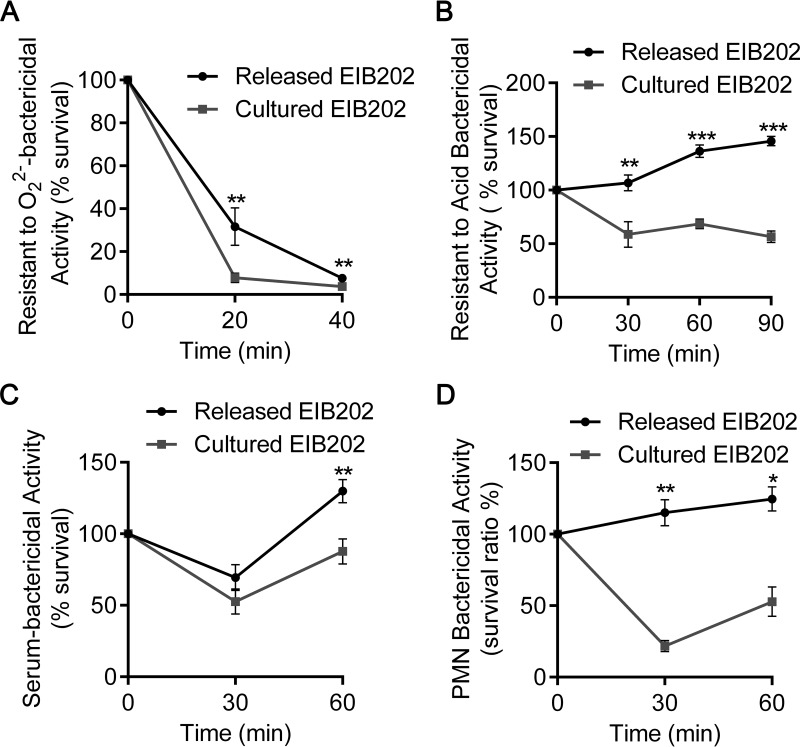
During infection, it is essential for bacteria to evade extensive host antimicrobial defenses (29, 30). Here, we investigated the capability of macrophage-released E. tarda and DMEM-cultured E. tarda to withstand multiple host defenses, including reactive oxygen species (ROS), acidified environments, serum factors, and polymorphonuclear leukocytes. First, we simulated a hostile intracellular environment by adding H2O2 or HCl to TSB. Significantly, more bacterial survivors were detected for released E. tarda than cultured E. tarda when they were incubated in the two simulated hostile environments (Fig. 5A and B). KEGG enrichment analysis of the RNA-seq data demonstrated that many genes potentially related to reactive oxygen species scavenging and acid tolerance were consistently upregulated after macrophage infection (see Table S1 in the supplemental material). These data indicate that macrophage-released E. tarda has stronger resistance to oxidative and acidic stresses. Next, we compared their abilities to resist extracellular host defenses, which typically include serum killing and neutrophil phagocytosis. As shown in Fig. 5C and D, released E. tarda displayed significantly enhanced viability when incubated with mouse serum or PMNs. These results demonstrate that macrophage-released E. tarda gains a stronger ability to counteract host immune defenses.
FIG 5.
Macrophage-released E. tarda revealed multiple resistances to host-killing effectors. Every 2 × 105 CFU of macrophage-released or DMEM-cultured E. tarda was incubated in 1 ml of TSB added with 0.75 mM H2O2 (A), 1 ml of TSB with a pH of 5.5 (B), 100 μl of mouse serum (C), or suspended PMNs at an MOI of 1:1 (D) at 30°C statically. The resistance to extensive host bactericidal activities, including O22−, acid, serum, and PMNs, was measured by counting the number of surviving E. tarda CFU at various points in time. Graphs show the means ± SD of results of triplicate cultures, and data are representative of at least three experiments. *, P < 0.05; **, P < 0.01. GO, Gene Ontology.
Macrophage infection contributes to the full virulence of E. tarda in vivo.
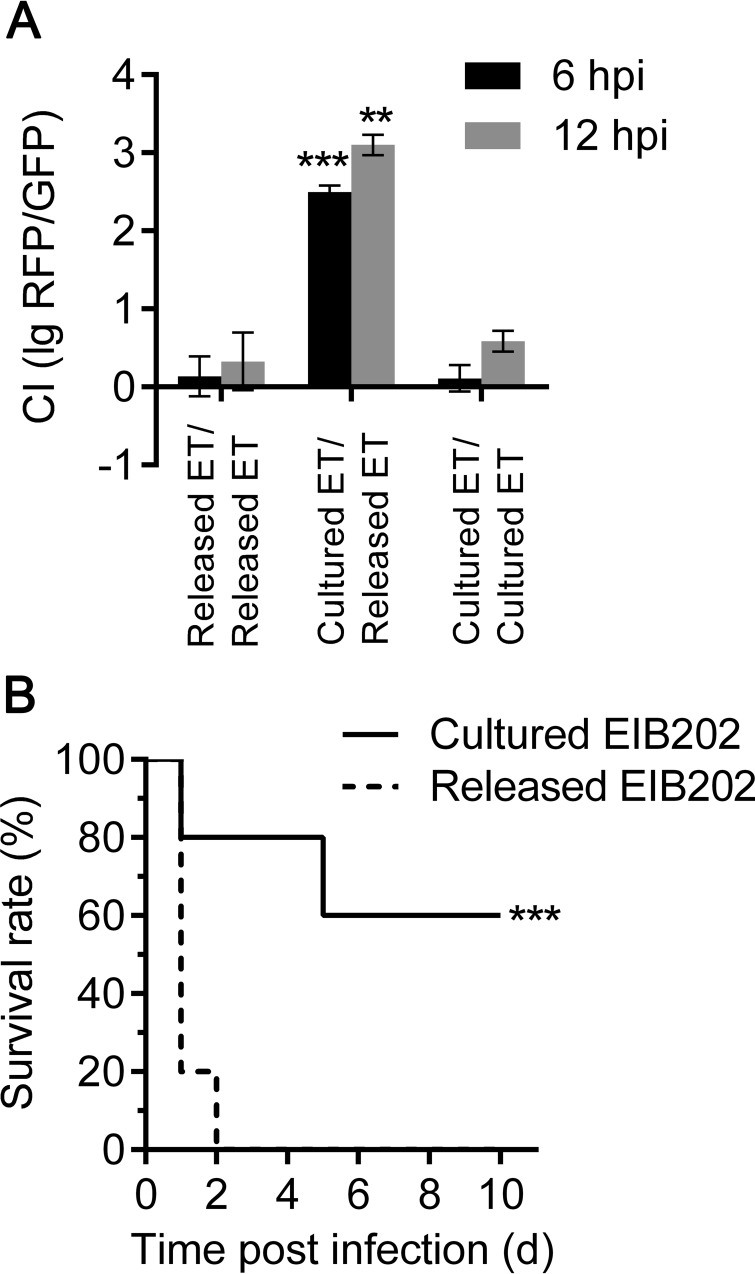
Given the evidence that macrophage-primed E. tarda presented both increased infectivity to epithelial cells and enhanced resistance to multiple host defenses in vitro, it remains important to determine whether macrophage infection strengthens its pathogenicity in vivo. In the competitive index (CI) assay, we used equal numbers of released and cultured E. tarda cells to infect each mouse and determined the bacterial loads in the livers of the infected mice on the basis of their differences in fluorescence (released E. tarda expressing RFP, cultured E. tarda expressing GFP). At 6 and 12 h after infection, we recovered 1 to 10 CFU of cultured E. tarda for every 1,000 CFU of released E. tarda, yielding a CI value of log10 (RFP/GFP) between 2 and 3 (Fig. 6A). In contrast, when we used RFP-labeled released E. tarda to compete with GFP-labeled released E. tarda or RFP-labeled cultured E. tarda to compete with GFP-labeled cultured E. tarda, we found roughly equal recoveries, with the CI near zero (Fig. 6A). Furthermore, when we challenged the wild-type mice with either released E. tarda or cultured E. tarda (Fig. 6B), the mice infected with released E. tarda succumbed to systemic infection within 2 days, whereas the mice injected with cultured E. tarda maintained a survival rate of 60%. Similarly, BMDM-released E. tarda also resulted in increased mortality in the mouse model compared with that in mice after infection with DMEM-cultured E. tarda (see Fig. S3 in the supplemental material). These data reveal that macrophage-released E. tarda exhibits significantly enhanced in vivo virulence over DMEM-cultured E. tarda.
FIG 6.
E. tarda released from macrophages exhibits enhanced virulence in vivo. (A) C57BL/6 mice were injected intraperitoneally with equal amounts of variously combined macrophage-released and DMEM-cultured E. tarda bacteria that were marked with constitutively expressed mCherry (RFP) or GFP, respectively. The bacterial burdens in the mouse livers were indicated by the number of red or green colonies on diluted TSB plates at 6 and 12 h p.i. (n = 5 mice per group). Results are presented as CIs [log10 (RFP-expressing bacteria/GFP-expressing bacteria)]; a CI of 2 corresponds to every 100 CFU of RFP-expressing bacteria for 1 CFU of GFP-expressing bacteria. Graphs show the means ± SD of results of triplicate cultures, and data are representative of at least three experiments. **, P < 0.01; ***, P < 0.001. (B) Survival rates of C57BL/6 mice that were intraperitoneally infected with macrophage-released or DMEM-cultured E. tarda (1.5 × 106 CFU/g, n = 5 mice per group). Data are representative of three experiments. ***, P < 0.001, log-rank test.
Involvement of T3SS and T6SS in macrophage-induced bacterial virulence.
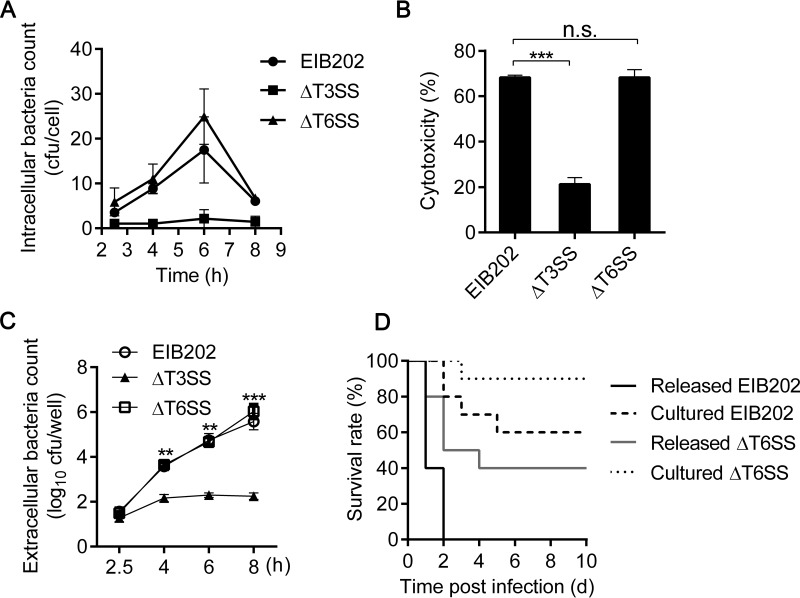
To determine whether the enhanced virulence of macrophage-released E. tarda is specifically correlated to the induced T3SS and T6SS genes, we compared the capabilities for intracellular replication, cytotoxicity, and bacterial escape among the ΔT3SS, ΔT6SS, and EIB202 strains. First, the ΔT3SS mutant showed an obvious defect in replication within J774A.1 cells, while the ΔT6SS mutant replicated at a level similar to that of the wild-type strain (Fig. 7A). Subsequently, significantly reduced cytotoxicity was observed in ΔT3SS mutant-infected cells but not in EIB202- or ΔT6SS mutant-infected cells (Fig. 7B). Furthermore, EIB202 and the ΔT6SS mutant efficiently escaped from infected cells over time, while the ΔT3SS mutant was detected in a very small amount in the supernatant during the whole infection process (Fig. 7C). These findings reveal that T3SS, but not T6SS, plays essential roles in bacterial replication and escape from macrophages. To further investigate the contributions of T6SS to enhanced bacterial virulence, the macrophage-released ΔT6SS mutant was collected and assessed for its virulence in mice (Fig. 7D). On one hand, a significant decrease in the survival rate of infected mice was observed with the macrophage-released ΔT6SS mutant compared to that with the DMEM-cultured ΔT6SS mutant (P = 0.018). This result demonstrates that intramacrophage infection reinforces bacterial virulence in the ΔT6SS mutant, which means that, besides T6SS, other unknown virulent factors may also participate in macrophage-induced bacterial virulence. On the other hand, the macrophage-released ΔT6SS mutant resulted in a higher survival rate of infected mice than did macrophage-released EIB202 (P = 0.009). This result illustrates that T6SS contributes at least partially to enhanced bacterial virulence in vivo. In brief, our data suggest that both T3SS and T6SS induced by macrophage infection actively promote the virulence of E. tarda.
FIG 7.
T3SS and T6SS are involved in macrophage-induced bacterial virulence. (A) J774A.1 cells were infected with E. tarda EIB202, the ΔT3SS mutant, or the ΔT6SS mutant at an MOI of 10 for 2 h and then treated as described in the legend to Fig. 1A. (B) The cytotoxicity of E. tarda-infected macrophages was assessed as the release of lactate dehydrogenase at 8 h. (C) Viable extracellular bacteria were counted by coating TSB plates with diluted supernatants of E. tarda-infected macrophages. Graphs show means ± SD of results of triplicate cultures, and data are representative of at least three experiments. (D) Survival rates of C57BL/6 mice that were intraperitoneally infected with macrophage-released or DMEM-cultured EIB202 or the macrophage-released or DMEM-cultured ΔT6SS mutant (1.5 × 106 CFU/g; n = 10 mice per group). Data are representative of three experiments. n.s., not significant.
DISCUSSION
The adaptation of bacteria to particular host niches depends on the activity of various adaptation factors; for pathogens, these are known as virulence factors. Particularly, pathogenic bacteria utilize types III (T3SS), IV (T4SS), and, in some instances, VI (T6SS) to transfer a broad arsenal of virulence factors, referred to as effectors, into host cells during infection (31–33). Salmonella enterica was reported to express a characteristic intracellular transcriptomic signature during infection, including the upregulation of Salmonella pathogenicity islands 1 and 2 (34, 35). A T6SS locus in Burkholderia pseudomallei was identified from in vivo-induced genes during invasion of macrophages (36). Here, our data demonstrate that the population of E. tarda, released from infected macrophages, displays a T3SS/T6SS-responsive transcriptional profile that plays an important role in macrophage-induced bacterial virulence. Although T3SS and T6SS are the most important components of virulence in E. tarda (24, 27), few T3SS or T6SS effectors have been identified so far. EseG was the first characterized Edwardsiella T3SS effector to have been proven to disassemble microtubule structures when overexpressed in mammalian cells (28). Recently, EseJ was found to be a novel T3SS effector that reduces bacterial adhesion to endothelial progenitor cells and facilitates intracellular bacterial replication (37). In contrast, although the E. tarda T6SS cluster was reported as early as 2004 (25), as the first reported T6SS locus in bacteria, none of its T6SS effectors has been identified to date. Given the evidence that bacterial effectors are often transcriptionally induced during the bacterial infection cycle, we propose that the infection-inducible gene collection uncovered in this work might serve as a core database for identifying new T3SS or T6SS effectors.
During infection, pathogens often have to counteract extracellular defenses, such as complement, antibody, and cationic antimicrobial peptides (38, 39), and some bacteria evade harsh extracellular defenses simply by entering host cells (40, 41). However, once inside an infected cell, these pathogens are further challenged by intense intracellular defense mechanisms, such as the acidification of endosomes, reactive oxygen species, nitric oxide, and a group of cytokines and chemokines. Correspondingly, infection-related signals often stimulate pathogens to generate multiple resistances to host-killing mechanisms. For instance, studies with Salmonella have shown that the bacterial effector was translocated into cytosol by sensing acidic pH (42). Other environmental conditions, such as ROS stress and nutrition deficiency, were also reported to modulate fitness gene expression (43, 44). Although it is easily recognized that bacterial virulence properties are enhanced following host-specific reprogramming of gene expression, few studies to date have provided direct evidence to verify the enhanced bacterial virulence both in vitro and in vivo. In this work, we actually correlate the altered gene expression pattern, especially induced T3SS and T6SS, with enhanced epithelial invasiveness, resistance to multiple host bactericidal effectors, and in vivo virulence for E. tarda. From multiple perspectives, this work gives direct evidence that macrophage infection reprograms the virulent gene expression profile of E. tarda and promotes bacterial pathogenesis in vivo.
Inflammasomes, as cytosolic multiprotein complexes, are assembled to drive activation of caspase-1, maturation of IL-1β and IL-18, and induction of an inflammatory cell death program termed pyroptosis (45). Miao et al. reported that pyroptotic cells can release bacteria from macrophages and expose them to neutrophils and other innate immune defenses. Thus, this caspase-1-induced pyroptosis has been implicated as a protective mechanism in host defenses against bacteria (20). Conversely, E. tarda has been found to replicate in macrophages for virulence priming and then escape from macrophages via the induction of caspase-1-dependent cell pyroptosis, resulting in enhanced bacterial virulence both in vivo and in vitro. Our data suggest that the extrusion from macrophages via cell death might facilitate E. tarda to establish systematic infection by allowing a surviving bacterial population to circumvent host immune surveillance and spread to neighboring cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China grants 31430090 and 31472308 and National High Technology Research and Development Program of China grant 2013AA093101.
We thank Ka Yin Leung for helpful discussions and suggestions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00978-15.
REFERENCES
- 1.Cossart P, Sansonetti PJ. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 2.Poole K. 2012. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol 20:227–234. doi: 10.1016/j.tim.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Liu XJ, Kang LQ, Liu YJ, Li H, Peng X. 2013. Characterization of the Edwardsiella tarda proteome in response to different environmental stresses. J Proteomics 80:320–333. doi: 10.1016/j.jprot.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Raspoet R, Appia-Ayme C, Shearer N, Martel A, Pasmans F, Haesebrouck F, Ducatelle R, Thompson A, Van Immerseel F. 2014. Microarray-based detection of Salmonella enterica serovar Enteritidis genes involved in chicken reproductive tract colonization. Appl Environ Microbiol 80:7710–7716. doi: 10.1128/AEM.02867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. 1999. Salmonella Typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun 67:5690–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gort AS, Miller VL. 2000. Identification and characterization of Yersinia enterocolitica genes induced during systemic infection. Infect Immun 68:6633–6642. doi: 10.1128/IAI.68.12.6633-6642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danelishvili L, Stang B, Bermudez LE. 2014. Identification of Mycobacterium avium genes expressed during in vivo infection and the role of the oligopeptide transporter OppA in virulence. Microb Pathog 76:67–76. doi: 10.1016/j.micpath.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe H, Kamitani S, Fukui-Miyazaki A, Shinzawa N, Nakamura K, Horiguchi Y. 2015. Detection of genes expressed in Bordetella bronchiseptica colonizing rat trachea by in vivo expressed-tag immunoprecipitation method. Microbiol Immunol 59:249–261. doi: 10.1111/1348-0421.12247. [DOI] [PubMed] [Google Scholar]
- 9.Karsi A, Gulsoy N, Corb E, Dumpala PR, Lawrence ML. 2009. High-throughput bioluminescence-based mutant screening strategy for identification of bacterial virulence genes. Appl Environ Microbiol 75:2166–2175. doi: 10.1128/AEM.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci U S A 103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behar SM, Divangahi M, Remold HG. 2010. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol 8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monack DM, Mecsas J, Bouley D, Falkow S. 1998. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J Exp Med 188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. 2012. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect 14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Mohanty BR, Sahoo PK. 2007. Edwardsiellosis in fish: a brief review. J Biosci 32:1331–1344. doi: 10.1007/s12038-007-0143-8. [DOI] [PubMed] [Google Scholar]
- 15.Okuda J, Arikawa Y, Takeuchi Y, Mahmoud MM, Suzaki E, Kataoka K, Suzuki T, Okinaka Y, Nakai T. 2006. Intracellular replication of Edwardsiella tarda in murine macrophage is dependent on the type III secretion system and induces an up-regulation of anti-apoptotic NF-kappaB target genes protecting the macrophage from staurosporine-induced apoptosis. Microb Pathog 41:226–240. doi: 10.1016/j.micpath.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Xie HX, Lu JF, Rolhion N, Holden DW, Nie P, Zhou Y, Yu XJ. 2014. Edwardsiella tarda-induced cytotoxicity depends on its type III secretion system and flagellin. Infect Immun 82:3436–3445. doi: 10.1128/IAI.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Wang L, Zhang L, Qu J, Wang Q, Zhang Y. 2015. An invasive and low virulent Edwardsiella tarda esrB mutant promising as live attenuated vaccine in aquaculture. Appl Microbiol Biotechnol 99:1765–1777. doi: 10.1007/s00253-014-6214-5. [DOI] [PubMed] [Google Scholar]
- 18.Campbell JA, Eppersimons CF, Kligerman AD, Petro AB, Sharief Y, Allen JW. 1986. Sister chromatid exchange analysis in cultured primary lung, liver, and kidney cells of mice following in vivo exposure to vinyl carbamate. In Vitro Cell Dev Biol 22:443–448. doi: 10.1007/BF02623444. [DOI] [PubMed] [Google Scholar]
- 19.Spinner JL, Cundiff JA, Kobayashi SD. 2008. Yersinia pestis type III secretion system-dependent inhibition of human polymorphonuclear leukocyte function. Infect Immun 76:3754–3760. doi: 10.1128/IAI.00385-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuda J, Kiriyama M, Yamanoi E, Nakai T. 2008. The type III secretion system-dependent repression of NF-kappaB activation to the intracellular growth of Edwardsiella tarda in human epithelial cells. FEMS Microbiol Lett 283:9–14. doi: 10.1111/j.1574-6968.2008.01147.x. [DOI] [PubMed] [Google Scholar]
- 22.Strauss EJ, Ghori N, Falkow S. 1997. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect Immun 65:3924–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasa RP, Lim TM, Leung KY. 2001. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect Immun 69:5689–5697. doi: 10.1128/IAI.69.9.5689-5697.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY. 2005. Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151:2301–2313. doi: 10.1099/mic.0.28005-0. [DOI] [PubMed] [Google Scholar]
- 25.Rao PS, Yamada Y, Tan YP, Leung KY. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol 53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Yang M, Xiao J, Wu H, Wang X, Lv Y, Xu L, Zheng H, Wang S, Zhao G, Liu Q, Zhang Y. 2009. Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS One 4:e7646. doi: 10.1371/journal.pone.0007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 28.Xie HX, Yu HB, Zheng J, Nie P, Foster LJ, Mok YK, Finlay BB, Leung KY. 2010. EseG, an effector of the type III secretion system of Edwardsiella tarda, triggers microtubule destabilization. Infect Immun 78:5011–5021. doi: 10.1128/IAI.00152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones CL, Napier BA, Sampson TR, Llewellyn AC, Schroeder MR, Weiss DS. 2012. Subversion of host recognition and defense systems by Francisella spp. Microbiol Mol Biol Rev 76:383–404. doi: 10.1128/MMBR.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 33.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 35.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 37.Xie HX, Lu JF, Zhou Y, Yi J, Yu XJ, Leung KY, Nie P. 2015. Identification and functional characterization of the novel Edwardsiella tarda effector EseJ. Infect Immun 83:1650–1660. doi: 10.1128/IAI.02566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben NA, Klimpel GR. 2008. Subversion of complement activation at the bacterial surface promotes serum resistance and opsonophagocytosis of Francisella tularensis. J Leukoc Biol 84:77–85. doi: 10.1189/jlb.0807526. [DOI] [PubMed] [Google Scholar]
- 39.Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 40.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. 2012. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog 8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang XS, Liu JH, Chen XJ. 2010. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol 10:230. doi: 10.1186/1471-2229-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meibom KL, Charbit A. 2010. Francisella tularensis metabolism and its relation to virulence. Front Microbiol 1:140. doi: 10.3389/fmicb.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao EA, Rajan JV, Aderem A. 2011. Caspase-1-induced pyroptotic cell death. Immunol Rev 243:206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.