Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) systems in bacteria and archaea target foreign elements, such as bacteriophages and conjugative plasmids, through the incorporation of short sequences (termed spacers) from the foreign element into the CRISPR array, thereby allowing sequence-specific targeting of the invader. Thus, CRISPR-Cas systems are typically considered a microbial adaptive immune system. While many of these incorporated spacers match targets on bacteriophages and plasmids, a noticeable number are derived from chromosomal DNA. While usually lethal to the self-targeting bacteria, in certain circumstances, these self-targeting spacers can have profound effects in regard to microbial biology, including functions beyond adaptive immunity. In this minireview, we discuss recent studies that focus on the functions and consequences of CRISPR-Cas self-targeting, including reshaping of the host population, group behavior modification, and the potential applications of CRISPR-Cas self-targeting as a tool in microbial biotechnology. Understanding the effects of CRISPR-Cas self-targeting is vital to fully understanding the spectrum of function of these systems.
INTRODUCTION
Bacteria and archaea are under constant threat of viral predation and have evolved numerous mechanisms to defend against infection (1, 2). One such mechanism is the clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) protein system, which provides adaptive immunity against viruses and plasmids, collectively referred to as mobile genetic elements (MGEs) (3–5). While many divergent CRISPR-Cas systems exist, currently divided into 6 distinct types (6, 7), their general function is conserved. A CRISPR array is composed of short repeat sequences flanking unique spacer inserts transcribed by a promoter found in an adjacent AT-rich sequence (termed the leader) into a long precursor RNA molecule known as pre-CRISPR RNA (pre-crRNA). This pre-CRISPR RNA transcript is processed into multiple RNA molecules, known as mature crRNA, through nucleolytic cleavage at specific sequences in the repeats, with one exception being the type II-C system, in which mature crRNA is generated through transcription from promoters in each CRISPR (8). The mature crRNA then associates with Cas proteins to form the targeting CRISPR ribonucleoprotein complex (9, 10). The crRNA molecule is critical in host defense against MGEs, since the transcribed spacer provides the specificity of its target, and once bound through Watson-Crick base pairing, results in either degradation (type I) or cleavage (types II-VI) of the target through Cas protein nuclease activity.
The incorporation of a short sequence (usually 30 to 40 bp) from the invading MGE into the CRISPR array as a new spacer is a process termed CRISPR adaptation, a key step in CRISPR adaptive immunity. Analysis of the sequences of spacers from a given CRISPR array can serve as a history of previous CRISPR-Cas interactions with invading MGEs. The first evidence that CRISPR-Cas systems function as an immune system came from such an analysis, when in 2005, three separate groups examined a variety of CRISPR arrays and found that spacers matched sequences found in phages and plasmids (11–13). However, in addition to the matches with MGEs, spacers were also found that target sites on the bacterial or archaeal genome. For example, in Yersinia spp., of 36 spacers analyzed, the majority were of bacteriophage origin, but 8 spacers matched sequences on the Yersinia chromosome (13). A similar pattern was observed in a broader analysis of 4,500 spacers across archaea and bacteria, in which 35% of the spacers that matched sequences in the NCBI database were derived from chromosomal DNA and apparently were not related to foreign elements or prophages (12). Since these early observations, self-targeting spacers have consistently been found in CRISPR arrays (14–19), demonstrating that the insertion of a self-targeting spacer is not a rare event.
Given that at least one of the six types of CRISPR-Cas systems are present in 84% and 45% of sequenced archaeal and bacterial genomes, respectively (20), it is clear that these systems are as widespread as they are diverse. As research on these systems expands, and since a surprisingly large percentage of CRISPR spacers have been shown to have sequence identity to bacterial chromosomal targets, it is becoming clear that CRISPR-Cas systems can play a role in biological functions beyond adaptive immunity (21) and that self-targeting spacers can, at least in part, drive these alternative functions. This review aims to summarize current research on CRISPR-Cas self-targeting in prokaryotes and the role these events can play in important biological functions.
SELF-TARGETING WITH 100% COMPLEMENTARITY CAN DRIVE EVOLUTION
The most likely outcome of a 100% complementary, self-targeting spacer is cell death (Fig. 1A); such events have been experimentally demonstrated in multiple CRISPR-Cas types (22–24). For this reason, there are mechanisms that prevent a spacer from targeting the CRISPR array from which it was transcribed, such as the requirement of a protospacer adjacent motif (PAM) in the type I, II, V, and VI systems (25) or inactivation of the targeting CRISPR ribonucleoprotein complex through base pairing between the 5′ handle of the crRNA and the CRISPR repeats in the type III system (26). However, these mechanisms would not prevent cell death if a spacer is acquired that targets the host chromosome, and accordingly, evidence from the type I-E system of Escherichia coli suggests that the bacteria have evolved to preferentially sample foreign DNA over chromosomal DNA during naive CRISPR adaptation. It was observed previously in CRISPR adaptation studies using E. coli as a model system that the cell is roughly 100 to 1,000 times more likely to incorporate plasmid DNA over chromosomal DNA into its CRISPRs after normalizing for their respective size (27, 28). Recent evidence suggests one such mechanism for this preferential incorporation of foreign DNA into CRISPR regions during naive CRISPR adaptation is the involvement of the RecBCD complex. During DNA replication, when a double-stranded break occurs, RecBCD participates in the generation of single-stranded DNA, which can serve as a substrate for spacer acquisition by the Cas1-Cas2 complex. Chi sites, which are 8-nucleotide motifs well represented in the E. coli genome, limit the extent of single-stranded DNA generated by the RecBCD complex, and it was found the CRISPR-Cas system avoids the acquisition of DNA near Chi sites (27). Therefore, the lower prevalence of Chi sites on plasmids versus the E. coli genome, along with the preference of RecBCD to degrade linear DNA, such as recently injected phage DNA, might explain the observed preferential incorporation of foreign DNA. Cas1 has been shown to physically interact with RecB and RecC (29), and recently, a second group confirmed the requirement of the RecBCD complex during naive CRISPR adaptation in E. coli (30), further supporting this model.
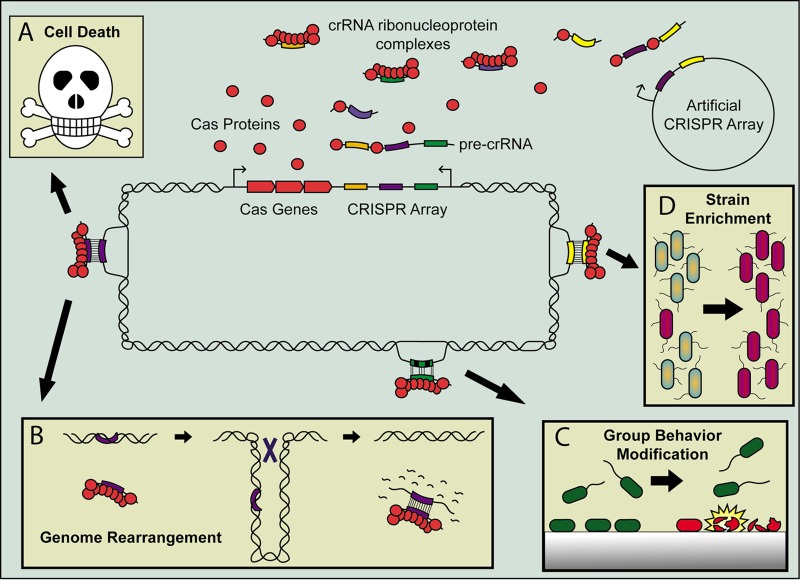
FIG 1.
Consequences of CRISPR self-targeting in the type I system. The self-targeting crRNA can be transcribed from either the native CRISPR array or an artificial array on a plasmid, and the crRNA associates with the Cas proteins expressed from the native Cas-encoding operon to form the self-targeting crRNA ribonucleoprotein complex. These self-targeting spacers include both 100% complementary spacers (purple and yellow), as well as partially matching spacers (green and black). The most likely outcome of a 100% complementary match is cell death (A) due to the nucleolytic activity of the CRISPR-Cas system; however, this results in strong selection for mutants that remove the targeted region, for example, by genome rearrangement, such as excision of pathogenicity island or curing of a prophage (B). A partially matching self-targeting spacer can modulate group behavior, such as biofilm formation, by triggering viable planktonic cells (green cells) to die (red cells) upon surface binding (C). Bacterial populations transformed with a plasmid containing an artificial self-targeting spacer can enrich for bacterial strains that do not contain the target on their genome (purple cells, D).
Nevertheless, the frequency at which self-targeting spacers are found in CRISPR arrays suggests that, although less common than the incorporation of foreign DNA, incorporation of a self-targeting spacer can occur, perhaps impacting the biology of the host bacterial cell. This idea was directly tested in Pectobacterium atrosepticum. The native CRISPR2 array of P. atrosepticum contains a self-targeting spacer 100% complementary to a chromosomal gene within an ∼100-kb horizontally acquired island named HAI2, but CRISPR lethality is abrogated due to a nonconsensus PAM (22). To assay the consequences of CRISPR self-targeting, P. atrosepticum was transformed with an inducible plasmid containing a truncated type I-F CRISPR1 leader, appropriate repeats, and an engineered spacer targeting the same gene in HAI2 as the native spacer but with the correct PAM. Induction of the self-targeting spacer-containing plasmid resulted in a cessation of bacterial growth and elongation of the bacterial cell indicative of DNA damage and the subsequent SOS response, confirming that a self-targeting spacer is cytotoxic (22). Interestingly, when the bacteria were left for 36 h, suppressor mutants arose in which CRISPR targeting was ablated, including deletions of the chromosomal target. The size of the deletion differed between the mutants and included excision of the entire ∼100-kb HAI2 island. A similar effect was observed when the spacer was designed to target a core region of the genome instead of HAI2, demonstrating that while a self-targeting spacer will normally result in cell death, it can also reshape the bacterial genome. Furthermore, these findings show that self-targeting is not dependent on the target being horizontally acquired, such as the integrative conjugative element HAI2.
A similar result was demonstrated using the native type II-A CRISPR-Cas system of Streptococcus thermophilus. When S. thermophilus was transformed with a plasmid carrying genes encoding artificial spacers targeting genomic islands within the transformed strain, >99% of the transformed bacteria were killed, but the surviving transformants contained large deletions of the targeted regions (31). The authors showed that the deletions were the result of recombination between insertion sequence elements (IS) on the S. thermophilus chromosome flanking the targeted region, and that the IS-dependent recombination events were occurring at a low frequency in the wild-type population. Taken together, these data indicate that the incorporation of a self-targeting spacer can drive evolution, but likely through selection of a small population of bacteria in which excision of the targeted region has occurred naturally (Fig. 1B).
SELF-TARGETING WITH 100% COMPLEMENTARITY CAN SELECT FOR PROPHAGE-CURED BACTERIA
When a bacterium is lysogenized by a particular phage, the phage genome is for all intents and purposes part of the bacterial genome. Thus, another potential benefit of a self-targeting spacer is its ability to incorporate a spacer targeting the prophage, which will reshape a bacterial population by selecting for cells that have excised the prophage from their chromosome (Fig. 1B). This notion was tested in an hns mutant strain of E. coli in which the native CRISPR system, normally heat-stable nucleoid-structuring (H-NS) silenced, was active. The E. coli strain harboring a lambda phage with a temperature-sensitive cI gene allowing controlled lytic induction was transformed with an inducible plasmid containing either an artificial spacer targeting the prophage or a control spacer with no target. Upon simultaneous induction of both the prophage and the artificial spacer-containing plasmid, the majority of cells (>99%) were killed; however, the survival frequency of cells transformed with the prophage-targeting spacer was 500-fold higher than the survival frequency of cells transformed with the control spacer (32). These data suggest that the CRISPR-Cas system, under the right circumstances, can select for cells that have excised the phage.
The ability to select for cells that have cured a prophage is likely not limited to E. coli. In Streptococcus pyogenes, 13 sequenced strains were assayed for CRISPR arrays, and of the 8 CRISPR-positive strains, 41 distinct spacers were identified, with 26 spacers found to target S. pyogenes prophages. However, no strain contained a spacer targeting a prophage on its own genome (33). Additionally, it was found the CRISPR-negative strains contained significantly more prophages than the CRISPR-positive strains, and furthermore, within the 8 CRISPR-positive S. pyogenes strains, an inverse correlation was found between the number of spacers per genome and the number of prophages within each genome. Similarly, recently, the CRISPR-Cas systems of the genus Bifidobacterium were analyzed, and 25 out of 32 characterized type I and type II CRISPR-Cas systems contained at least one spacer sequence targeting a Bifidobacterium lysogenic prophage, including two instances in which the Bifidobacterium species included a spacer targeting a prophage on its own genome (16). Similar to S. pyogenes, a positive correlation was found between strains lacking a CRISPR-Cas system and the number of times prophages were found on the chromosome targeted by spacers in other Bifidobacterium CRISPR-Cas systems. For example, Bifidobacterium angulatum contains 26 unique spacers that target prophages on 11 other Bifidobacterium species genomes while harboring no spacer-targeted prophages on its own genome.
Additionally, a bioinformatic analysis of 365 Pseudomonas aeruginosa CRISPR-Cas systems from 672 sequenced P. aeruginosa strains found that 55% of the 2,823 unique spacers matched sites in the P. aeruginosa genome, the vast majority of which are predicted to target the P. aeruginosa accessory genome, including potential prophages (17). The P. aeruginosa strains lacking CRISPR-Cas systems were on average 300 kbp larger than those with a CRISPR-Cas system. Thus, it is possible the CRISPR-Cas systems of S. pyogenes, Bifidobacterium spp., and P. aeruginosa are either actively preventing the uptake of MGEs (including prophages) or incorporating self-targeting spacers that select for cells that have purged these MGEs in a manner analogous to the experimentally demonstrated removal of genomic regions in P. atrosepticum (22) and S. thermophilus (31).
SELF-TARGETING WITH 100% COMPLEMENTARITY IN THE TYPE III-A SYSTEM CAN HELP MICROBES TOLERATE TEMPERATE PHAGES BY BLOCKING THE ENTRY OF PROPHAGES INTO THE LYTIC CYCLE
While the presence of a lysogenic phage on a bacterial chromosome can be detrimental to bacterial fitness (34), it has been well established that prophages can provide beneficial functions to, and increase the fitness of, the bacterial host (35, 36). For this reason, curing a phage from the bacterial chromosome can have a selective disadvantage. Unlike the type I and II systems discussed above, a type III-A/B CRISPR-Cas array can potentially include a self-targeting spacer without associated cytotoxicity due to general transcription-dependent targeting of the type III-A/B systems (37–39).
A recent study of Staphylococcus aureus showed that when spacers targeted lytic genes silenced during lysogeny, no CRISPR interference through the type III-A system was detected. CRISPR interference could only be achieved when the target gene was expressed; this finding was demonstrated by integrating a CRISPR-targeted gene under the control of a tightly regulated inducible promoter on the S. aureus chromosome. This strain was then transformed with a plasmid expressing the spacer targeting the integrated gene, and CRISPR interference was detected only upon induction of the chromosomal promoter driving expression of the targeted gene. Additionally, only spacers targeting the nontemplate strand of the targeted gene can generate CRISPR interference, illustrating transcription-dependent targeting, and furthermore, distinguishing the targeting requirements of the type III system from the type I and II systems (37, 40). The implication of these findings is that bacteria with spacers capable of self-targeting a prophage can tolerate a 100% complementary self-match and reap the putative benefits of a prophage while also preventing phage-mediated lysis.
SELF-TARGETING WITH PARTIAL COMPLEMENTARITY CAN DRIVE GROUP BEHAVIOR
The research discussed so far has focused on the interaction of a spacer that is 100% complementary with a DNA target on (or associated with) the host chromosome, but a partially matching spacer can still elicit an effect without the lethality typically associated with a 100% complementary spacer. Generally, partially matching spacers are thought to be important in CRISPR priming, in which the presence of a spacer partially matching an MGE will greatly increase the likelihood of incorporating new spacers against the MGE, and particularly sequences that match regions adjacent to the partially matched target (41–45). Additionally, it was recently found that spacers with a mismatch in regions considered indispensable for CRISPR interference, such as the seed sequence or PAM, may still be capable of functioning directly in CRISPR-mediated interference (46). Therefore, self-targeting spacers with partial complementarity can likely still drive biological functions.
The best example of a partially complementary spacer impacting microbial behavior is the modification of group behavior by the type I-F CRISPR-Cas system in P. aeruginosa. When P. aeruginosa is lysogenized by the bacteriophage DMS3, the interaction of a partially complementary spacer encoded by the P. aeruginosa CRISPR2 array with a target on the DMS3 prophage inhibits biofilm formation and swarming motility, two group behaviors important for virulence (47, 48). These altered biofilm and swarming phenotypes are dependent on the nickase activity of the effector protein Cas3, since a mutation in the catalytic region of the nuclease domain of Cas3 fully restores biofilm formation and swarming motility (48). The 2 mismatches at the +9 and +11 positions on the spacer prevent a lethal self-targeting event but promote sufficient CRISPR targeting activity to induce RecA, which in turn directly induces the expression of SOS-regulated, phage-related autolysis genes that induce cell death upon the bacteria attaching to a surface. Induction of the phage-related autolysis genes effectively inhibits biofilm and swarming motility through death of the surface-associated bacteria while having no noticeable effect on the planktonic population (Fig. 1C) (49). Given that numerous P. aeruginosa Mu-like phages contain the same protospacer target as DMS3 (50, 51), it is unclear if this interaction was directly selected or simply a side effect of CRISPR-Cas immunity. Nevertheless, the demonstration that a partially matching spacer can elicit such a strong biological response, in combination with the frequency of self-targeting spacers present in CRISPR arrays across both bacteria and archaea, suggests the potential of other partial matching interactions impacting important aspects of bacterial biology. It is unlikely that the biological impact of partial complementarity of a CRISPR spacer is limited to P. aeruginosa.
SELF-TARGETING CAN BE EXPLOITED FOR USE IN BIOTECHNOLOGY
CRISPR-Cas research has received a great deal of attention, largely for use of the type II system (52, 53) and the recently described type V system (54), in the genetic engineering of eukaryotic systems. Recent work has demonstrated that CRISPR-Cas technology is not limited to the engineering of eukaryotic systems and can be used in bacterial and archaeal systems as well. For example, for the type I-E system, it has been shown that E. coli lacking the effector enzyme Cas3 can be transformed with plasmids harboring self-targeting spacers without lethal effects, and designing these spacers to target the promoter of a particular gene will result in silencing of that particular gene due to the binding of the Cascade complex (55, 56). Exploiting CRISPR-Cas systems for such engineering approaches is likely not limited to the type I-E system and would potentially allow the endogenous type I systems of any number of bacteria to be coopted for CRISPR-mediated genetic control. Such approaches may be particularly useful in nonmodel organisms.
As another example, it has been shown that the cytotoxic effects of CRISPR self-targeting, such as those described earlier (22), can be used to remove particular strains of bacteria from a population. A mixed but equal population of E. coli K-12 and E. coli B, which share 99% sequence identity, was transformed with plasmids harboring self-targeting spacers unique to either sequences of strain K-12 or B (Fig. 1D), and the resulting transformants were composed almost entirely of the nontargeted strain (>99.9%) due to CRISPR interference from the self-targeting spacers (57). This cytotoxicity is the basis of “CRISPR antimicrobials,” in which antibiotic strains of a particular bacterial species, including E. coli (58) and S. aureus (59), are selected out of a population through CRISPR self-targeting of antibiotic resistance genes.
CONCLUSIONS AND FUTURE PERSPECTIVES
While the incorporation of a self-targeting spacer into a CRISPR array is typically a lethal event for a microbe, the surprisingly high frequency of self-targeting spacers identified from sequenced CRISPR arrays indicates that the occurrence of CRISPR-Cas self-targeting is not rare in nature. Self-targeting spacers with 100% complementarity in a type I or II system present the microbe with the challenge of avoiding CRISPR-induced lethality, which can reshape bacterial populations by selecting for bacteria in which genome rearrangement has occurred, such as excising pathogenicity islands or curing of a prophage. There are several examples in bacteria in which CRISPR-positive strains have a smaller genome than that of CRISPR-negative strains, and it is unclear if the reduction in genome size (specifically MGEs) is the result of the CRISPR-Cas system actively preventing the acquisition of MGEs or if incorporation of self-targeting spacers is selecting for bacteria in the population that have purged their genome of any of these invading sequences. It is important to note that microbes containing a type III CRISPR system are more tolerant of self-targeting spacers, since target transcription is required for CRISPR interference, allowing the microbe to tolerate a prophage when the targeted gene is silenced, which is the case for most phage genes during lysogeny.
Self-targeting is not limited to 100% complementary spacers; a self-targeting spacer can still have meaningful biological consequences even with multiple mismatches between the spacer and its genomic target, such as group behavior modification in P. aeruginosa. This is a largely unexplored area of research, and with the sheer number of potential partially matching spacers already identified in bacterial genomes, it is tempting to speculate that other microbial behaviors can be attributed to CRISPR self-targeting, especially in light of recent studies demonstrating functional CRISPR interference even with multiple mutations disrupting spacer-protospacer complementarity (46, 60). This is a largely unexplored area of research.
Overall, we are only beginning to understand the role of CRISPR self-targeting, and with the diversity of CRISPR-Cas types and variety of biological consequences of a self-targeting spacer that were already demonstrated, chromosomal-targeting spacers are gaining attention. It will be exciting to see future advances in our understanding of the biological functions and continued applications of CRISPR self-targeting in biotechnology.
ACKNOWLEDGMENTS
This study is supported by National Institutes of Health grants R37 AI83256-06 and R01-AI097307 and NSF grant MCB-9984521to G.A.O.
REFERENCES
- 1.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, Marraffini LA. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. 2015. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. 2013. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeks J, Naismith JH, White MF. 2013. CRISPR interference: a structural perspective. Biochem J 453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 12.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 13.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 14.Brodt A, Lurie-Weinberger MN, Gophna U. 2011. CRISPR loci reveal networks of gene exchange in archaea. Biol Direct 6:65. doi: 10.1186/1745-6150-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleckaityte M, Zilnyte M, Zvirbliene A. 2012. Insights into the CRISPR/Cas system of Gardnerella vaginalis. BMC Microbiol 12:301. doi: 10.1186/1471-2180-12-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briner AE, Lugli GA, Milani C, Duranti S, Turroni F, Gueimonde M, Margolles A, van Sinderen D, Ventura M, Barrangou R. 2015. Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS One 10:e0133661. doi: 10.1371/journal.pone.0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Belkum A, Soriaga LB, LaFave MC, Akella S, Veyrieras JB, Barbu EM, Shortridge D, Blanc B, Hannum G, Zambardi G, Miller K, Enright MC, Mugnier N, Brami D, Schicklin S, Felderman M, Schwartz AS, Richardson TH, Peterson TC, Hubby B, Cady KC. 2015. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. mBio 6(6):e01796-15. doi: 10.1128/mBio.01796-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westra ER, Buckling A, Fineran PC. 2014. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol 12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 22.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aklujkar M, Lovley DR. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol Biol 10:230. doi: 10.1186/1471-2148-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 26.Marraffini LA, Sontheimer EJ. 2010. Self vs. non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díez-Villaseñor C, Guzman NM, Almendros C, García-Martínez J, Mojica FJ. 2013. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol 10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV, Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivančić-Baće I, Cass SD, Wearne SJ, Bolt EL. 2015. Different genome stability proteins underpin primed and naive adaptation in E. coli CRISPR-Cas immunity. Nucleic Acids Res 43:10821–10830. doi: 10.1093/nar/gkv1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selle K, Klaenhammer TR, Barrangou R. 2015. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A 112:8076–8081. doi: 10.1073/pnas.1508525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R, Qimron U. 2010. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J Bacteriol 192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. 2011. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One 6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeBardeleben HK, Lysenko ES, Dalia AB, Weiser JN. 2014. Tolerance of a phage element by Streptococcus pneumoniae leads to a fitness defect during colonization. J Bacteriol 196:2670–2680. doi: 10.1128/JB.01556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brüssow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602, table of contents. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cumby N, Davidson AR, Maxwell KL. 2012. The moron comes of age. Bacteriophage 2:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldberg GW, Jiang W, Bikard D, Marraffini LA. 2014. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 39.Jiang W, Samai P, Marraffini LA. 2016. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg GW, Marraffini LA. 2015. Resistance and tolerance to foreign elements by prokaryotic immune systems–curating the genome. Nat Rev Immunol 15:717–724. doi: 10.1038/nri3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 42.Fineran PC, Gerritzen MJ, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. 2014. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A 111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. 2014. Priming in the type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res 42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. 2013. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol 10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS One 7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue C, Seetharam AS, Musharova O, Severinov K, J Boruns SJ, Severin AJ, Sashital DG. 2015. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res 43:10831–10847. doi: 10.1093/nar/gkv1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cady KC, O'Toole GA. 2011. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193:3433–3445. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heussler GE, Cady KC, Koeppen K, Bhuju S, Stanton BA, O'Toole GA. 2015. Clustered regularly interspaced short palindromic repeat-dependent, biofilm-specific death of Pseudomonas aeruginosa mediated by increased expression of phage-related genes. mBio 6(3):e00129-15. doi: 10.1128/mBio.00129-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. 2004. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J Bacteriol 186:6560–6574. doi: 10.1128/JB.186.19.6560-6574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cazares A, Mendoza-Hernández G, Guarneros G. 2014. Core and accessory genome architecture in a group of Pseudomonas aeruginosa Mu-like phages. BMC Genomics 15:1146. doi: 10.1186/1471-2164-15-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terns RM, Terns MP. 2014. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet 30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. [DOI] [PubMed] [Google Scholar]
- 54.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo ML, Mullis AS, Leenay RT, Beisel CL. 2015. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res 43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rath D, Amlinger L, Rath A, Lundgren M. 2015. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 57.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. 2014. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5(1):e00928-13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maniv I, Jiang W, Bikard D, Marraffini LA. 2016. Impact of different target sequences on type III CRISPR-Cas immunity. J Bacteriol 198:941–950. doi: 10.1128/JB.00897-15. [DOI] [PMC free article] [PubMed] [Google Scholar]