Abstract
During a long-term, large network study of the ecology of plant endophytes in native habitats, various nematodes have been found. Two poplar species, Populus angustifolia (narrowleaf cottonwood) and Populus trichocarpa (black cottonwood), are important ecological and genomic models now used in ongoing plant–pathogen–endophyte interaction studies. In this study, two different aphelenchid nematodes within surface-sterilized healthy leaves of these two Populus spp. in northwestern North America were discovered. Nematodes were identified and characterized microscopically and molecularly with 28S ribosomal RNA (rRNA) and 18S rRNA molecular markers. From P. angustifolia, Aphelenchoides saprophilus was inferred to be closest to another population of A. saprophilus among sequenced taxa in the 18S tree. From P. trichocarpa, Laimaphelenchus heidelbergi had a 28S sequence only 1 bp different from that of a Portuguese population, and 1 bp different from the original Australian type population. The 28S and 18S rRNA trees of Aphelenchoides and Laimaphelenchus species indicated L. heidelbergi failed to cluster with three other Laimaphelenchus species, including the type species of the genus. Therefore, we support a conservative definition of the genus Laimaphelenchus, and consider these populations to belong to Aphelenchoides, amended as Aphelenchoides heidelbergi n. comb. This is the first report of these nematode species from within aboveground leaves. The presence of these fungal-feeding nematodes can affect the balance of endophytic fungi, which are important determinants of plant health.
Keywords: nematode ecology, phylogeny, ribosomal DNA, systematics, taxonomy
During a long-term, large network study of the ecology of plant endophytes in native habitats, various nematodes have been found. These include fungal-feeding Paraphelenchus acontioides Taylor and Pillai, 1967 (Carta et al., 2011) and bacterial-feeding Panagrolaimus artyukhovskii Blinova and Mishina, 1975 (Baynes et al., 2012) within cheatgrass stems from Colorado. Two other species of fungal-feeding nematodes were isolated from within dicots, specifically from the leaves of two species of poplar (P. angustifolia and P. trichocarpa) originating in Utah and western Washington, respectively. These plants are important ecological and genomic models, respectively, used in ongoing plant–pathogen–endophyte interaction studies (Busby et al., 2013). Here, we characterize these two fungal-feeding nematodes microscopically and with two DNA markers for identification and phylogenetic analysis. Their potential ecological impact is also discussed.
Materials and Methods
Microscopy:
Nematodes from leaves surface-sterilized with 70% ethyl alcohol were imaged at ×40-60 on an Olympus BX51 microscope equipped with polarization optics and with a DP71 camera (Olympus America Inc., Center Valley, PA). Measurements in micrometers were taken with an ocular micrometer on a Zeiss Ultraphot II compound microscope (Carl Zeiss, Inc., Jena, Germany)with Nomarski optics before formalin fixation, and images were also measured with the calibrated measuring tool in the imaging program CellSens version 1.6 (Olympus America Inc.). Fixed specimens were processed for permanent slides according to the formalin–glycerine method (Golden, 1990). Measurements in micrometers and morphometrics were calculated on an Excel spreadsheet.
DNA analysis:
Specimens were mechanically disrupted in 20 µl of extraction buffer (Thomas et al., 1997), then stored in polymerase chain reaction (PCR) tubes at −80°C until needed. Extracts were prepared from thawed pools by incubating the tubes at 60°C for 60 min, followed by 95°C for 15 min to deactivate proteinase K. Two microliter of the extract was used for each 25-µl PCR reaction.
The ribosomal 28S D2-D3 expansion segment was amplified with primers D2A 5′-ACAAGTACCGTGAGGGAAAGTTG-3′ and D3B 5′TCGGAAGGAACCAGCTACTA-3′ (Nunn, 1992) using a previously published amplification procedure (Ye et al., 2007). The sequence is listed in Table 1 with other 28S sequences from GenBank.
Table 1.
Species, strains, localities, 28S ribosomal DNA sequence lengths, and GenBank accession numbers for sequences included in phylogenetic analysis.
For 18S sequence reaction components, per 25 µl reaction: 17.55 µl H2O, 2.5 µl 10× PCR buffer, 0.5 µl dNTP mix (10 mM each dNTP), 0.75 µl MgCl2, 50 mM, 0.75 µl 18S-G18S4 primer, 10 µM, 0.75 µl 18S-18P primer, 10 µM, 0.2 µl Taq (1 unit Invitrogen platinum, Carlsbad, CA), 23 µl of the above mix + 2 µl template DNA. Cycling conditions were 94°C for 2 min; 94°C for 30 sec, 50°C for 30 sec, 68°C for 2 min, repeating 40 times; 68°C for 10 min; hold at 4°C. Primers of Thomas et al., 1997 were used for PCR and sequencing.
28S rRNA gene PCR products were visualized with UV illumination after ethidium bromide staining. DNA was excised from the gels and purified with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Clean PCR products were directly sequenced by a local vendor (Genewiz, Inc., Germantown, MD and South Plainfield, NJ). PCR products were visualized and purified within the Lonza FlashGel™ DNA system (VWR International, Radnor, PA), and sequences were generated with an ABI BigDye Terminator v3.1 kit with sample sequence data analyzed on an ABI 3130XL Automated DNA sequencer (Applied Biosystems, Foster City, CA). The 28S sequence was determined on both strands using D2A and D3B primers.
The DNA sequences for the following genes were deposited in GenBank: A. cf. saprophilus 18S rDNA (KT884899), A. heidelbergi (n. comb.) (= original Laimaphelenchus) 28S rDNA (KT884898), and Aphelenchoides cf. parietinus 18S rDNA (KU525689).
Phylogenetic analysis:
DNA sequences were analyzed using BLASTN of nematode sequences contained in the EBI-EMBL parasite sequence database (http://www.ebi.ac.uk/blast2/parasites.html). Nematode sequences with highest e-values for BLAST similarity and morphologically close nematode species that had GenBank sequences were aligned in ClustalW (Thompson et al., 1994) using default parameters, as implemented in Geneious version 7.1.7 (BioMatters, Auckland, New Zealand). Markov chain was run for 1,100,000 generations, with the first 100,000 trees discarded as burn-in. The Bayesian likelihood tree was inferred using the MrBayes plugin (Huelsenbeck and Ronquist, 2001) as also implemented in Geneious. GenBank sequences (Table 1 for 18S rDNA, Table 2 for 28S rDNA) were aligned with those determined in this study.
Table 2.
18S rDNA sequence numbers, lengths, and localities from GenBank for Aphelenchoides and Laimaphelenchus sequences included in phylogenetic analysis.
Results
Systematics
Aphelenchoides cf. saprophilus Franklin, 1957
(Fig. 1)
Fig. 1.
Aphelenchoides cf. saprophilus male. A. Body, lateral view. B. Pharynx with stylet (S), median bulb, and pharyngeal gland (G). C. Tail with spicule. D. Tail tip with mucro, endophytic from within healthy, surface-sterilized narrow leaf cottonwood leaves, Populus angustifolia.
Voucher description
Female (n = 4):
L = 521 ± 34 (495–567) µm, V% = 70 ± 1 (69–72), a = 26.6 ± 3.1 (22.1–28.9), b = 10.0 ± 1.0 (8.9–10.9), c = 16.5 ± 2.7 (12.6–18.3), c′ = 3.2 ± 0.2 (2.8–3.3) µm, stylet = 10.8 1.2 (9.3–12.1) µm, body width (BW) = 20 ± 2 (18–22), tail = 32.8 ± 6.2 (28.8–42.0) µm, postvulval sac (PVS) = 57 ± 0 (56–57) µm, PVS/vulval anal distance (VAD) % = 46 ± 4 (41–51).
Male (n = 4):
L = 534 ± 24 (509–567) µm, a = 30.0 ± 3.44 (26.1–34.3), b = 9.9 ± 0.57 (9.3–10.5), c = 15.8 ± 1.74 (14.5–18.3), c′ = 3.02 ± 0.37 (2.52–3.40), stylet = 10.3 µm, tail = 34.0 ± 4.0 (28.8–38.4) µm, spicule = 21.8 ± 1.6 (20.0–23.0).
Locality and host:
Ogden, UT, along Weber River, P. angustifolia (narrowleaf cottonwood), cultured on Trichoderma sp. endophytic from within P. angustifolia.
Specimen designation and deposition:
Seven slides (G-21529 to G-21535) with 10 females and 20 males were deposited in the U.S. Department of Agriculture Nematode Collection (USDANC).
Differential diagnosis:
The stylet demonstrates a wider range than the type population of A. saprophilus (9.3–12.1 vs. 11 µm for original description where n = 4), and two of the four lateral incisures are very faint, but this diagnosis fits better than Aphelenchoides pinusi Bajaj and Walia, 2000, a species with two lateral incisures and a larger spicule size (>25 µm) than A. saprophilus.
Aphelenchoides heidelbergi (Zhao, Davies, Riley, and Nobbs, 2007) n. comb. (syn. Laimaphelenchus heidelbergi Zhao, Davies, Riley, and Nobbs, 2007)
(Fig. 2)
Fig. 2.
Laimaphelenchus heidelbergi/Aphelenchoides heidelbergi n. comb. female from within healthy, surface-sterilized black cottonwood leaves, Populus trichocarpa. A. Body, lateral view with vulva (arrow). B. Vulva, ventral view (arrow). C. Offset tail tip with single tubercle.
Voucher description
Female (n = 3):
L = 397 ± 68 (349–445) µm, a = 38.1 ± 1.57 (37.0–39.2), b = 7.0 ± 0.08 (6.9–7.0), b′ = 4.42 ± 0.29 (4.22–4.62), c = 19.0 ± 5.26 (15.3–22.7), c′ = 3.55 ± 0.71 (3.04–4.05), V = 0.67 ± 0.01 (0.67–0.68), stylet = 8.7 ± 0.6 (8.2–9.1) µm, tail = 22.2 ± 9.7 (15.3–29.1) µm, PVS = 35 ± 7 (30–40) µm, PVS/BW = 3.3 ± 0.3 (3.1–3.5), PVS/VAD% = 36 ± 4 (33–42).
Male (n = 3):
L = 445 ± 213 (264–679) µm, w = 11 ± 3 (8–14) µm, e = 55 ± 12 (41–61) µm, testis = 266 ± 173 (133–462) µm, t = 30.8 ± 4.6 (25.6–34.4) µm, a = 40.8 ± 14.33 (24.4–50.9), b = 8.3 ± 3.55 (4.3–11.1), b′ = 4.4 ± 1.7 (2.4–5.7), c =14.4 ± 5.9 (8.1–9.7), c′ = 3.5 ± 1.5 (2.5–4.5), stylet = 10.9 ± 0.6 (10.2–11.2) µm, spicule = 14.9 ± 1.3 (14.0–15.8) µm.
Locality and host:
An alluvial floodplain of the Nisqually River west of the crest of the Cascade Mountains in western Washington (latitude and longitude: 46.84808 N, −122.31585 W), P. trichocarpa.
Specimen designation and deposition:
Four slides (G-21546 to G-21549) with five females and four males (one juvenile) were deposited in the USDANC.
Differential diagnosis:
Both the stylet and “b” ratio are slightly smaller in this population than in Australian (Zhao et al., 2007) or Portuguese (Maleita et al., 2014) populations of L. heidelbergi.
Molecular alignment and phylogeny:
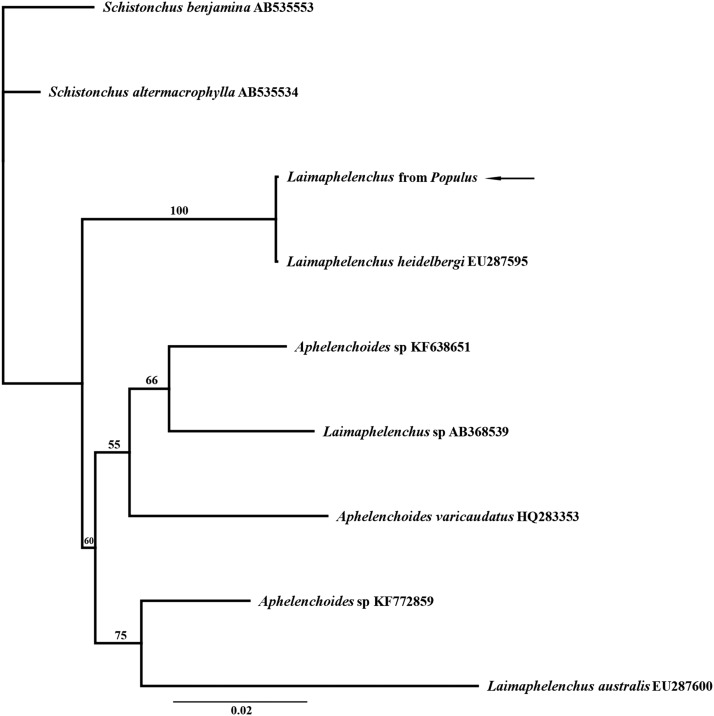
28S rDNA: The sequence from this population was 99.7% identical to that of Australian L. heidelbergi Zhao, Davies, Riley, and Nobbs, 2007 from pine (EU287585; Zhao et al., 2008), with G-A transitions at 106 and 305/688 alignment pair positions. The sequence differed from that of a Portuguese cork bark population (KJ564293) only at the first position (99.8% similar). In the tree (Fig. 3), L. heidelbergi did not share a clade with Laimaphelenchus sp. and L. australis Zhao, Davies, Riley and Nobbs, 2006, a species having the tail and vulval features of the Laimaphelenchus type species Laimaphelenchus penardi Steiner, 1914.
Fig. 3.
Bayesian phylogenetic tree of Aphelenchoides and Laimaphelenchus species inferred from an 806-bp fragment of 28S ribosomal DNA (rDNA). Sequences were aligned in ClustalW. In analysis, Markov chain was run for 1,100,000 generations, with first 100,000 trees discarded as burn-in. LnL mean/run = −4184.6. Node support is given above branches to left of nodes. Sequence generated in this work is indicated by an arrow.
18S rDNA:
In the tree (Fig. 4), L. heidelbergi did not share a clade with three other Laimaphelenchus species, including the type species of the genus. Instead, it was inferred to be much closer to A. cf. parietinus Franklin, 1957 (slightly lower a ratio and higher c′ ratio than the type population of A. parietinus) from Idaho Pinus strobus var. monticola taproots with galls of undetermined origin (USDANC G-21546 to G-21549). The relatively short sequence for A. cf. saprophilus had 94.1% similarity (536/570 bp similarity of aligned nucleotides) compared to the 1,660-bp sequence for A. saprophilus isolate 2140 (FJ040408) from the laboratory at Wageningen, The Netherlands (Holterman et al., 2006).
Fig. 4.
Bayesian phylogenetic tree of Aphelenchoides and Laimaphelenchus species inferred from an 1846-bp fragment of 18S rDNA. Sequences were aligned in ClustalW. Sequences were analyzed under a Tamura–Nei model. Markov chain was run for 1,100,000 generations, with first 100,000 trees discarded as burn-in. LnL mean/run = −13413.144, −13413.594. Node support is given above branches to left of nodes. Sequences generated in this work are indicated by arrows.
Discussion
Laimaphelenchus spp. are polyphyletic among Aphelenchoides and Schistonchus in molecular phylogenetic trees of ribosomal and cytochrome oxidase I genes (Kanzaki et al., 2009; Zeng et al., 2010; Wang et al., 2013), consistent with our findings (Figs. 3,4). This isolate of putative L. heidelbergi does not have the four-pronged tail, the sclerotized vagina, or the vulval flap present in L. penardi, the type species of the genus. Therefore, we support the conservative definition of Hirling (1982, 1986) for Laimaphelenchus, and we amend species L. heidelbergi to A. heidelbergi n. comb. sensu Hirling, 1986. The divergent position of L. heidelbergi from other Laimaphelenchus species that are more congruent with the original genus description was also noted in other phylogenetic analyses of Aphelenchida (Oro, 2015). Laimaphelenchus heidelbergi is not closely related to poplar leaf endophyte A. saprophilus (i.e., a nonpathogenic nematode that lives inside surface-sterilized plant leaves), but the latter species is closely related to another potential root endophyte, A. cf. parietinus (Fig. 4). Therefore, the association with leaves does not seem to be limited to close phylogenetic relatives. Possibly all Aphelenchoides are capable of living endophytically, but further sampling and experimentation are necessary to support this hypothesis.
Laimaphelenchus and Aphelenchoides have been found in lichens (Hirling, 1982), moss, and algae from coniferous trees (Hirling, 1986), from twigs and bark (on and under) of trees (Negi and Ye, 2011), and insect frass from trees (Bajaj and Walia, 2000). However, to our knowledge, there have been no reports of L. heidelbergi or A. saprophilus from within leaves independent of soil. Likewise, neither nematode species has been reported to cause damage to plants. The presence of fungal-feeding nematodes undoubtedly affects the balance of endophytic fungi within a leaf community, and incidence of disease can depend as much or more on composition of endophyte species than on plant genotype (Raghavendra and Newcombe, 2013). Furthermore, the ability of an Aphelenchoides species to cause plant disease may depend on the composition of different endophytic fungi within a given host. Although epiphytic organisms were not the focus of this paper, and thus were not sampled, based on our finding here it would not be surprising to find Aphelenchoides living epiphytically as well, especially in humid habitats. Further characterization of nematode abundance, geographical distribution, variability, life history, and culturability of these nematodes is worth pursuing to better understand plant–microbe relationships within the poplar microbiome (Hacquard and Schadt, 2015).
Literature Cited
- Bajaj HK, Walia KK. A new species of Laimaphelenchus Fuchs, 1937 (Nematoda : Aphelenchina) from Kalesar forest, Haryana, India. Indian Journal of Nematology. 2000;30:88–90. [Google Scholar]
- Baynes MA, Russell DM, Newcombe G, Carta LK, Rossman AY, Ismaiel A. A mutualistic interaction between a fungivorous nematode and a fungus within the endophytic communityof Bromus tectorum. Fungal Ecology. 2012;5:610–623. [Google Scholar]
- Busby PE, Zimmerman N, Weston DJ, Jawdy SS, Houbraken J, Newcombe G. Leaf endophytes and Populus genotype affect severity of damage from the necrotrophic leaf pathogen, Drepanopeziza populi. Ecosphere. 2013;4:125. [Google Scholar]
- Carta L, Skantar AM, Handoo ZA, Baynes MA. Supplemental description of Paraphelenchus acontioides (Tylenchida: Paraphelenchidae), with ribosomal DNA trees and a morphometric compendium of female Paraphelenchus. Nematology. 2011;13:887–899. [Google Scholar]
- Franklin MT. Aphelenchoides composticola n. sp. and A. saprophilus n. sp. from mushroom compost and rotting plant tissues. Nematologica. 1957;2:306–313. [Google Scholar]
- Golden AM. 1990. Preparation and mounting nematodes for microscopic observation. Pp. 197–205 in B. M. Zuckerman, W. F. Mai, and L. R. Krusberg, eds. Plant Nematology Laboratory Manual. Amherst, MA: University of Massachusetts Agricultural Experiment Station.
- Hacquard S, Schadt CW. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytologist. 2015;205:1424–1430. doi: 10.1111/nph.13133. [DOI] [PubMed] [Google Scholar]
- Hirling W. A survey of the species of the nematode genera Laimaphelenchus Fuchs, 1937 and Ruidosaphelenchus Laumond and Carle 1971, with keys to species and three new species of Laimaphelenchus (Nematoda: Tylenchida) Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz. 1982;89:30–42. [Google Scholar]
- Hirling W. 1984/1985, publ. 1986. Laimaphelenchus penardi (Nematoda: Tylenchida) from the type locality and contribution on the identity, biology and distribution of this nematode and of three related species, L. montanus, L. sylvaticus and L. praepenardi n. sp. Zoologische Beiträge 29:349–375.
- Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, Bakker J, Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Molecular Biology and Evolution. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kanzaki N, Giblin-Davis RM, Scheffrahn RH, Center BJ, Davies KA. Pseudaphelenchus yukiae n. gen., n. sp. (Tylenchina: Aphelenchoididae) associated with Cylindrotermes macrognathus (Termitidae: Termitinae) in La Selva, Costa Rica. Nematology. 2009;11:869–881. [Google Scholar]
- Maleita C, Costa SR, Abrantes I. First report of Laimaphelenchus heidelbergi (Nematoda: Aphelenchoididae) in Europe. Forest Pathology. 2014;45:76–81. [Google Scholar]
- Negi S, Ye J-R. 2011. Occurrence of nematode species associated with pine wood in Jiangsu, Zhejiang and Gansu provinces in China. Journal of Nanjing Forestry University (Natural Sciences Edition) 35:149–150.
- Nunn G. 1992. Nematode molecular evolution. An investigation of evolutionary patterns among nematodes based upon DNA sequences. Ph.D. dissertation, University of Nottingham, UK.
- Oro V. Description of Laimaphelenchus belgradiensis sp. nov. (Nematoda: Aphelenchoididae) and its phylogenetic and systematic position within Aphelenchoidoidea. European Journal of Plant Pathology. 2015;142:13–23. [Google Scholar]
- Raghavendra AKH, Newcombe G. The contribution of foliar endophytes to quantitative resistance to Melampsora rust. New Phytologist. 2013;197:909–918. doi: 10.1111/nph.12066. [DOI] [PubMed] [Google Scholar]
- Thomas WK, Vida JT, Frisse LM, Mundo M, Baldwin JG. DNA sequences from formalin-fixed nematodes: Integrating molecular and morphological approaches to taxonomy. Journal of Nematology. 1997;29:250–254. [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang P, Gu J-F, Wang J-L, Li H-M. Description of Aphelenchoides xui sp. n. (Nematoda: Aphelenchoididae) in packaging wood from South Africa. Nematology. 2013;15:279–289. [Google Scholar]
- Ye W, Giblin-Davis RM, Davies KA, Purcell MF, Scheffer SJ, Taylor GS, Center TD, Morris K, Thomas WK. Molecular phylogenetics and the evolution of host plant associations in the nematode genus Fergusobia (Tylenchida: Fergusobiinae) Molecular Phylogenetics and Evolution. 2007;45:123–141. doi: 10.1016/j.ympev.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Zeng Y-S, Ye WM, Giblin-Davis RM, Li C-H, Du Z-J, Zhao C. Schistonchus hirtus n. sp. (Nematoda: Aphelenchoididae), an associate of Ficus hirta in China. Nematology. 2010;12:543–556. [Google Scholar]
- Zhao ZQ, Davies KA, Riley IT, Nobbs JM. Laimaphelenchus heidelbergi sp. nov. (Nematoda: Aphelenchina) from Victoria, Australia, and emendment of the diagnosis of the genus. Transactions of the Royal Society of South Australia. Incorporated. 2007;131:182–191. [Google Scholar]