Abstract
MircroRNA-217 (miR-217) has been showed to involve in the initiation and development of human cancers, and is recognize as a tumor suppressor miRNA in several tumors. However, the clinical significance and its underlying role in human glioma remain unclear. Herein, we found that the expression of miR-217 was significantly down-regulated in glioma tissues as compared with adjacent normal brain tissues. Clinical association analysis disclosed that low-expression of miR-217 was evidently negative associated with advanced tumor stage (grade III + IV) in glioma. Further function assays showed that miR-217 inhibited proliferation, colony formation, invasion and migration of glioma cells. Notably, runt-related transcription factors 2 (Runx2) was identified as a functional target of miR-217 in glioma. Furthermore, an inverse correlation between miR-217 and Runx2 expression was observed in glioma tissues. Downregulation of Runx2 has similar with inhibition effect of overexpression of miR-217, and upregulation of Runx2 reversed the effects of overexpressing of miR-217. Taken together, these results suggest a critical role of miR-217 in suppressing proliferation, migration, and invasion of glioma by targeting Runx2.
Keywords: Glioma, miR-217, proliferation, invasion, Runx2
Introduction
Glioma represents the most common tumors in the central nervous system with high morbidity and mortality, and accounts for about 80 % of malignant brain tumors [1]. Despite the advances in therapeutic intervention, such as glioma surgery, radiotherapy, chemotherapy, gene therapy, immunotherapy and other novel biological therapies, the median survival duration of patients with glioblastoma multiforme (GBM) have not been significantly improved because of their invasiveness and the low sensitivity to radio-/chemo- therapeutic agents [2]. Thus, it is quite urgent to understand the molecular mechanisms by which glioma initiates, progresses, invades, and recurs to develop novel and effective therapeutic strategies for glioma.
MicroRNAs (miRNAs) are an 18 to 25 nucleotide-long, single-stranded, noncoding RNA molecules that negatively regulate gene expression at a post-translational level by binding to regions of sequence complementarity to the 3’ untranslated region (3’UTR) of mRNAs, leading to either the degradation or translational inhibition of the mRNA [3]. It was found that miRNAs involve in various cellular processes, including cell proliferation, apoptosis, cell cycle, migration and invasion, and stem cell renewal [4,5]. Accumulating evidence has been showed that miRNAs were aberrantly expressed in human various cancers, including glioma and it played oncogenic or tumor suppressive roles in various human cancers, including in glioma [6-8].
As a member of the miRNAs family, mircroRNA-217 (miR-217) has been showed to involve in the initiation and development of human cancers, and function as tumor suppressor in majority cancers, such as ovarian cancer [9], osteosarcoma [10], esophageal squamous cell carcinoma cells [11], hepatocellular carcinoma [12], pancreatic ductal adenocarcinoma [13], lung cancer [14] and colorectal cancer [15]. However, the clinical significance of miR-217 in glioma and its underlying molecular pathways involved in the initiation and development of glioma have not been investigated. Therefore, the primary aim of this study was to investigate miR-217 clinical diagnosis significance in patients with glioma and its role and underlying molecular mechanism in glioma procession.
Materials and methods
Clinical samples and cell lines
36 samples of glioma tissues were obtained from the Department of Neurosurgery, the first of Hospital of Jilin University during July 2012 to December 2014. Twenty normal brain tissue samples used as controls were obtained from internal decompression of patients who underwent surgical operation for cerebral injury and cerebral hemorrhage in the First of Hospital of Jilin University. All specimens were confirmed by pathological diagnosis and classified. Ten of the 36 gliomas were classified as low-grade (4 WHO I, 6 WHO II), and 26 gliomas were classified as high-grade (12 WHO III, 14 WHO IV) gliomas according to the WHO criteria. The glioma tissues and normal tissues were snap-frozen in liquid nitrogen immediately after resection and stored at -80°C until use. All samples were used after obtaining informed consent. The study was approved by the Medical Ethics Committee of Jilin University (Changchun, China).
Primary normal human astrocytes (NHA) and four human glioma cell lines (U251, U87, U118 and LN18) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and were routinely cultured in complete Dulbecco’s modified Eagle medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) with 100 units/mL penicillin and 100 mg/mL streptomycin (Sigma, St- Louis, MO, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative real-time PCR
Cultured cells and frozen tumor specimens were subject to total RNA extraction using Trizol reagent (Invitrogen, California, USA). The concentration and purity of all RNA were detected by NanoDrop ND-2000 Spectrophotometer (NanoDrop Technologies, Houston, TX, USA). To detect miR-217 expression, complementary DNA(cDNA) were synthesized using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Amplification and detection of miR-217 and U6 were performed using a TaqMan Human MiRNA Assay Kit (Applied Biosystems) under ABI PRISM 7900 Sequence Detection System (Applied Biosystems). The primes of miR-217 and U6 were obtained from Applied Biosystems. For detect Runx2 mRNA expression, cDNA was synthesized using M-MLV reverse transcriptase (Takara, Dalian, China) according to the supplier’s protocol. Quantitative real-time PCR (qRT-PCR) was performed with the 7300 sequence detection system (Applied Biosystems/Roche) using SYBR Green Master Mix (Applied Biosystems). The primes of Runx2 and GAPDH were used in this study as described previously [16]. The mRNA and miRNA expression levels were normalized to those of GAPDH and U6, respectively, using the 2-ΔΔCt method.
Cell transfection
miR-217 mimic (miR-217) and corresponding negative control (miR-NC), the siRNAs targeting human Runx2 (si-Runx2) and corresponding negative control (si-NC) were brought form GenePharma Co., Ltd (Shanghai, China). Runx2 overexpressed plasmids were obtained from Ribobio Co. (Guangzhou, China). These molecular productions were transfected into U87 cells when cells were grown to 80-90% confluence, using the Lipofectamine 2000 (Invitrogen) according to the instructions provided by the manufacturer.
Western blotting
Cells were harvested and lysed in ice-cold RIPA buffer (Beyotime, Jiangsu, China) according to the manufacturer’s instructions, then Cells were lysed using ice-cold RIPA buffer and centrifuged at 14,000 × g 4°C for 5 min. Protein concentrations were determined using the BCA protein assay kit (Beyotime). Prepared protein samples (20 μg each lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Millipore, Wisconsin, USA). The membrane was blocked with 5% skimmed milk in TBST and incubated with the antibody against Runx2 (1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and GAPDH (1:5000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight 4°C. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5000 dilution, Santa Cruz, USA) for 2 h at room temperature. Protein band were visualized using the enhanced chemiluminescence Kit (PerkinElmer) on Bio-Rad ChemiDocXRS (Bio-Rad Laboratories, Hercules, CA, USA).
Cell proliferation and colony formation assay
Cell proliferation was measured by CCK8 assay. In briefly, the 1 × 103 transfected cells were seeded into 96-well plates. At indicated times (24 h, 48 h, 72 h, 96 h), 20 μl of Cell Counting Kit-8 (Beyotime) were added to each well and incubated for 2 hours at 37°C. Optical density (OD) was detected at the wavelength of 450 nm on a Synergy 2 microplate reader (Biotek, USA).
For colony formation assay, the 1,000 transfected cells were seeded in 6-well plates and cultured for 14 days at DMEM medium containing 10% FBS. Then the colonies were washed three times with PBS, and fixed with 75% ethanol for 10 min, dried and stained with 0.1% crystal violet solution (Sigma, USA) for 10 min. The colonies were taken pictures and counted under the light microscope (Olympus, Tokyo, Japan).
Cell migration and invasion
Wound healing assays were performed to detect cell migration. In briefly, the transfected cells were seeded in 6-well plates and cultured overnight, and an artificial wound was created using a 200 μl pipette tube. The wound closure was observed after 24 h and imaged under a light microscope (Olympus, Tokyo, Japan). To assess the migration rate, we measured the fraction of cell coverage across the line.
Cell invasion was evaluated using a transwell assay with Matrigel (8-μm pore; BD Biosciences). In briefly, transfected cells (5 × 104 cell/well) in free-serum DMEM medium were seeded into the upper well of the matrigel-coated invasion chamber. The lower chamber was filled with DMEM medium with 10% FBS as chemoattractant. After cells had been cultured for 48 h at 37°C, non-invading cells were removed from the top well, while cells that had migrated to the lower surface of the membrane were fixed with 70% ethanol for 30 min and stained with 0.1% crystal violet for 10 min. The invaded cells were photographed and counted in five randomly selected fields for each well using light microscope (Olympus).
Luciferase reporter assay
The candidate target genes of miR-217 were predicted using miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) and TargetScan (http://www.Targetscan.org/). The wild-type 3’-UTR segment of the Runx2 mRNA (not the full length of Runx2 3’-UTR) containing miR-217 binding sites was amplified and cloned into the dual-luciferase reporter vector pGL3 (Promega, Madison, WI, USA) termed as: Wt-Runx2-3’UTR. A mutant construct in miR-217 binding sites of Runx2 3’UTR region also was generated by Quick Change Site-Directed Mutagenesis Kit (Agilent, Roseville City, CA), and subcloned into pGL3-control vector (Ambion), and termed as Mut-Runx2-3’UTR. For dual-luciferase reporter assay, U87 cells were transfected with miR-217 or miR-NC for 24 hours and then the cells were transfected with Wt/Mut-Runx2-3’UTR reporter plasmid using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, Luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (Promega, Madison, USA) according to the instructions provided by manufacture. Renilla-luciferase was used for normalization.
Statistical analysis
All data are presented as the mean ± standard deviation (SD) from at least three times independently experiment. The SPSS software package (version 16.0, SPSS Inc; Chicago, IL, USA) was used to perform the statistical analysis. Student’s t-test or ANOVA was employed to determine statistical significance as appropriate. P values less than 0.05 were considered to be statistically significant.
Results
miR-217 expression was downregulated in glioma tissues and cell lines
To investigate the potential biological significance of altered miR-217 expression in glioma progression, we evaluated miR-217 expression in 36 glioma tissues and 20 normal brain (NB) tissues by quantitative RT-PCR (qRT-PCR). As shown in Figure 1A, the expression of miR-217 was significantly decreased in glioma tissue compared with NB tissues. In addition, we found that miR-217 expression in high-grade (III + IV) glioma was significantly lower than those of low-grade (I + II) glioma (Figure 1B), suggesting low-expression of miR-217 was evidently negative associated with advanced tumor stage (grade III + IV) in glioma (Figure 1B). In addition, we also assessed the expression of miR-217 in four human glioma cell lines (U251, U87, U118 and LN18) and normal human astrocytes (NHA) by qRT-PCR. Our results showed that the expression level of miR-217 was decreased in four glioma cell lines compared with NHA (Figure 1C). The U87 cell line has lowest expression of miR-217, thus, it was selected for next study.
Figure 1.
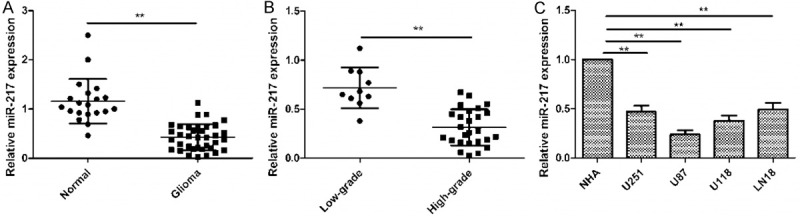
miR-217 expression is down-regulated in CRC samples and cell lines. A. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of miR-217 expression in glioma tissues and normal brain tissues. **P<0.01 versus Normal brain tissue. B. qRT-PCR analysis of miR-217 expression in low-grade glioma and high-grade glioma. Low-grade glioma refers to World Health Organization (WHO) grade I and WHO grade II, high-grade glioma refers to WHO grade III and WHO grade IV. **P<0.01 versus Low-grade. C. qRT-PCR analysis of miR-217 expression in four human glioma cell lines (U251, U87, U118 and LN18) and normal human astrocytes (NHA). *P<0.05, P<0.01 versus NHA.
miR-217 inhibits cell proliferation, colony formation, migration and invasion in human glioma cells
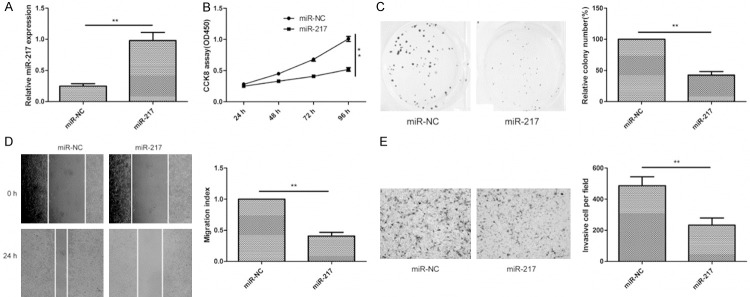
To examine the role of miR-217 in the growth and metastasis of glioma cells, U87 cells were transfected with a miR-217 mimic or miR-NC, then cell proliferation, colony formation, migration and invasion were determined at indicated time. Increased expression of miR-217 in U87 cells transfected with the miR-217 mimic was confirmed using qRT-PCR (Figure 2A). CCK8 assay showed that overexpression of miR-217 significantly decreased the proliferation of cells, compared with negative control cells (Figure 2B). Consistent with this result, overexpression of miR-217 significantly inhibited colony formation in glioma cells (Figure 2C). In addition, wound healing and transwell invasion assays also showed that overexpression of miR-217 significantly inhibited migration (Figure 2D) and invasion (Figure 2E) of glioma cells. These results indicate that miR-217 exerts tumor suppressor role in glioma cells.
Figure 2.
miR-217 inhibits cell proliferation, colony formation, migration and invasion in human glioma cells. A. The expression level of miR-217 were detected in miR-217 mimics transfected cells by qRT-PCR. B-E. cell proliferation, colony formation, migration and invasion were determined in U87 cells transfected with miR-217 mimics or miR-NC. *P<0.05, P<0.01 versus miR-NC.
Runx2 is a target of miR-217 in glioma cells
Bioinformatics soft (miRTarBase and TargetScan) was used to identify the target of miR-217 in glioma cells. It was found that Runx2 3’UTRs has a binding sequences for miR-217 at position (3386-3393) (Figure 3A). To verify Runx2 as a direct target of miR-217, luciferase activity assay was performed. Here, we found that miR-217 significantly inhibited the luciferase activity of 3’-UTR of Runx2 in U87 cells (Figure 3B). Moreover, overexpression of miR-217 significantly suppressed levels of both Runx2 mRNA and protein these levels (Figure 3C and 3D). These data suggested that Runx2 is a direct target of miR-217.
Figure 3.
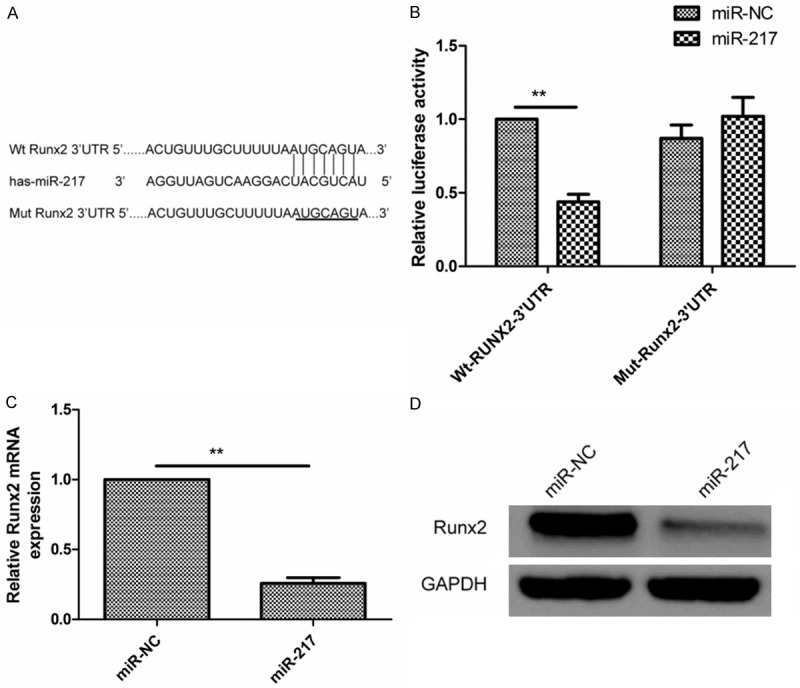
Runx2 is a target of miR-217 in glioma cells. A. Runx2 have binding sequences for miR-217 at position (3386-3393). B. The luciferase activity of U87 cells was determined after co-transfection with the miR-217 mimics or miR-NC and wide-type or mutant-type report plasmid. C. qRT-PCR was performed to detect the expression of Runx2 in U87 cells transfected with miR-217 mimics or miR-NC. The GAPDH was used as an internal control. D. Western blotting analyzed the protein level of Runx2 in U87 cells transfected with miR-217 mimics or miR-NC. The GAPDH was used as an internal control. *P<0.05, P<0.01 versus miR-NC.
Runx2 expression was upregulated and inversely correlated with miR-217 expression in glioma tissues
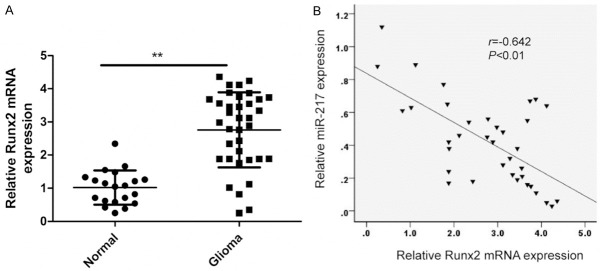
Further experiments were performed to investigate the expression of Runx2 in glioma tissues and normal brain tissues. The result of qRT-PCR showed that the mRNA expression of Runx2 in glioma tissues was upregulated compared with normal brain tissues. In addition, the relationship between miR-217 and Runx2 expression in patients with brain glioma was also investigated. Spearman’s correlation analysis showed a reversed correlation between miR-217 expression levels and Runx2 mRNA levels in glioma tissues (r = -0.642, P<0.01) (Figure 4).
Figure 4.
Runx2 expression was upregulated and inversely correlated with miR-217 expression in glioma tissues. A. Runx2 mRNA expression were determined in glioma tissues and normal brain tissues by qRT-PCR. The GAPDH was used as an internal control. **P<0.01 versus Normal brain tissue. B. Reverse relationship between Runx2 and miR-217 expression was explored by Spearman’s correlation in glioma tissues.
Downregulation of Runx2 exhibited similar effect with miR-217 overexpression in glioma cells
To investigate the biological functions of Runx2 in glioma cells, endogenous expression of Runx2 was knockdown in U87 cells with specific siRNA against Runx2 (si-Runx2). We found that the expression Runx2 on mRNA level (Figure 5A) and protein level (Figure 5B) was significantly inhibited in U87 cells by si-XIAP. Downregulation of Runx2 in U87 cells significantly inhibited cell proliferation (Figure 5C), colony formation (Figure 5D), migration (Figure 5E), and invasion (Figure 5F). These results suggested that the inhibition of Runx2 had similar effect with miR-217 overexpression in glioma cells.
Figure 5.
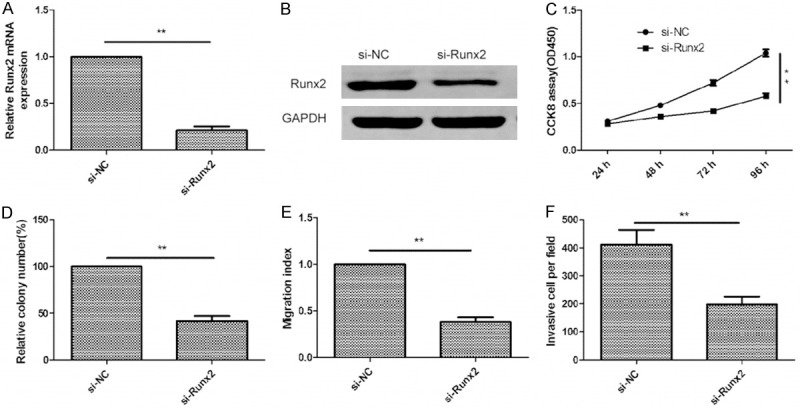
Downregulation of Runx2 exhibited similar effect with miR-217 overexpression in glioma cells. A and B. Runx22 expression on mRNA level and protein level were measured in U87 cells transfected with si-Runx2 or si-NC by qRT-PCR and western blot, respectively. GAPDH was used as an internal control. C-F. Cell proliferation, colony formation, migration and invasion were determined in U87 cells after transfected with si-Runx2 or si-NC. *P<0.05, **P<0.01 versus si-NC.
Overexpression of Runx2 rescues the effects of miR-217 in glioma cells
To investigate the functional relevance of Runx2 targeting by miR-217, we assessed whether Runx2 overexpression could rescue the inhibitory effects of miR-217 on glioma cell proliferation, colony formation, migration and invasion. U87 cells were co-transfected with miR-217 or miR-NC and overexpression Runx2 plasmid. qRT-PCR and Western blot analysis was used to validate the Runx2 expression in the rescue experiment (Figure 6A and 6B). In addition, our results also showed that the forced expression of Runx2 rescued the inhibition effect of miR-217 on cell proliferation, colony formation, migration and invasion (Figure 6C-F). These data indicated that miR-217 exerts inhibition effect on glioma growth and metastasis partially by repressing Runx2.
Figure 6.
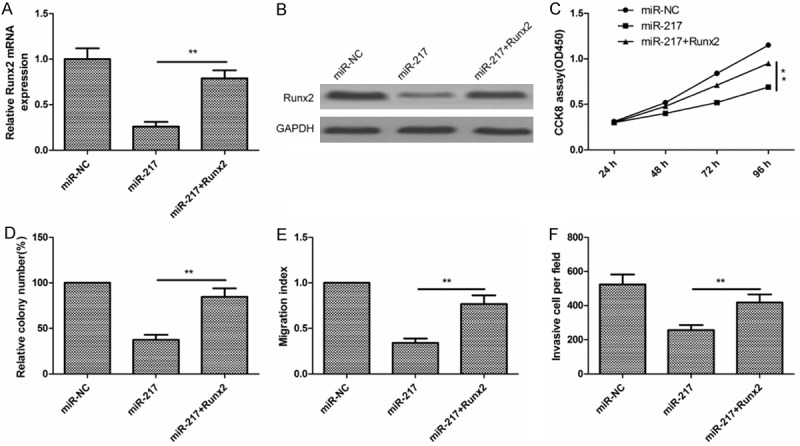
Overexpression of Runx2 rescues the effects of miR-217 in glioma cells. A and B. Runx2 expression on mRNA level and protein level in U87 cells co-transfected with Runx2 overexpression plasmid and miR-217 mimic or miR-NC by qRT-PCR and western blot, respectively. GAPDH was used as an internal control. C-F. Cell proliferation, colony formation, migration and invasion were determined in U87 cells transfected with miR-217 mimic with/without Runx2 overexpression plasmid. *P<0.05, **P<0.01 versus miR-217.
Discussion
Accumulating evidence showed that miRNAs involved in tumorigenesis and metastasis of various human cancers, including glioma [6-8]. Here, we found that miR-217 was down-regulated in glioma tissues and cell lines, compared with that in normal brain tissues and primary normal human astrocytes (NHA). Moreover, we demonstrated that restoration of miR-217 inhibited proliferation, colony formation, migration and invasion of glioma cancer cells. These results suggested that miR-217 might play a key role in glioma procession and likely would be applied as novel therapeutic agents.
Aberrant expression of miR-217 has been found in various human cancers. For ovarian cancer [9], osteosarcoma [10], esophageal squamous cell carcinoma cells [11], hepatocellular carcinoma [12], pancreatic ductal adenocarcinoma [13], lung cancer [14] and colorectal cancer [15], miR-217 expression is frequent downregulated and act as a tumor suppressor, while it overexpressed and function as an oncogene in breast cancer [17], B-cell lymphomas [18]. However, the function and relevant mechanisms of miR-217 in glioma have not been identified. In this study, we found that the expression of miR-217 was significantly down-regulated in glioma tissues and cell lines, and its expression was evidently negative associated with advanced tumor stage (grade III + IV) in glioma. We also demonstrated that restoration of miR-217 in glioma cells inhibited proliferation, colony formation, invasion and migration. These results suggested that miR-217 functioned as a tumor suppressor in glioma.
To investigate the molecular mechanism of the tumor suppressor role of miR-217 in glioma, we used two bioinformatics soft ( miRTarBase and TargetScan) to identify the target of miR-217 in glioma cells. Runx2 3’UTRs were found to have a binding sequences for miR-217 at position (3386-3393). luciferase activity assay, qRT-PCR and western blot assay further confirmed that RUNX2 is a target gene of miR-217. Runx2 is an important member of runt-related transcription factor (RUNX) family, which form the core binding factor (CBF) complex and bind DNA to either activate or repress gene transcription [19]. Growing evidence has been demonstrated that Runx2 expression was upregulated in various cancer, including gloima [20]. Runx2 over-expression was closely related to the poor prognosis of patients [21], and could induce epithelial-mesenchymal transition [22], promote cancer cell motility and invasion in vitro [23], tumor growth in vivo [24], while silencing or inhibiting RUXN2 activity was able to suppress tumor growth and metastasis [25,26], suggesting that Runx2 function as oncegene in various cancers. In the present study, we found that Runx2 expression was upregulated and inversely correlated with miR-217 expression in glioma tissues. In addition, we also found that downregulation of Runx2 had similar effect with miR-217 overexpression in glioma cells, and overexpression of Runx2 rescued the effects of miR-217 in glioma cells. These results suggested that miR-217 exerted tumor suppressor role in glioma, at least in part, by repressing Runx2 expression.
In summary, the present study first demonstrated that miR-217 expression are down-regulated in glioma cell lines and tissues, its expression was negative associated with advanced tumor stage (grade III + IV), and that miR-217 inhibited proliferation, colony formation, migration and invasion of glioma cells by repressing Runx2. Therefore, targeting to the miR-217/Runx2 axis may be a new therapeutic application to treat patients with glioma in the future.
Disclosure of conflict of interest
None.
References
- 1.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 5.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–2315. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tivnan A, McDonald KL. Current progress for the use of miRNAs in glioblastoma treatment. Mol Neurobiol. 2013;48:757–768. doi: 10.1007/s12035-013-8464-0. [DOI] [PubMed] [Google Scholar]
- 8.Karsy M, Arslan E, Moy F. Current Progress on Understanding MicroRNAs in Glioblastoma Multiforme. Genes Cancer. 2012;3:3–15. doi: 10.1177/1947601912448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Li D, Zhang W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol Rep. 2016;35:1671–9. doi: 10.3892/or.2015.4498. [DOI] [PubMed] [Google Scholar]
- 10.Wei R, Deng Z, Su J. miR-217 targeting Wnt5a in osteosarcoma functions as a potential tumor suppressor. Biomed Pharmacother. 2015;72:158–164. doi: 10.1016/j.biopha.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Su J, Wang Q, Liu Y, Zhong M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol Cell Biochem. 2014;392:289–296. doi: 10.1007/s11010-014-2039-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 13.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Shen ZL, Jiang KW, Zhao G, Wang CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, Ye YJ, Wang S. MicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanism. BMC Cancer. 2015;15:437. doi: 10.1186/s12885-015-1438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D, Zheng X, Shan W, Shan Y. The overexpression of miR-30a affects cell proliferation of chondrosarcoma via targeting Runx2. Tumour Biol. 2015 doi: 10.1007/s13277-015-4454-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Yuan Y, Cui J, Xiao T, Jiang D. MiR-217 Promotes Tumor Proliferation in Breast Cancer via Targeting DACH1. J Cancer. 2015;6:184–191. doi: 10.7150/jca.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Yebenes VG, Bartolome-Izquierdo N, Nogales-Cadenas R, Perez-Duran P, Mur SM, Martinez N, Di Lisio L, Robbiani DF, Pascual-Montano A, Canamero M, Piris MA, Ramiro AR. miR-217 is an oncogene that enhances the germinal center reaction. Blood. 2014;124:229–239. doi: 10.1182/blood-2013-12-543611. [DOI] [PubMed] [Google Scholar]
- 18.Slattery ML, Lundgreen A, Herrick JS, Caan BJ, Potter JD, Wolff RK. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and riskof colon and rectal cancer: additional support for a TGF-beta-signaling pathway. Carcinogenesis. 2011;32:318–326. doi: 10.1093/carcin/bgq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vladimirova V, Waha A, Luckerath K, Pesheva P, Probstmeier R. Runx2 is expressed in human glioma cells and mediates the expression of galectin-3. J Neurosci Res. 2008;86:2450–2461. doi: 10.1002/jnr.21686. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Xu S, Lin S, Zhao W. Overexpression of runt-related transcription factor-2 is associated with advanced tumor progression and poor prognosis in epithelial ovarian cancer. J Biomed Biotechnol. 2012;2012:456534. doi: 10.1155/2012/456534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu DF, Kondo T, Nakazawa T, Oishi N, Kawasaki T, Mochizuki K, Yamane T, Katoh R. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Lab Invest. 2012;92:1181–1190. doi: 10.1038/labinvest.2012.84. [DOI] [PubMed] [Google Scholar]
- 22.Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS, Nathan SS, Dobson JR, Lian JB, Ito Y, Voorhoeve PM, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 2010;12:R89. doi: 10.1186/bcr2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra S, Vaughn AD, Devore DI, Roth CM. Delivery of siRNA silencing Runx2 using a multifunctional polymer-lipid nanoparticle inhibits osteogenesis in a cell culture model of heterotopic ossification. Integr Biol (Camb) 2012;4:1498–1507. doi: 10.1039/c2ib20200j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Lu H, Wang S, Chen B, Liu Z, Ke X, Liu T, Fu J. The anthraquinone derivative Emodin inhibits angiogenesis and metastasis through downregulating Runx2 activity in breast cancer. Int J Oncol. 2015;46:1619–1628. doi: 10.3892/ijo.2015.2888. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Chen B, Zou R, Tu X, Tan S, Lu H, Liu Z, Fu J. Codonolactone, a sesquiterpene lactone isolated from Chloranthus henryi Hemsl, inhibits breast cancer cell invasion, migration and metastasis by downregulating the transcriptional activity of Runx2. Int J Oncol. 2014;45:1891–1900. doi: 10.3892/ijo.2014.2643. [DOI] [PubMed] [Google Scholar]