Abstract
Background: The expression pattern and regulatory effect of microRNA-375 (miR-375) in human pancreatic cancer was explored. Methods: Gene expression of miR-375 was compared between pancreatic tumors and non-tumorous pancreatic tissues, as well as pancreatic cancer cell lines and normal epithelial cells. MiR-375 was downregulated in pancreatic cancer cell lines, Capan-1 and PANC-1 cells, to assess possible tumor suppressive effects on cancer proliferation, migration, cisplatin chemosensitivity and in vivo growth of tumor explant. The regulation of miR-375 on its target gene, homeobox B3 (HOXB3) gene, was assessed though luciferase activity assay and qRT-PCR. HOXB3 was also downregulated in Capan-1 and PANC-1 cells to assess its functional correlation with miR-375 on cancer regulation. Results: MiR-375 was upregulated in pancreatic tumors and pancreatic cancer cell lines. MiR-375 downregulation had tumor suppressive effects in Capan-1 and PANC-1 cells by reducing cancer proliferation & migration, increasing cisplatin sensitivity and inhibiting in vivo tumor explant growth. HOXB3 was directly bound by miR-375, and was negatively regulated by miR-375 in pancreatic cancer cells. Subsequent HOXB3 downregulation reversed the suppression of miR-375 downregulation on cancer proliferation, migration and cisplatin chemosensitivity in pancreatic cancer. Conclusion: MiR-375 is an oncogene in pancreatic cancer. Deregulation of miR-375 is inhibitory to the development of pancreatic cancer, and reversely regulated by HOXB3.
Keywords: Pancreatic cancer, miR-375, HOXB3, cancer proliferation, cancer migration, cisplatin
Introduction
Pancreatic ductal adenocarcinoma, or pancreatic cancer is one of the most malignant tumors for both men and women worldwide [1,2]. Due to the heterogeneous nature of carcinogenesis and pathology, early diagnosis and prevention of pancreatic cancer are very difficult [3,4]. For patients diagnosed with pancreatic cancer, postoperative tumor reoccurrence is often the case and 5-year survival rate is below 10% [1,3]. During past decades, though many signaling pathways have been identified to play important roles in pancreatic cancer development and progression, little progress has made on the clinical side to identify efficient biomarkers and targeted genes for pancreatic cancer diagnosis and therapy [4,5].
MicroRNAs (miRNAs) are groups of small noncoding RNAs that bind the 3’-untranslated regions (3’-UTRs) of targeted genes to suppress gene and protein production, thus exerting various regulatory functions on tissue and organ development in both animal and humans [6,7]. In pancreatic cancer, miRNAs have been found to be aberrantly expressed in tumor tissues, and subsequently play critical roles in carcinogenesis, cancer development and metastasis through multiple signaling pathways [8-10]. Among many of the tumor-associated miRNAs, microRNA-375 (miR-375) has been mostly identified as a tumor suppressor. In cervical cancer, hepatocellular carcinoma, liver cancer or gastric cancer, miR-375 was found to be downregulated in human tumors or tumor cell lines, and upregulation of miR-375 exerted inhibitory effects on tumor development, progression, metastasis and apoptosis [11-14]. Interestingly, a recent microarray study showed an opposite expression pattern of miR-375 in pancreatic cancer, as miR-375 was upregulated in human pancreatic tumors than in normal pancreatic tissues [15]. This suggests that miR-375 may have distinctly different functional roles in pancreatic cancer than in other cancers.
In this study, we decided to follow up on miR-375 to elucidate its expression and functions in pancreatic cancer. We firstly used quantitative method, qRT-PCR, to assess the expression level of miR-375 in both in vivo pancreatic tumors and in vitro pancreatic cancer cell lines. We then downregulated endogenous miR-375 in pancreatic cancer cell lines to investigate its effect on cancer proliferation, migration, cisplatin chemosensitivity and in vivo growth of tumor explant. We then took further steps to explore the downstream target gene of miR-375 in pancreatic cancer, using the methods including luciferase activity assay and siRNA-mediated gene downregulation. The results of this study would undoubtedly further our understandings on the molecular mechanisms of microRNA regulation pancreatic cancer.
Materials and methods
Pancreatic tumor samples and cell lines
Paired pancreatic tumor samples (T), and their adjacent non-tumorous pancreatic epithelial tissues (ANT) were extracted from 22 patients at Changzheng and Changhai Hospitals between Jun 2014 to December 2015. All patients signed consent forms. All procedures were approved by the Ethic Committees at Changzheng and Changhai Hospitals in Shanghai, China, and conducted in accordance with the regulations of Declaration of Helsinki. All clinical samples, upon retrieval, were immediately snap-frozen in liquid nitrogen and started at -80°C until further processed.
Ten pancreatic cancer cell lines were used in this study. HS-766T, Capan-1, PANC-1, Mia PaCa-2 were obtained from ATCC (American Type Culture Collection, USA). HPAC, AsPC-1 BxPC-3, Capan-2, CFPAC-1 and SW1990 were obtained from Cell bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). A control human pancreatic duct epithelial-like cell line, hTERT-HPNE was obtained from ATCC. All cells were cultured in RPMI-1640 medium (ThermoFisher Scientific, USA) supplemented with 10% fetal calf serum (FCS, Sigma Aldrich, USA), and maintained in an atmosphere of 95/5% O2/CO2 at 37°C.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA of pancreatic tumors samples or cell lines was extracted using a mirVana miRNA Isolation Kit (Ambion, USA) according to the manufacture’s recommendation. RNA quality was determined by A Nano-Drop ND-3000 UV-spectrophotometer (ThermoFisher Scientific, USA) according to manufacturer’s recommendation. Quantitative real-time PCR (qRT-PCR) was performed on an ABI Prism 7300 Sequence Detector System (Applied Biosystems, USA) to determine the gene expression levels of hsa-miR-375 and HOXB3. For hsa-miR-375, a Taqman MicroRNA Assays (Applied Biosystems, USA) was carried out using U6 snRNA as loading standard. For HOXB3, a miScript SYBR Green PCR Kit (Qiagen, USA) was carried out using 18s as loading standard. Fold changes of gene expression was determined by 2-∆∆Ct method, where Ct stands for the cycle number and ΔCt was calculated by subtracting the Ct of loading standards (U6 or 18s) from the Ct of genes of interest (hsa-miR-375 or HOXB3).
MiR-375 downregulation assay
Endogenous miR-375 expressions were downregulated in two pancreatic cell lines, Capan-1 and PANC-1 cells through lentiviral transduction. Lentiviruses containing the synthetic oligonucleotides of hsa-miR-375 inhibitor (L-miR375-In), and non-specific control miRNA (L-miR-C) were purchased from RiboBio (RiboBio Biotech, China). Capan-1 and PANC-1 cells were transduced with L-miR375-In or L-miR-C, and 8 μg/mL polybrene, at multiplicity of infection (MOI) of 15~20 for 12 h. After that, cells were cultured with freshly prepared medium without lentiviruses for another 72 h. Transduction efficiency was then checked by qRT-PCR.
Proliferation assay
Capan-1 and PANC-1 cells were seeded in 96-well plate (5 × 103 cells/well). Cancer proliferation was determined by a Vybrant® Cell MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazoliumbromide) Proliferation Assay (ThermoFisher Scientific, USA) at 1, 2, 3, 4 and 5 days after seeding. The absorbance was measured by a spectrophotometer reader (Bio-Tek, USA) at 490 nm.
Migration assay
In a transwell migration assay (ThermoFisher Scientific, USA), Capan-1 and PANC-1 cells were seeded in matrigel pre-coated inserts (5 × 104 cells/insert) in RPMI-1640 medium with 2% FCS. The lower chambers were filled with RPMI-1640 medium with 10% FCS as chemoattractant. 24 h after seeding, the inserts were removed. Cells invaded into lower chambers were fixed by methanol, stained with crystal violet, and visualized under a Zeiss inverted light microscope (LSM700, Zeiss, Germany). In each well, cells within five random fields were counted. The average number of migrated cells under each experimental condition was then calculated, and normalized to the averaged numbers under control conditions to assess relative migration capability.
Cisplatin assay
Capan-1 and PANC-1 cells were seeded in 96-well plate (5 × 103 cells/well). When cells reached ~80% confluence, cisplatin (0.01~100 μM) was added into wells for 24 h, followed by MTT assay. For each experimental condition, relative percentage of viable cells was determined by normalizing the MTT assay absorbances to the absorbance of 0.01 μM cisplatin treatment under control condition.
In vivo tumor explant assay
With lentiviral transfection, Capan-1 cells were subcutaneously inoculated into left flanks of athymic female nude mice (1 × 106 cells/mouse). The weekly growth-curve of the size of in vivo tumor explants was plotted as by measuring the lengths (L, mm) and widths (W, mm) of explants and using the equation, L*W*W/2 (mm3). Five weeks after the in vivo assay, mice were sacrificed and tumor explants were retrieved for inspection.
Luciferase activity assay
Human HOXB3 3’-UTR, including the complimentary binding site of hsa-miR-375, was inserted downstream of the firefly cassette of pmirGLO dual-luciferase miRNA target expression vector (Promega, USA) to make HOXB3-3UTR luciferase plasmid. The binding site on HOXB3 3’-UTR was then mutated by a site-directed gene mutagenesis kit (Beyotime, China). The mutated HOXB3 3’-UTR was also cloned into pmirGLO firefly cassette to make HOXB3-3UTR (Mu) luciferase plasmid. Synthetic oligonucleotides of miR-375 mimics (miR375-mimics) and scramble-miRNA (miR-S) were purchased from SunBio (SunBio Tech, Guangzhou, China). In HEK293T cells, co-transfection of HOXB3-3UTR, HOXB3-3UTR (Mu) or Renilla luciferase plasmid, and miR375-mimics or miR-S, was performed using DharmaFECT 4 transfection reagent (Dharmacon, USA). 48 h after co-transfection, a luciferase activity assay (Promega, USA) was performed. Firefly luciferase activities were normalized to Renilla luciferase activities.
HOXB3 downregulation assay
HOXB3 specific siRNA (siRNA-HOXB3) and a non-specific siRNA (siRNA-C) were purchased from SunBio (SunBio Tech, Guangzhou, China). Capan-1 and PANC-1 cells were transfected with 100 nM siRNA-HOXB3 or siRNA-C for 24 h. Transfection efficiency was checked by qRT-PCR.
Statistical analysis
All experiments were performed by at least three independent repeats. Data were presented as means ± SEM. Statistical analysis was done using SPSS 11.0 software (SPSS, USA). Unpaired two-tail student’s t-test was used to analyze differences between two groups of data. One-way ANOVA with post-hoc test was used to analyze differences between three or more groups of data. P values of <0.05 were considered as significantly different.
Results
MiR-375 was upregulated in pancreatic tumors and pancreatic cancer cell lines
We used qRT-PCR to examine miR-375 expression in both clinical samples of pancreatic tumors and in vitro pancreatic cancer cell lines. Paired tumor tissues (T) and adjacent non-tumorous epithelial tissues (ANT) were obtained from 22 patients diagnosed with pancreatic cancer. Analysis showed that in all patients, miR-375 was significantly upregulated in tumors than in non-tumorous pancreatic epithelial tissues (Figure 1A, *P < 0.05). In addition, miR-375 expression was compared between a human pancreatic duct epithelial-like cell line hTERT-HPNE and 10 well-defined in vitro pancreatic cancer cell lines, HS-766T, Capan-1, PANC-1, Mia PaCa-2, HPAC, AsPC-1 BxPC-3, Capan-2, CFPAC-1 and SW1990. Analysis of qRT-PCR demonstrated that, similar to the expression pattern in pancreatic tumors, miR-375 were also significantly upregulated in all pancreatic cancer cell lines (Figure 1B, *P < 0.05).
Figure 1.
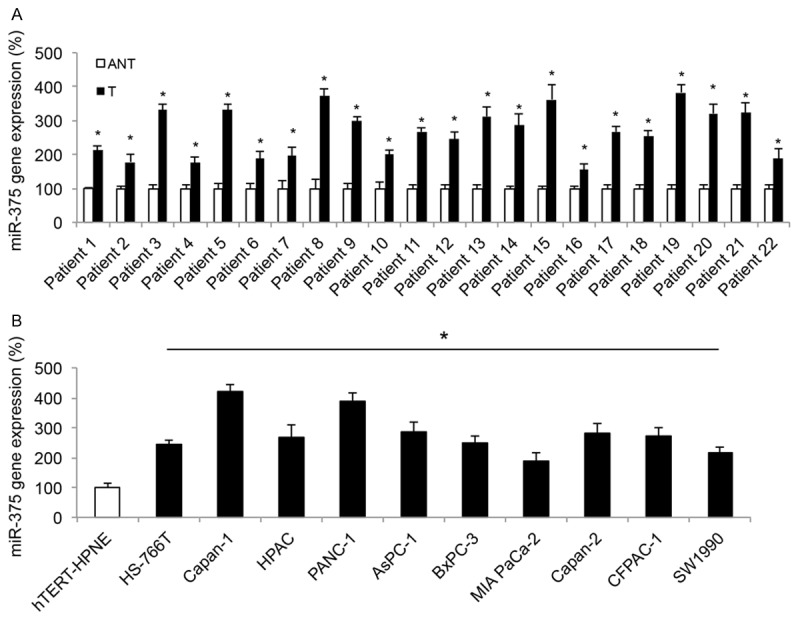
Expression of miR-375 in pancreatic tumors and cell lines. A. In clinical samples retrieved from 22 patients with pancreatic cancer, qRT-PCR was performed to compare endogenous miR-375 expression between pancreatic tumors (T) and their adjacent non-tumorous pancreatic epithelial tissues (ANT) (*P < 0.05, Student’s t-test). B. Endogenous miR-375 expression was also compared between a human pancreatic duct epithelial-like cell line hTERT-HPNE, and ten pancreatic cancer cell lines, HS-766T, Capan-1, PANC-1, Mia PaCa-2, HPAC, AsPC-1 BxPC-3, Capan-2, CFPAC-1 and SW1990 (*P < 0.05, Student’s t-test).
MiR-375 downregulation reduced pancreatic cancer proliferation and migration
In order to understand the underlying mechanism of miR-375 in pancreatic cancer, we transduced lentivirus of miR-375 inhibitor (L-miR375-In) into pancreatic cell lines Capan-1 and PANC-1 to create pancreatic cancer cells with stable miR-375 inhibition. Analysis of qRT-PCR confirmed that, Capan-1 or PANC-1 cells transduced with L-miR375-In had significant low level of endogenous miR-375 expressions than cells transduced with a control miRNA lentivirus, L-miR-C (Figure 2A, *P < 0.05).
Figure 2.
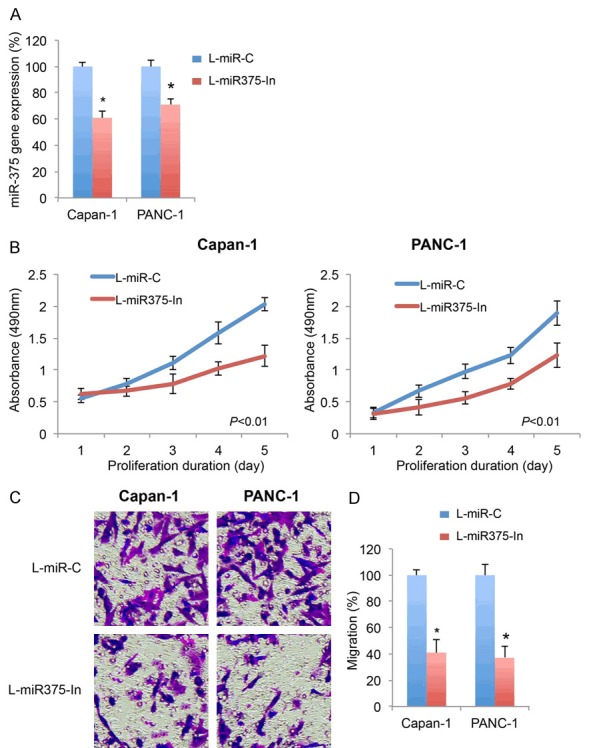
Effect of miR-365 downregulation on pancreatic cancer proliferation and migration (A) Two pancreatic cancer cell lines, Capan-1 and PANC-1 cells were transduced with miR-375 inhibitor lentivirus (L-miR375-In), or a non-specific control miRNA lentivirus (L-miR-C). After transduction was stabalized, transduction efficiency was examined by qRT-PCR on endogenous expressions of miR-375 in Capan-1 and PANC-1 cells (*P < 0.05, Student’s t-test). (B) In lentivirus-transduced Capan-1 and PANC-1 cells, an MTT assay was performed for 5 days. Absorbances at 490 nm were measured and compared between pancreatic cancer cells transduced with L-miR375-In and cells transduced with L-miR-C (P < 0.01, one-way ANOVA). (C) In lentivirus-transduced Capan-1 and PANC-1 cells, a transwell migration assay was performed. Light images were shown for migrating cells transduced with L-miR375-In and cells transduced with L-miR-C. (D) Relative migrations were quantified and compared between cells transduced with L-miR375-In and cells transduced with L-miR-C (*P < 0.05, Student’s t-test).
We then explored the cancer-related effects of miR-375 downregulation in lentivirus-transduced Capan-1 and PANC-1 cells. Firstly, an MTT-assay was performed. After 5 days, it showed miR-375 downregulation significantly reduced cancer proliferation in both Capan-1 and PANC-1 cells (Figure 2B, P < 0.01, one-way ANOVA). Secondly, a transwell migration assay was performed. The light images showed that, significantly less cells migrated into lower chambers while miR-375 was downregulated in Capan-1 or PANC-1 cells (Figure 2C). Using quantitative assessment, we confirmed that pancreatic cancer migration capabilities were significantly reduced by miR-375 downregulation (Figure 2D, *P < 0.05).
MiR-375 downregulation increased pancreatic cancer cisplatin sensitivity and inhibited in vivo growth of tumor explant
We also explored the effect of miR-375 downregulation on chemosensitivity in pancreatic cancer cells. After lentivirus transduction, Capan-1 and PANC-1 cells were incubated with cisplatin at concentrations of 0.01, 0.1, 1, 10, 50 and 100 μM for 24 h. The viable cells were then measured by MTT assay and compared between cells transduced with L-miR375-In and cells transduced with L-miR-C. It showed that miR-375 downregulation significantly increased cisplatin sensitivity in Capan-1 and PANC-1 cells (Figure 3A, P < 0.01, one-way ANOVA).
Figure 3.
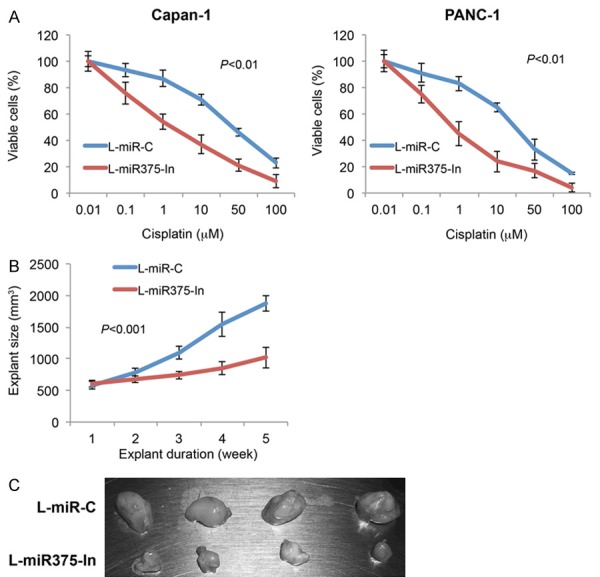
Effect of miR-375 downregulation on pancreatic cancer chemosensitivity and in vivo tumor explants. A. In lentivirus-transduced Capan-1 and PANC-1 cells, cisplatin, at concentrations of 0.01, 0.1, 1, 10, 50 and 100 µM, were added into culture for 24 h. Cell viability was assessed using MTT assay and compared between cells transduced with L-miR375-In and cells transduced with L-miR-C (P < 0.01, one-way ANOVA). B. Capan-1 cells, transduced with L-miR375-In or L-miR-C, were injected into left flanks of adult null mice. The lengths (L, mm) and widths (W, mm) of explants were measured weekly and explant sizes were calculated using the equation, L*W*W/2 (mm3). The in vivo tumor explant assay was carried out for 5 weeks (P < 0.01, one-way ANOVA). C. At the end in vivo assay, mice were sacrificed. Tumor explants transduced with L-miR375-In and tumor explants transduced with L-miR-C were showed.
We then investigated the effect of miR-375 downregulation on in vivo growth of pancreatic tumor explant. Lentivirus-transduced Capan-1 cells, either with L-miR375-In or L-miR-C, were subcutaneously inoculated into left flanks of athymic female nude mice. The in vivo transplantation was carried out for 5 weeks. Every week, explant sizes were compared between tumors transduced with L-miR375-In and tumors transduced with L-miR-C. It demonstrated that miR-375 downregulation significantly inhibited the in vivo growth of pancreatic tumor explants (Figure 3B, P < 0.01, one-way ANOVA). The tumor explants were extracted at the end of in vivo assay. It showed that tumors transduced with L-miR375-In were significantly smaller than tumors transduced with L-miR-C (Figure 3C).
HOXB3 is targeted by miR-375 in pancreatic cancer
We then studied several online algorithms, including TargetScan (www.targetscan.org) and PicTar (pictar.mdc-berlin.de) to search the downstream target gene of miR-375 in pancreatic cancer. It brought to our attention that, HOXB3 gene contained a gene sequence at 3’-UTR that complimentarily binds miR-375 (Figure 4A, upper panel). We thus generated two HOXB3 3’-UTR specific luciferase plasmids, with one containing the miR-375 binding sequence (HOXB3-3UTR) and the other containing the mutated miR-375 binding sequence (HOXB3-3UTR (Mu) (Figure 4A, lower panel). We then performed a luciferase activity assay by co-transfecting HEK273T cells with HOXB3-3UTR, HOXB3-3UTR (Mu) or a Renilla luciferase plasmid, and miR-375 mimics or a scrambled miRNA (miR-S), 48 h after co-transfection, the measurement of luciferase assay showed that miR-375-mimics associated relative luciferase activities were significantly suppressed by co-transfection of HOXB3-3UTR (Figure 4B, *P < 0.05), but not by HOXB3-3UTR (Mu) (Figure 4B, ∆P > 0.05), indicating that HOXB3 gene was directly targeted by miR-375.
Figure 4.
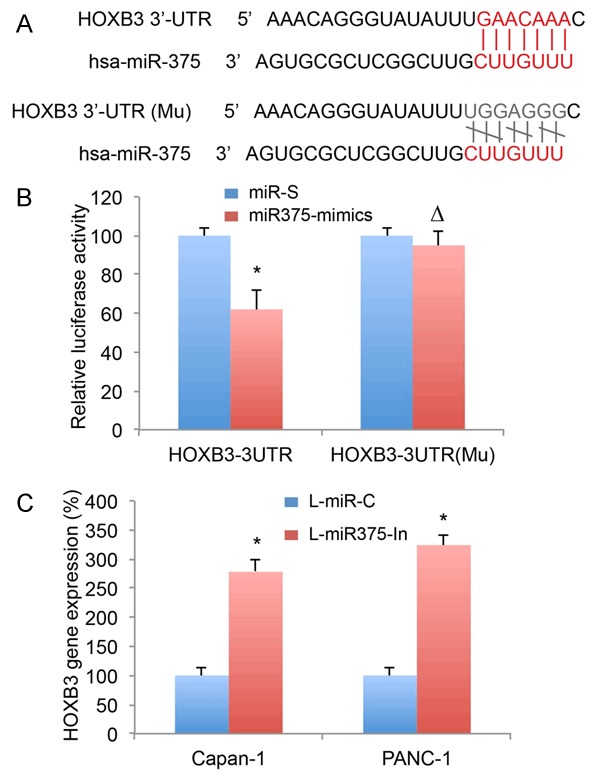
Association of miR-375 with HOXB3 gene in pancreatic cancer. A. Diagrams were shown for human HOXB3 gene 3’-UTR with binding sites of miR-375 (Upper panel), and a mutated (Mu) OXB3 3’-UTR without binding site of miR-375 (Lower panel). B. In HEK293T cells, HOXB3-3UTR, HOXB3-3UTR (Mu) or a Renilla luciferase plasmid, were co-transfected with miR375-mimics or scrambled miRNA (miR-S). 48 h after co-transfection, a luciferase activity assay (Promega, USA) was performed (*P < 0.05, ∆P > 0.05, Student’s t-test). C. In lentivirus-transduced Capan-1 and PANC-1 cells, qRT-PCR was used to compare HOXB3 expression between cells transduced with L-miR375-In and cells transduced with L-miR-C (*P < 0.05, Student’s t-test).
To verify the direct targeting of miR-375 on HOXB3 in pancreatic cancer, we assessed the gene expressions of HOXB3 in Capan-1 cells or PANC-1 cells with miR-375 downregulation. It showed that HOXB3 was significantly upregulated in cells transduced with L-miR375-In than in cells transduced with L-miR-C, indicating that HOXB3 was inversely regulated by miR-375 downregulation in pancreatic cancer (Figure 4C, *P < 0.05).
HOXB3 inversely correlated miR-375 mediation on cancer proliferation migration and chemosensitivity in pancreatic cancer
In miR-375 downregulated Capan-1 and PANC-1 cells, we used HOXB3-specific siRNA (siRNA-HOXB3) to downregulate endogenous HOXB3 gene. In parallel control transfection, a non-specific siRNA (siRNA-C) was used. 24 h after transfection, qRT-PCR confirmed that HOXB3 mRNA levels were significantly downregulated in cells transfected with siRNA-HOXB3 than in cells transfected with siRNA-C (Figure 5A, *P < 0.05).
Figure 5.
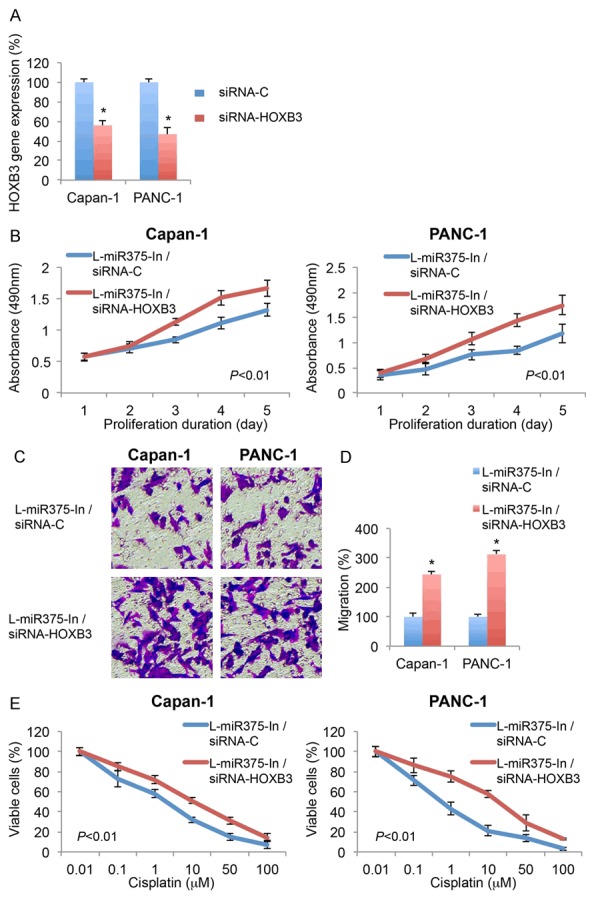
Effect of HOXB3 downregulation on miR-375 downregulated pancreatic cancer cells. A. Capan-1 and PANC-1 cells, after transduction of L-miR375-In, were transfected with HOXB3 specific siRNA (siRNA-HOXB3, 100 nM) or a scrambled siRNA (siRNA-C, 100 nM) for 24 h. After that, qRT-PCR was performed to assess the transfection efficiency (*P < 0.05, Student’s t-test). B. After siRNA transfection, an MTT assay was performed for 5 days. Absorbances at 490 nm were measured and compared between pancreatic cancer cells transfected with siRNA-HOXB3 and cells transfected with siRNA-C (P < 0.01, one-way ANOVA). C. A transwell migration assay was also performed. Light images were shown for migrating cells transfected with siRNA-HOXB3 and cells transfected with siRNA-C. D. Relative migrations were quantified and compared between cells transfected with siRNA-HOXB3 and cells transfected with siRNA-C (*P < 0.05, Student’s t-test). E. Also after siRNA transfection, Capan-1 and PANC-1 cells were treated with cisplatin, at concentrations of 0.01, 0.1, 1, 10, 50 and 100 µM, for 24 h. Cell viability was assessed using MTT assay and compared between cells transfected with siRNA-HOXB3 and cells transfected with siRNA-C (P < 0.01, one-way ANOVA).
Then, those cells were re-seeded, and the effects of HOXB3 downregulation on miR-375 mediated pancreatic cancer cell modulation were re-examined through functional assays. Firstly, 5-day MTT assay demonstrated that, in miR-375 downregulated Capan-1 and PANC-1 cells, HOXB3 downregulation induced significant pancreatic cancer proliferations (Figure 5B, P < 0.01. one-way ANOVA). Secondly, transwell assay showed that HOXB3 downregulation significantly increased pancreatic cancer migrations (Figure 5C, 5D, *P < 0.05). Thirdly, cisplatin assay demonstrated HOXB3 downregulation dramatically reduced pancreatic cancer cisplatin chemosensitivity (Figure 5E, P < 0.01. one-way ANOVA). Therefore, the results of proliferation, migration and cisplatin assays all indicated that HOXB3 downregulation reversed miR-375 downregulation induced cancer inhibition in pancreatic cancer.
Discussions
In many of the human cancers, microRNA-375 was found to be aberrantly downregulated, and acting as a tumor suppressor by inhibiting cancer proliferation and metastasis [11-14]. However, in this study, we discovered that miR-375 expression was much different in pancreatic cancer than in other cancers. The analysis of qRT-PCR on clinical samples from 22-pancreatic cancer patients and 10 pancreatic cancer cell lines demonstrated miR-375 was significantly upregulated in pancreatic cancer. These results are in line with a previous microarray study [15], not only confirming the overexpressing pattern of miR-375, but also suggesting a much different role of miR-375 in regulating pancreatic cancer development.
This hypothesis was then examined by down-regulating endogenous miR-375 in two pancreatic cancer cell lines, Capan-1 and PANC-1 cells, through lentiviral transduction. Our data on MTT assay, transwell assay, chemosensitivity assay and in vivo explant assay demonstrated that miR-375 downregulation significantly inhibited pancreatic cancer proliferation, migration, cisplatin chemoresistance and in vivo explant growth. Thus, it is very clear that miR-375 is acting as a tumor oncogene, rather a tumor suppressor, in pancreatic cancer.
Also in this study, we identified that HOXB3 is acting as a downstream target gene of miR-375 in pancreatic cancer. We demonstrated, through a luciferase activity assay, that miR-375 could bind the complimentary site on 3’-UTR of human HOXB3 gene. We then showed that gene expression of HOXB3 was significantly upregulated while miR-375 was downregulated in pancreatic cancer Capan-1 and PANC-1 cells. Most importantly, after using siRNA technology to knock down endogenous HOXB3 in miR-375-downregulated pancreatic cancer cells, we found the HOXB3 downregulation reversely impacted pancreatic cancer proliferation, migration and cisplatin sensitivity, than miR-375 downregulation. In a previous study, HOXB3, along with HOXB1, was shown to be the target of miR-10a, as well as regulated by retinoic acid receptor signaling pathway in regulating metastasis pancreatic cancer [16]. It is not clear whether miR-10a or by retinoic acid receptor signaling pathway has cross-link effect on miR-375 regulation in pancreatic cancer. Future experiment would certainly help to elucidate the intricate correlation of different molecular pathways in regulating cancer development in pancreatic cancer.
Overall, this study provided solid evidence of miR-375 acting as a tumor oncogenic regulator, very likely through the inverse association with tumor suppressor gene HOXB3, in human pancreatic cancer. The discovery of miR-375 and HOXB3 signaling pathway in regulating cancer development may potentially be used as a therapeutic approach to treat patients with pancreatic cancer.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Rossi ML, Rehman AA, Gondi CS. Therapeutic options for the management of pancreatic cancer. World J Gastroenterol. 2014;20:11142–11159. doi: 10.3748/wjg.v20.i32.11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari D, Gustafsson A, Andersson R. Update on the management of pancreatic cancer: surgery is not enough. World J Gastroenterol. 2015;21:3157–3165. doi: 10.3748/wjg.v21.i11.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammed S, Van Buren G 2nd, Fisher WE. Pancreatic cancer: advances in treatment. World J Gastroenterol. 2014;20:9354–9360. doi: 10.3748/wjg.v20.i28.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mihaljevic AL, Michalski CW, Friess H, Kleeff J. Molecular mechanism of pancreatic cancer--understanding proliferation, invasion, and metastasis. Langenbecks Arch Surg. 2010;395:295–308. doi: 10.1007/s00423-010-0622-5. [DOI] [PubMed] [Google Scholar]
- 6.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 7.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gayral M, Jo S, Hanoun N, Vignolle-Vidoni A, Lulka H, Delpu Y, Meulle A, Dufresne M, Humeau M, Chalret du Rieu M, Bournet B, Selves J, Guimbaud R, Carrere N, Buscail L, Torrisani J, Cordelier P. MicroRNAs as emerging biomarkers and therapeutic targets for pancreatic cancer. World J Gastroenterol. 2014;20:11199–11209. doi: 10.3748/wjg.v20.i32.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitkara D, Mittal A, Mahato RI. miRNAs in pancreatic cancer: therapeutic potential, delivery challenges and strategies. Adv Drug Deliv Rev. 2015;81:34–52. doi: 10.1016/j.addr.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Shi M, Xie D, Gaod Y, Xie K. Targeting miRNAs for pancreatic cancer therapy. Curr Pharm Des. 2014;20:5279–5286. doi: 10.2174/1381612820666140128210443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–187. e178. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 14.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 15.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 16.Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137:2136–2145. e1–7. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]


