Abstract
Recent findings indicate that microRNAs (miRNAs) play a crucial role in lung cancer development, progression and regression. In our previous study, we identified miR-326 is down-regulated in lung cancer. However, the role of miR-326 hasn’t been revealed yet. The aim of the current study is to investigate the function and regulation mechanism of miR-326 in lung cancer. MTT assays, Transwell migration assays and xenograft model in nude mice were carried to detect the effects of miR-326 on cell proliferation, migration and tumor growth in nude mice. Flow cytometry was used to analyze the effects of miR-326 on cell cycle and apoptosis. By using siRNAs and luciferase assays, we also demonstrated that Phox2a is a functional target of miR-326, and that miR-326 is regulated by long non-coding RNA HOTAIR through silencing HOTAIR. Enforced expression of miR-326 inhibited cell proliferation and migration in vitro and tumor growth in nude mice, decreased proportion of cells in S phase and increased cell apoptosis in both A549 and H838 cells. In addition, we found miR-326 bound to 3’UTR of Phox2a but not KLF3, and enforced expression of miR-326 decreased accumulation of Phox2a in both A549 and H838. Moreover, exogenous expression of Phox2a compromised inhibitory effects of miR-326 on cell proliferation and migration. We also found silencing of HOTAIR caused increased expression of miR-326. miR-326 regulates cell proliferation and migration in lung cancer by targeting Phox2a and is regulated by HOTAIR.
Keywords: miR-326, cell proliferation, migration, lung cancer, Phox2a, HOTAIR
Introduction
Lung cancer is one of the most aggressive malignancies in worldwide. The current standard of care for lung cancer is surgical resection followed by adjuvant radiation therapy and chemotherapy. Intensive efforts have been made to investigate the initiation and development of lung cancer. However, the molecular mechanisms of carcinogenesis remain unclear. Recent findings indicate that microRNAs (miRNAs) plays a crucial role in the development, progression and regression of lung cancer.
MiR-326 was first reported as miRNA expressed in neurons [1]. Expression of miR-326 associates with multiple pathological process, including multiple sclerosis [2], brain tumor [3], lung cancer [4], pulmonary fibrosis [5] and type 1 diabetes [6]. Recent studies indicate miR-326 regulates TH-17 differentiation [2], carcinogenesis [3], metastasis [7] and invasion [8] of tumor, adipogenic differentiation of adipose-derived stem cells [9], chemotherapy resistance [10]. One of the most important mechanisms underlying function of miR-326 is that miR-326 binds to the 3’ un-coding regions of mRNA complementarily. Studies in multiple fields suggest that miR-326 regulates multiple targets in different pathological context. Several targets of miR-326 have been identified, including pyruvate kinase type M2 (PKM2) [11], multidrug resistance-associated protein 1 (ABCC1) [12], nin one binding protein (NOB1) [13] and Bcl-xL [14].
Paired-like homeobox 2a (phox2a) is first identified in differentiating neurons and essential for neurogenesis [15,16]. Phox2a is a transcription factor that regulates transcription of dopamine beta-hydroxylase (DBH) [17] and alpha3 nicotinic receptor subunit [18]. Mutation of Phox2a associates with congenital fibrosis of the extraocular muscles type 2 [19-21]. Single point mutations in the homeodomain of Phox2a resulted in a failure to transactivate the DBH [22]. Phox2a is overexpressed in neuroblastoma tumors [23]. Kruppel-like factors (KLFs) are a large class of basic transcription element-binding protein in eukaryotrs, and KLF3 is a stronger transcriptional repressor of KLFs family. KLF3 could increase maturation toward the marginal zone B cells and bind to the promoters of β-globin [24,25]. In this study, Phox2a and KLF3 are predicted the target genes of miR-326 through using online program TargetScan.
LncRNA HOX transcript antisense intergenic RNA (HOTAIR) is overexpressed in a variety of malignancy, and is linked closely with increase in cancerous metastasis. HOTAIR is up-regulated by tumor-promoting Col-1 in NSCLC cells [26]. More importantly, HOTAIR could sponge miRNAs and function as a competing endogenous RNA, to regulate the expression of HER2 in GC cells [27]. Despite HOTAIR plays an important role in tumor growth and metastasis, the mechanism in lung cancer is still unclear.
In this study, we report the inhibitory role of miR-326 in tumor growth and metastasis of lung cancer. Furthermore, we demonstrate that Phox2a is a functional target of miR-326 and miR-326 is regulated by long non-coding RNA HOTAIR. Taken together, our data establish HOTAIR-miR-326-Phox2a axis as regulator of lung cancer with potential therapeutic implications.
Materials and methods
Cell culture and transfection
Human lung cancer cell lines A549, 95D, NCI-H460, HLamp and H838 were obtained from Shanghai Cell Institute Country Cell Bank, (Shanghai, China). A549 cells were grown in DMEM medium with 10% fetal bovine serum (FBS) (GIBCo/BRL, MD), supplemented with 100 U/ml penicillin G and 100 μg/ml streptomycin (Sigma-Aldrich Corp., St. Louis, MO). 95D, NCI-H460, HLamp and H838 cells were grown in 1640 medium with 10% fetal bovine serum (FBS) (GIBCo/BRL, MD), supplemented with 100 U/ml penicillin G and 100 μg/ml streptomycin (Sigma-Aldrich Corp., St. Louis, MO). Cells were maintained at 37°C in a humidified 5% CO2 incubator.
For miRNA and siRNA transfection, A549 and H838 cells plated in triplicate in 6-well plates. 100 nM mimics of miR-326 or siRNA of HOTAIR were transfected into lung cancer cells using lipofectamine 2000 (invitrogen) according to manufacturer’s protocol.
Plasmids
The wild type 3’UTR of Phox2a and KLF3, and mutant 3’UTR of Phox2a and KLF3were amplified and inserted into the downstream of psiCHECK2 vector (Promega) All the primers were listed in Table 1. The sequences of all constructs were confirmed by DNA sequencing. siRNA of HOTAIR, and scrambled HOTAIR were synthesized by Jima Biotech (Shanghai, China), the primers of siRNA were listed in Table 2.
Table 1.
Primers for luciferase reporter constructs
| ID | Sequence (5’-3’) |
|---|---|
| psiCHECK-KLF3-XhoI F | CCGctcgagCCCACTCACCTGGCTCTCT |
| psiCHECK-KLF3-NotI R | ATAAGAATgcggccgcTCCATACTCTTACCCGAAA |
| psiCHECK-KLF3-3’UTR-mut-F | ATCCAGACTGCTAGTCCTTGATGCCTACATTCTTACCTCTGCCCTG |
| psiCHECK-KLF3-3’UTR-mut-R | CAGGGCAGAGGTAAGAATGTAGGCATCAAGGACTAGCAGTCTGGAT |
| psiCHECK-PHOX2A-XhoI F | CCGctcgagCCTCTGGAGGCTCCGAGC |
| psiCHECK-PHOX2A-NotI R | ATAAGAATgcggccgcGAAAAAGGACAACCAAAAAAAT |
| psiCHECK-Phox2a-3’UTR-mut-F | CTGGAGGCTCCGAGCCTGCCATTGTCTACGTCCCTGCCCCTCCAGGA |
| psiCHECK-Phox2a-3’UTR-mut-R | TCCTGGAGGGGCAGGGACGTAGACAATGGCAGGCTCGGAGCCTCCAG |
| psiCHECK-326-XhoI-F | CCGctcgagCTCATCTGTCTGTTGGGCTGGA |
| psiCHECK-326-NotI-R | ATAAGAATgcggccgcTGAATCCGCCTCGGGGCT |
Table 2.
siRNA for HOTAIR
| ID | Sequence (5’-3’) |
|---|---|
| scrambled siRNA | GAACGGAGCGAGCAGACCUUU |
| HOTAIR siRNA | GAACGGGAGUACAGAGAGAUU |
RNA extraction and quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted from cultured cells using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA samples were then reverse transcribed into cDNA using a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa) in a total volume of 20 μL according to the manufacturer’s protocol. Equal amounts of cDNA samples were used as a template for real-time PCR to detect the level of Phox2a or KLF3 expression. Quantitative PCR was performed using a LightCycler480 Real-Time PCR system and a SYBR Premix Ex TaqTMII PCR Kit (Takara); GAPDH was used as an endogenous reference, and each sample was normalized to its GAPDH content. All experiments were performed in duplicate and repeated twice. Results are represented as fold induction using the 2-ΔΔCt method. Primers for quantitative PCR are shown in Table 3.
Table 3.
Primers for quantitative real-time RT-PCR
| ID | Sequence (5’-3’) |
|---|---|
| hsa-mir-326 | CCTCTGGGCCCTTCCTCCAG |
| hsa-mir-326-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTGGAGGA |
| hsa-mir-326-F | ACACTCCAGCTGGGCCTCTGGGCCCT |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| All-R | CTCAACTGGTGTCGTGGA |
| HOTAIR-F | TGCTACTTGTGTAGACCCAG |
| HOTAIR-R | AGCAAAGGCTGGACCTTTGCT |
| GAPDH-F | ACACCCACTCCTCCACCTTT |
| GAPDH-R | TTACTCCTTGGAGGCCATGT |
MTT assays
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assays were performed according to the manufacturer’s protocol. Briefly, 5000 cells/well were plated in triplicate in 96-well plates. The MTT reagent was prepared at 5 mg/ml in Phosphate Buffered Saline (PBS). This MTT stock solution was then added to each well at a 1:10 dilution. Cells were incubated for 4 h, and the resulting crystals were dissolved in 100 µl Dimethyl sulfoxide (DMSO). The absorbance at 490 nm was measured using a multi-well plate reader.
Transwell assays
The assay was done by using chambers with polycarbonate filters (pore size, 8 µm) (Becton Dickinson Labware). Twenty-four hours after miRNA transfection, the cells were harvested and 5×104 transfected cells in 200 µL of 0.1% serum medium were placed in the upper chamber. The lower chamber was filled with 10% fetal bovine serum medium (600 µL). After 24 h incubation and removal of the cells on the upper chamber of the filter with a cotton swab, the cells on the underside were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet in 20% ethanol, and counted in five randomly selected fields under phase contrast microscope. The migrated cells were monitored by photographing at 400× magnification with LEICA Microscope. The assays were performed in triplicate.
Xenograft model in nude mice
Six-week-old female athymic nude mice were subcutaneously injected in the right armpit region with 1×107 cells in 0.1 mL of PBS. Two groups of mice (n = 6/group) were tested. Group 1 (NC) was injected with A549 or H838 cells transfected with NC; and group 2 (miR-326) was injected with A549 or H838 cells transfected with mimics of miR-326. The tumor sizes were measured every 7 days with calipers. The tumor volume was calculated with the formula: (L×W2)/2, where L is the length and W is the width of the tumor. After the mice were killed at five weeks, the weights of the tumors were measured. All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 80-23, revised 1996) and were performed according to the institutional ethical guidelines for animal experiments.
Cell cycle analysis
For cell cycle analysis, 2×105 cells were seeded in a 6-well culture plate and grown for 24 h. Twenty-four hours after miRNA transfection the cells were trypsinized, washed twice with cold PBS and fixed with cold 70% ethanol at 4°C overnight. The cells were then washed twice with PBS and incubated with 10 mg/ml RNase A, 400 mg/ml propidium iodide (Sigma-Aldrich) and 0.1% Triton-X in PBS at room temperature (RT) for 30 min. Cells were subsequently analyzed by flow cytometry.
Apoptosis assays
KGI Annexin V-FITC apoptosis detection kit was used to detect apoptosis in A549 and H838 cells. According to manufacturer’s instruction manual, the cells were digested with trypsin, and centrifuged on 2000 rpm for 5 min. After collection, the cells were washed twice with PBS, and centrifuged at 2000 rpm for 5 min, 1-5×105 cells were collected and suspensed with 500 μl Binding Buffer. 5 μl Annexin V-FITC and 5 μl Propidium Iodide was respectively added and mixed on room temperature, and away from light for 15 min. Within 1 hour, the cells were detected by flow cytometry. The excitating wavelength (Ex) was 488 nm and emission wavelength (Em) was 530 nm. Green fluorescence of Annexin V-FITC was detected through FITC channel (FL1); PI red fluorescence was detected by PI channel (FL2 or FL3). Fluorescent compensation adjustment: normal cells without apoptosis-inducing treatment were used as controls for fluorescence compensation settings adjustment.
Luciferase assays
A549 cells were seeded in 96-well plates at 6,000 cells per well the day before transfection. A mixture of 100 ng luciferase reporter constructs (psiCHECK-Phox2a-WT or psiCHECK-KLF3-WT, and psiCHECK-Phox2a-mutant or psiCHECK-KLF3-mutant) and 200 ng of NC or miR-326 mimics was transfected into A549 cells with Lipofectamine 2000. Forty-eight hours later, Firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter System (Promega) according to the manufacturer’s protocol.
Western blot analysis
Western blot analysis was performed according to standard Western blot procedures as previously described [28]. Briefly, proteins were separated by 10% SDS-PAGE and then transferred to nitrocellulose membrane (Bio-Rad). After blocking in 5% nonfat milk, the membranes were incubated with the following primary antibodies: mouse anti-KLF3 monoclonal antibody (mAb; 1:300; Abcam), mouse anti-Phox2a monoclonal antibody (mAb; 1:300; Abcam), mouse anti-GAPDH mAb (1:1,000; Abcam). The proteins were visualized with enhanced chemilumin escence reagents (Pierce).
Statistical analysis
The experimental results were shown as mean ± standard deviation (SD) unless otherwise noted; the Student’s t-test and one-way analysis were used for statistical analysis. A P value of <0.05 was considered statistically significant.
Results
Effects of miR-326 on proliferation and migration in vitro and tumor growth in vivo
We investigated the effects of miR-326 on proliferation by MTT assays, and migration by transwell assays. Exogenous expression of miR-326 was achieved by transfected miR-326 mimics into H838 and A549 cells, in which expression of miR-326 was relatively low. NC was served as negative control. Efficiency of transfection was verified by significant increase of miR-326 expression which was determined by QPCR (Figure 1A). We found exogenous expression of miR-326 inhibited proliferation and migration of A549 and H838 cells (Figure 1B and 1C).
Figure 1.
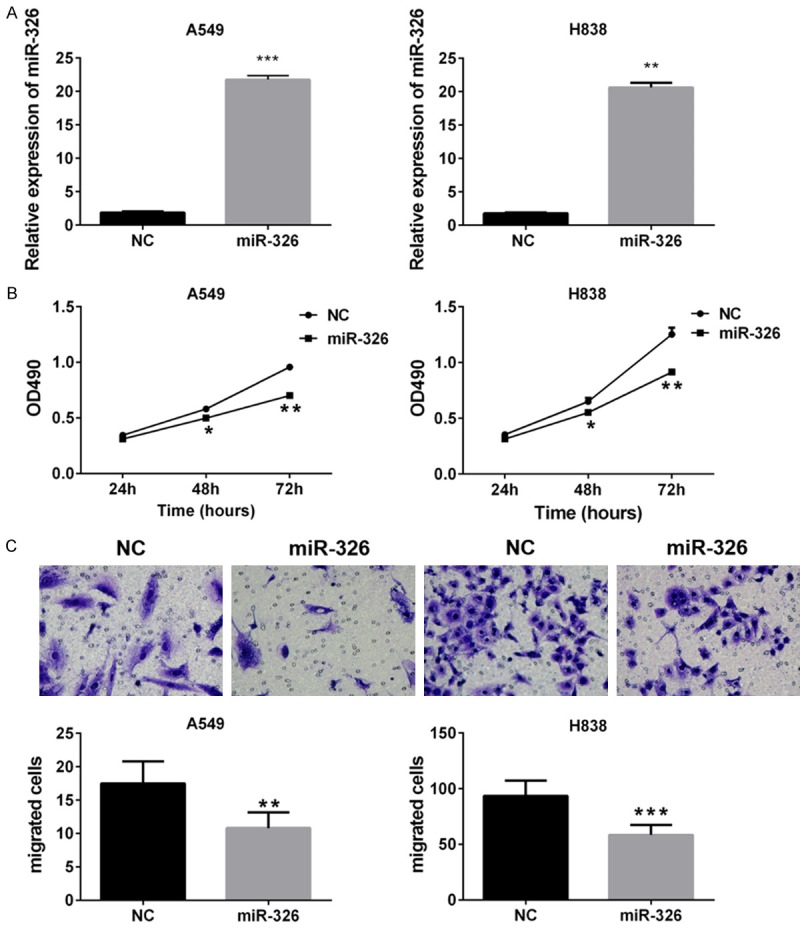
Effects of miR-326 on cell transfection, proliferation and migration in nude mice. A. QPCR assay was used to examine the efficiency of transfection in A549 and H838 cells. B. MTT assay of A549 and H838 cells. *P<0.05 vs NC. **P<0.01 vs NC. C. Transwell migration assays of A549 and H838 cells. Up, representative images from three independent experiments. Down, graph. Column, means; bar, SD. **P<0.01 vs NC. ***P<0.001 vs NC.
In order to verify our findings in vivo, we examined the effects of miR-326 on tumor growth in nude mice. NC or mimics of miR-326 transfected A549 or H838 cells were injected subcutaneously into nude mice. NC was served as control. Tumor sizes were recorded twice a week. We found nude mice injected with miR-326 transfected cells generated much smaller tumors than those injected with NC transfected cells (Figure 2A), which indicated miR-326 inhibits tumor growth in nude mice. The decreased tumor volume also demonstrated it (Figure 2C). We also observed the tumor cell from the nude mice in miR-326 treatment group exerted a loose arrangement a large necrotic region with H&E staining (Figure 2B).
Figure 2.
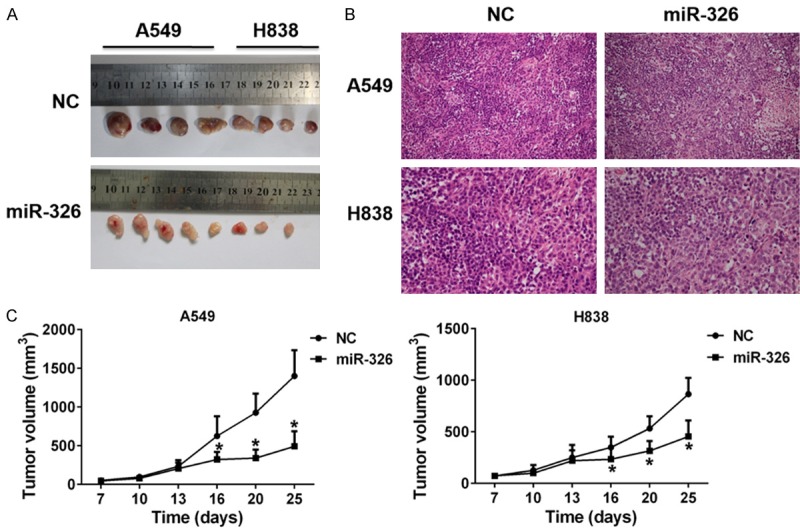
Effects of miR-326 on tumor growth in nude mice. A. Representative images of tumors isolated from nude mice that had been injected with A549 or H838 cells transfected with NC and mimics of miR-326. B. Representative hematoxylin and eosin stained sections of the tumor tissues isolated from nude mice. C. Growth curves of tumors in nude mice. *P<0.05 vs NC.
Effects of miR-326 on apoptosis and cell cycle
In order to study the mechanisms underlying the effects of miR-326 on promoting proliferation of lung cancer cells, the effects of miR-326 on cell cycle and apoptosis were examined by flow cytometry. We found enforced expression of miR-326 could increase the cells in G1 phase and decrease the cells in S phase (Figure 3A), which indicated miR-326 could induce G1 arrest in both A549 and H838 cells. Cell apoptosis assays also indicated that enforced expression of miR-326 could increase cellular apoptosis in both A549 and H838 cells (Figure 3B).
Figure 3.
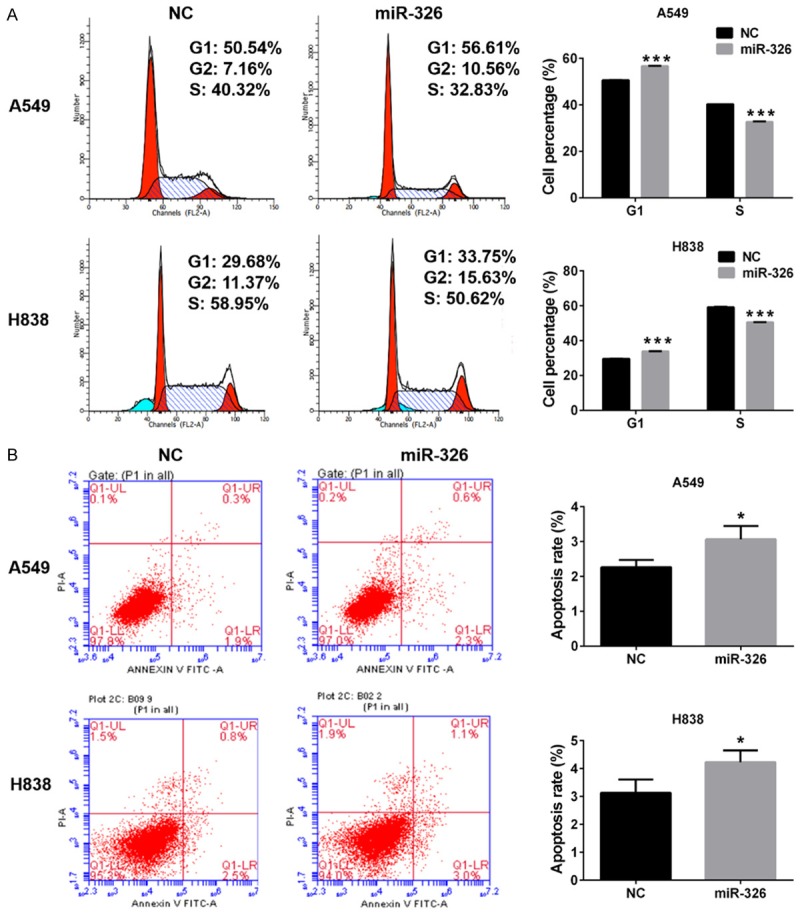
Effects of miR-326 on cell cycle and apoptosis. A. Cell cycle analysis of A549 and H838 cells. B. Cell distribution of A549 and H838 cells transfected with miR-326 or NC. Column, means; bar, SD. **P<0.01 vs NC. C. Cell apoptosis analysis of A549 and H838 cells transfected with miR-326 or NC. D. Apoptosis rate of A549 and H838 cells. Column, means; bar, SD. **P<0.01 vs NC.
miR-326 modulate the expression of Phox2a
miRNAs modulates cell proliferation by down-regulating expression of downstream targets. The mechanism underlying down-regulating effects of miRNAs on gene expression is that miRNAs could bind to 3’UTR of mRNA, which leads to consequential degradation or inhibit translation of mRNA. We identified several potential targets of miR-326 using online program TargetScan (data not shown). We speculated Phox2a and KLF3, which are well established transcription factor and main targets of miR-326. miR-326 binding sites were found in position 395-401 of KLF3 3’UTR and position 31-38 of Phox2a 3’UTR (Figure 4A). We constructed the wild type and mutant KLF3 and Phox2a to transfect the A549 and H838 cells. Luciferase assays were performed after 48 h transfection. Data of luciferase assays showed significant decreased relative luciferase activities in miR-326 and Phox2a transfected cells, while no significant changes were found in miR-326 and KLF3 transfected cells (Figure 4B). These data indicated miR-326 could bind to the 3’UTR of Phox2a but not the 3’UTR of KLF3.
Figure 4.
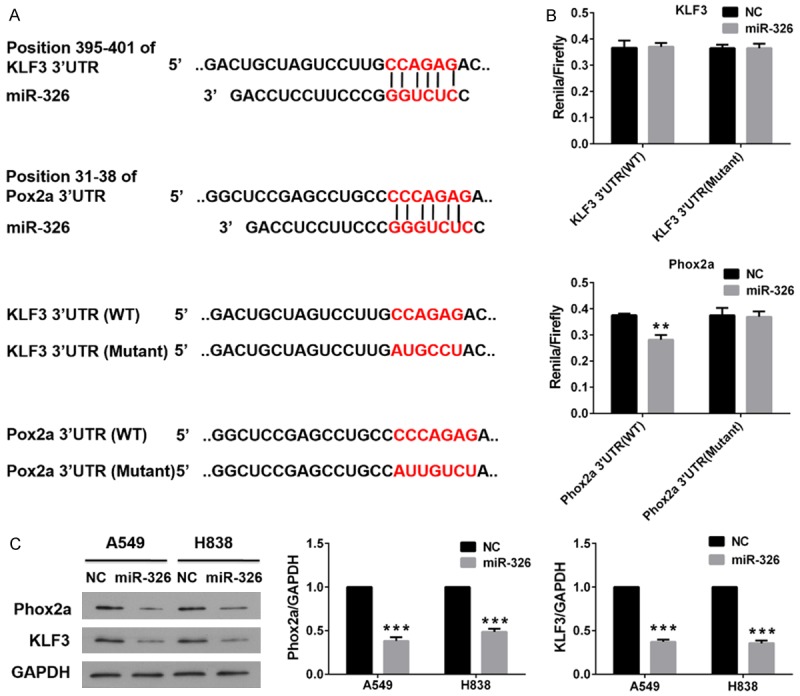
miR-326 negatively regulates Phox2a by binding to the Phox2a 3’UTR. A. Schematic of putative miR-326 binding sites and mutated binding sites in Phox2a and KLF3 3’UTR. Putative miR-326 binding site and mutated binding sites is indicted with red character. B. Luciferase reporter assay of A549 and H838 cells. Column, means; bar, SD. **P<0.01 vs NC. C. Protein expression of Phox2a and KLF3 in tumors isolated from nude mice that had been injected with A549 cells transfected with NC or mimics of miR-326. GAPDH served as an internal control. Column, means; bar, SD. ***P<0.001 vs NC.
The complementary binding of miRNAs with 3’UTR of mRNA could cause consequential degradation of target gene. Now that we had proved miR-326 bound to 3’UTR of Phox2a, we speculated miR-326 could down-regulate the expression of Phox2a. We examined the expression of Phox2a with western blot assay. Our data showed enforced expression of miR-326 caused decreased accumulation of Phox2a (Figure 4C). Interestingly, we also found exogenous expression of miR-326 also could decrease endogenous expression of KLF3, although miR-326 didn’t bind to 3’UTR of KLF3 (Figure 4C).
Phox2a compromises effects of miR-326 on cell proliferation, cell cycle and apoptosis. We had proved miR-326 binds to 3’UTR of Phox2a and regulated the expression of Phox2a, which indicated Phox2a may be a downstream target of miR-326. We speculated miR-326 could modulate cell proliferation, cell cycle and apoptosis by regulating expression of Phox2a. In order to verify our hypothesis, we investigated whether the expression of Phox2a could exert an impact o on cell proliferation, invasive capability, cell cycle and apoptosis. Results of MTT assays showed enforced expression of Phox2a could alleviate the inhibitory effects of miR-326 on cell proliferation of lung cancer cells (Figure 5A). Data also showed that enforced expression of Phox2a could significantly increase the invasive capability of lung cancer cells (Figure 5B). The results of flow cytometry suggested enforced expression of Phox2a could reverse the cells in S phase in lung cancer cells (Figure 6A). Apoptosis assays also indicated that enforced expression of Phox2a could decrease the apoptosis rates on apoptosis of lung cancer induced by miR-326 (Figure 6B).
Figure 5.

Enforced expression of Phox2a compromised effects of miR-326 on cell proliferation, migration. A. MTT assay of A549 and H838 cells. *P<0.05 vs NC. B. Transwell migration assays of A549 and H838 cells. Left, representative images from three independent experiments. Right, graph. Column, means; bar, SD. *P<0.05 vs NC.
Figure 6.
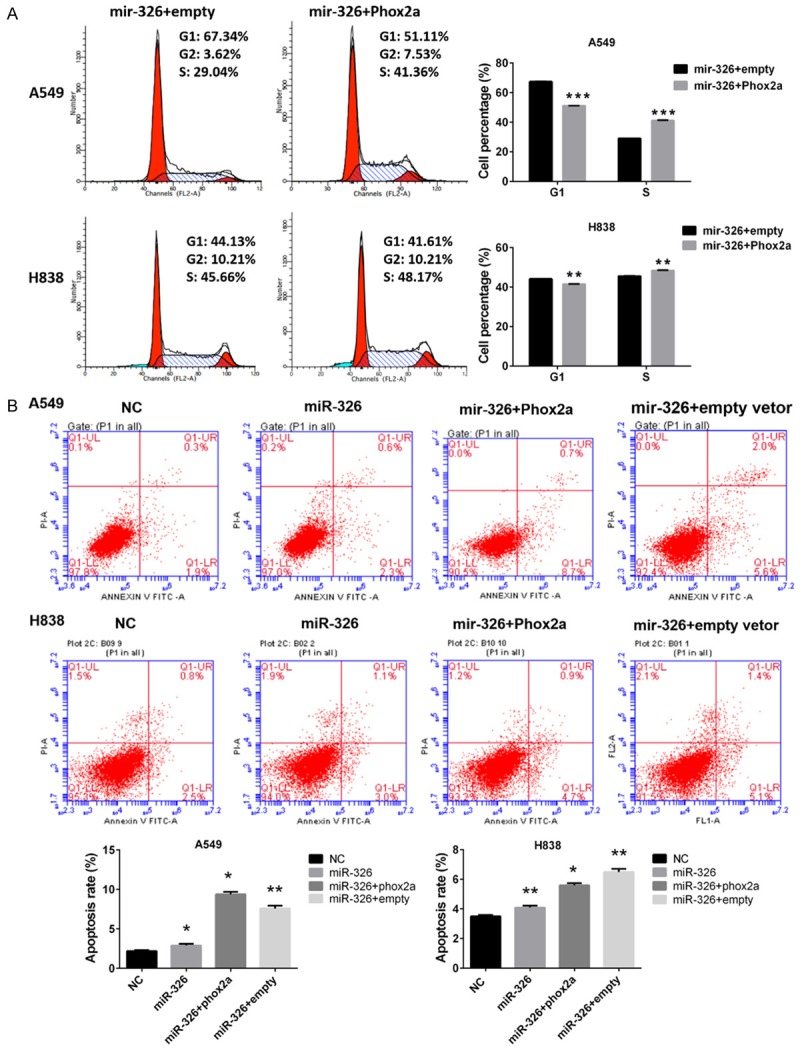
Enforced expression of Phox2a compromised effects of miR-326 on cell cycle and apoptosis. A. Cell cycle analysis of A549 and H1838 cells. Up, representative images from three independent experiments. Down, graph. Column, means; bar, SD. **P<0.01 vs NC. B. Apoptosis assays of A549 and H838 cells. Up, representative images from three independent experiments. Down, graph. Column, means; bar, SD. *P<0.05 vs NC.
HOTAIR regulates expression of miR-326
Using online tools we had identified the binding sites of miR-326 in HOTAIR (Figure 4A), which indicated HOTAIR could interact with miR-326. We investigated whether HOTAIR could regulate the expression of miR-326. QPCR was performed to examine the expression of HOTAIR and miR-326in lung cancer cells, and the cells were transfected with siRNAs to target HOTAIR. Our data showed silencing HOTAIR with siRNAs caused reduced expression of HOTAIR and increased accumulation of miR-326 in lung cancer cells (Figure 7A and 7B), which indicated HOTAIR could be a negative regulator of miR-326.
Figure 7.
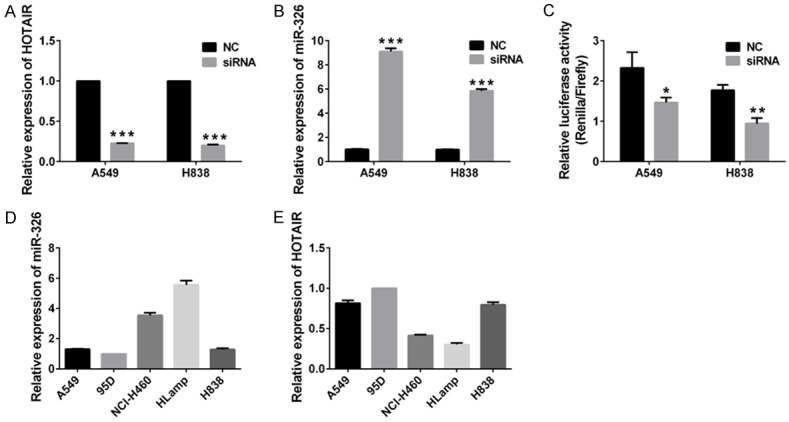
HOTAIR regulates expression of miR-326. (A and B) expression of HOTAIR and miR-326 in A549 and H838 transfected with siRNA targeted HOTAIR. NC served as control. Expression of miR-326 and HOTAIR was normalized with U6 and GAPDH, respectively. Column, means; bar, SD ***P<0.001 vs NC. (C) Relative luciferase activities of A549 and H838 transfected with siRNA targeted HOTAIR and miR-326 luciferase reporter construct psiCHECK-326. NC served as control. Column, means; bar, SD. *, P<0.05 vs NC. **P<0.01 vs NC. (D, E) Expression of miR-326 and HOTAIR in lung cancer cells. mRNA expression of miR-326 and HOTAIR in human lung cancer cell lines A549, 95D, H460, HLamp and H838 were examined with QPCR. Expression of miR-326 and HOTAIR was normalized with U6 and GAPDH, respectively. Column, means; bar, SD.
In order to further verify our finding, we constructed dual-luciferase reporters, which contained the predicted seed sequence in the 3’UTR of siRNA of HOTAIR. The empty vector of psiCHECK-2 was used as a negative control. After all vectors transfection, the luciferase activities were examined in lung cancer cells. Results showed that decreased relative luciferase activities were observed in lung cancercells (Figure 7C). Because silencing HOTAIR could increase accumulation of miR-326, and then miR-326 binds to reverse complementary sequence of pre-miR-326 in psiCHECK-326, which results in decreased luciferase activities of A549 cells. This result proves again that HOTAIR is a negative regulator of miR-326.
We also examined mRNA the expression of miR-326 and HOTAIR by QPCR in human lung cancer cell lines, including A549, 95D, H460, HLamp and H838 cells. Expression of miR-326 and HOTAIR was normalized with U6 and GAPDH, respectively. Our data showed higher expression of miR-326 was found in HLamp and H460 cells, while the expression of HOTAIR was relatively low. Lower expression of miR-326 and higher expression of HOTAIR were observed in 95D, H838 and A549 cells (Figure 7D and 7E). These findings supported our theory that HOTAIR could be a negative regulator of miR-326.
Discussion
Dysregulation of miR-326 is associated with various physical process and diseases. Du and colleges report that miR-326 regulated TH-17 differentiation, and is associated with multiple sclerosis [2]. Sebastiani and colleagues report that miR-326 is expressed at a higher level in type 1 diabetic patients with ongoing islet autoimmunity, and correlated with disease severity [6]. Our previous study indicates miR-326 is down-regulated in lung cancer, which inspire us that miR-326 may play an important role in lung cancer. However, the function of miR-326 in lung cancer remains unclear. In the current study, we report miR-326 regulates cell proliferation, migration, cell cycle and apoptosis in both A549 and H838 cells and inhibits tumor growth in nude mice (Figures 1 2 and 3). These data suggest miR-326 functions as tumor suppressor in lung cancer.
Phox2a plays a central role in development of the autonomic nervous system [19,22]. It regulates the expression of tyrosine hydroxylase [29], dopamine beta-hydroxylase [30,31], and two catecholaminergic biosynthetic enzymes which are essential for the differentiation and maintenance of the noradrenergic neurotransmitter phenotype. Phox2a also regulate the transcription of the alpha3 nicotinic acetylcholine receptor gene [18]. Mutations in this gene have been associated with autosomal recessive congenital fibrosis of the extraocular muscles [19-21,32]. Phox2a is regulated by sp1 and Phox2b. Phox2b directly binds to 5’ regulatory region of Phox2a, and sp1 biding site in promoter of Phox2a to stimulate transcription of Phox2a, respectively [33]. ERK1/2 regulate phosphorylation of Arix/Phox2a homeodomain protein, and results in decreased ability to interact with target genes [34]. Our data show miR-326 binds to 3’UTR of Phox2a, which leads to decreased expression of Phox2a in lung cancer cells (Figure 4). Our results also showed that enforced expression of Phox2a could make a difference on cell proliferation, cell cycle and apoptosis in lung cancer cells (Figures 5 and 6). Moreover, enforced expression of Phox2a compromises effects of miR-326 on cell proliferation, metastasis, cell cycle and apoptosis of lung cancer cells (Figure 7). Our findings indicate miR-326 is a negative regulator of Phox2a, thus provide new information for the regulatory mechanisms of Phox2a.
HOTAIR is a cancer-related long non-coding RNA and promotes carcinogenesis metastasis and invasion in various types of cancers [35-38]. HOTAIR negatively regulates miRNA-130a through directly binding to miRNA-130a [39]. In this study, we found that silencing of HOTAIR increases accumulation of miR-326 in both A549 and H838 cells (Figure 7B), which indicates HOTAIR is a negative regulator of miR-326. Moreover, decreased relative luciferase activities were observed in A549 cells transfected with psiCHECK-326 and siRNA of HOTAIR (Figure 7C). That’s because silencing HOTAIR increased accumulation of miR-326, and then miR-326 binds to reverse complementary sequence of pre-miR-326 in psiCHECK-326, which results in decreased luciferase activities of A549 cells. This result indicates again that HOTAIR is a negative regulator of miR-326.
Conclusion
The results of this study reveal that miR-326 regulates cell proliferation and metastasis in lung cancer by targeting Phox2a and is regulated by HOTAIR.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No: 81502513), the Jiangsu Province Natural Science Foundation of China (Grant No: BK20151028), the Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center For Cancer Personalized Medicine, Nanjing Medical University, the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801). The funding organizations had no role in study design, data collection, analysis, decision to publish and manuscript preparation.
Disclosure of conflict of interest
None.
Authors’ contribution
RW, YQS and WD conceived the design of the study, performed the experimental work and coordination. XFC, TPX, RX, LH and WMC participated in the experiments and contributed to analysis and interpretation of data. The manuscript was written by RW and YQS. All authors read and approved the final manuscript.
Abbreviations
- miR-326
microRNA 326
- Phox2a
paired-like homeobox 2a
- KLF3
Kruppel-like factor 3
- NC
negative control
- 3’UTR
3’ untranslated regions
- qRT-PCR
real-time reverse transcription PCR
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
References
- 1.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 3.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, Jiang J, diPierro CG, Li Y, Chiocca EA, Lee J, Fine H, Abounader R, Lawler S, Purow B. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–15168. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Chen X, Shu Y. Prediction of non-small cell lung cancer metastasis-associated microRNAs using bioinformatics. Am J Cancer Res. 2014;5:32–51. [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, Ghosh B. MicroRNA-326 regulates profibrotic functions of transforming growth factor-beta in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev. 2011;27:862–866. doi: 10.1002/dmrr.1262. [DOI] [PubMed] [Google Scholar]
- 7.Valencia K, Martin-Fernandez M, Zandueta C, Ormazabal C, Martinez-Canarias S, Bandres E, de la Piedra C, Lecanda F. miR-326 associates with biochemical markers of bone turnover in lung cancer bone metastasis. Bone. 2013;52:532–539. doi: 10.1016/j.bone.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Kim H, Park H, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. miR-326-histone deacetylase-3 feedback loop regulates the invasion and tumorigenic and angiogenic response to anti-cancer drugs. J Biol Chem. 2014;289:28019–28039. doi: 10.1074/jbc.M114.578229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang YF, Zhang Y, Li XY, Li C, Tian W, Liu L. Expression of miR-31, miR-125b-5p, and miR-326 in the adipogenic differentiation process of adipose-derived stem cells. OMICS. 2009;13:331–336. doi: 10.1089/omi.2009.0017. [DOI] [PubMed] [Google Scholar]
- 10.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, Wagar N, Yoon Y, Cho HT, Scala S, Shim H. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79:817–824. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 11.Kefas B, Comeau L, Erdle N, Montgomery E, Amos S, Purow B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–1112. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z. Distinct roles of DNMT1-dependent and -independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol. 2015;16:115. doi: 10.1186/s13059-015-0685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L, Hui H, Wang LJ, Wang H, Liu QF, Han SX. MicroRNA-326 functions as a tumor suppressor in colorectal cancer by targeting the nin one binding protein. Oncol Rep. 2015;33:2309–2318. doi: 10.3892/or.2015.3840. [DOI] [PubMed] [Google Scholar]
- 14.Yu S, Huang H, Deng G, Xie Z, Ye Y, Guo R, Cai X, Hong J, Qian D, Zhou X, Tao Z, Chen B, Li Q. miR-326 Targets Antiapoptotic Bcl-xL and Mediates Apoptosis in Human Platelets. PLoS One. 2015;10:e0122784. doi: 10.1371/journal.pone.0122784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 16.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem. 1998;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]
- 18.Benfante R, Flora A, Di Lascio S, Cargnin F, Longhi R, Colombo S, Clementi F, Fornasari D. Transcription factor PHOX2A regulates the human alpha3 nicotinic receptor subunit gene promoter. J Biol Chem. 2007;282:13290–13302. doi: 10.1074/jbc.M608616200. [DOI] [PubMed] [Google Scholar]
- 19.Nakano M, Yamada K, Fain J, Sener EC, Selleck CJ, Awad AH, Zwaan J, Mullaney PB, Bosley TM, Engle EC. Homozygous mutations in ARIX(PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat Genet. 2001;29:315–320. doi: 10.1038/ng744. [DOI] [PubMed] [Google Scholar]
- 20.Yazdani A, Chung DC, Abbaszadegan MR, Al-Khayer K, Chan WM, Yazdani M, Ghodsi K, Engle EC, Traboulsi EI. A novel PHOX2A/ARIX mutation in an Iranian family with congenital fibrosis of extraocular muscles type 2 (CFEOM2) Am J Ophthalmol. 2003;136:861–865. doi: 10.1016/s0002-9394(03)00891-2. [DOI] [PubMed] [Google Scholar]
- 21.Bosley TM, Oystreck DT, Robertson RL, al Awad A, Abu-Amero K, Engle EC. Neurological features of congenital fibrosis of the extraocular muscles type 2 with mutations in PHOX2A. Brain. 2006;129:2363–2374. doi: 10.1093/brain/awl161. [DOI] [PubMed] [Google Scholar]
- 22.Seo H, Hong SJ, Guo S, Kim HS, Kim CH, Hwang DY, Isacson O, Rosenthal A, Kim KS. A direct role of the homeodomain proteins Phox2a/2b in noradrenaline neurotransmitter identity determination. J Neurochem. 2002;80:905–916. doi: 10.1046/j.0022-3042.2002.00782.x. [DOI] [PubMed] [Google Scholar]
- 23.Wilzen A, Nilsson S, Sjoberg RM, Kogner P, Martinsson T, Abel F. The Phox2 pathway is differentially expressed in neuroblastoma tumors, but no mutations were found in the candidate tumor suppressor gene PHOX2A. Int J Oncol. 2009;34:697–705. doi: 10.3892/ijo_00000196. [DOI] [PubMed] [Google Scholar]
- 24.Alles M, Turchinovich G, Zhang P, Schuh W, Agenes F, Kirberg J. Leukocyte beta7 integrin targeted by Kruppel-like factors. J Immunol. 2014;193:1737–1746. doi: 10.4049/jimmunol.1302613. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Wang B, Jiang F, Wang D, Liu H, Yan Y, Dong H, Wang F, Gong B, Zhu Y, Dong L, Yin H, Zhang Z, Zhao H, Wu Z, Zhang J, Zhou J, Yu J. A feedback loop consisting of microRNA 23a/27a and the beta-like globin suppressors KLF3 and SP1 regulates globin gene expression. Mol Cell Biol. 2013;33:3994–4007. doi: 10.1128/MCB.00623-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, Xue YX. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, Ma G, Chu H, Tong N, Zhu H, Wang M, Qiang F, Zhang Z. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Fu Y, Yang F, Yang Y, Liu T, Zheng W, Zeng L, Chen T. Gracilaria lemaneiformis polysaccharide as integrin-targeting surface decorator of selenium nanoparticles to achieve enhanced anticancer efficacy. ACS Appl Mater Interfaces. 2014;6:13738–13748. doi: 10.1021/am5031962. [DOI] [PubMed] [Google Scholar]
- 29.Young HM, Ciampoli D, Hsuan J, Canty AJ. Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut. Dev Dyn. 1999;216:137–152. doi: 10.1002/(SICI)1097-0177(199910)216:2<137::AID-DVDY5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Xu H, Firulli AB, Zhang X, Howard MJ. HAND2 synergistically enhances transcription of dopamine-beta-hydroxylase in the presence of Phox2a. Dev Biol. 2003;262:183–193. doi: 10.1016/s0012-1606(03)00361-0. [DOI] [PubMed] [Google Scholar]
- 31.Adachi M, Browne D, Lewis EJ. Paired-like homeodomain proteins Phox2a/Arix and Phox2b/NBPhox have similar genetic organization and independently regulate dopamine beta-hydroxylase gene transcription. DNA Cell Biol. 2000;19:539–554. doi: 10.1089/104454900439773. [DOI] [PubMed] [Google Scholar]
- 32.Khan AO, Khalil DS, Al-Sharif LJ, Al-Tassan NA. Mutations in KIF21A and PHOX2A are absent in 16 patients with congenital vertical incomitant strabismus. Ophthalmic Genet. 2009;30:206–207. doi: 10.3109/13816810903183613. [DOI] [PubMed] [Google Scholar]
- 33.Flora A, Lucchetti H, Benfante R, Goridis C, Clementi F, Fornasari D. Sp proteins and Phox2b regulate the expression of the human Phox2a gene. J Neurosci. 2001;21:7037–7045. doi: 10.1523/JNEUROSCI.21-18-07037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh MM, Lupas G, Rychlik J, Dziennis S, Habecker BA, Lewis EJ. ERK1/2 is a negative regulator of homeodomain protein Arix/Phox2a. J Neurochem. 2005;94:1719–1727. doi: 10.1111/j.1471-4159.2005.03333.x. [DOI] [PubMed] [Google Scholar]
- 35.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 37.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, Zheng S. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emadi-Andani E, Nikpour P, Emadi-Baygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135. doi: 10.4103/2277-9175.133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, Qin YY, Gong W, Quan ZW. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]


