Abstract
Lung cancer is the most common cancer worldwide. However, no specific biomarker has been found in diagnosis and evaluation of therapeutic efficacy for lung cancer. The human lung-specific X protein gene (LUNX) was recently identified with a feature of lung tissue specificity. We applied the fluorescent quantitative polymerase chain reaction method to examine LUNX mRNA in plasma and peripheral blood mononuclear cells (PBMC) in patients with non-small cell lung cancer (NSCLC), benign lung diseases, extrapulmonary tumors, and healthy subjects. The results showed that LUNX mRNA in both of plasma and PBMC were significantly higher in lung cancer patients compared to other groups. In plasma, there were higher sensitivity and negative predictive value of LUNX mRNA than in PBMC. Patients with III~IV stages of lung cancer had more LUNX mRNA in plasma than the early stage of lung cancer sufferers. After a period of therapy, significant reductions of plasma LUNX mRNA in patients with I and II stages of lung cancer were found. Levels of plasma LUNX mRNA in patients who had succeeded to respond to therapy decreased compared to prior treatment. On the other hand, the post-treatment level was obviously increased in patients that had failed to respond to therapy. Patients with negative plasma LUNX mRNA after therapy displayed a favorable prognosis and survival rate. These preliminary data suggested that cell-free LUNX mRNA in plasma as a non-invasive biomarker, is superior to peripheral intracellular LUNX mRNA, and plays a critical role in specific diagnosis and prognostic prediction of non-small cell lung cancer.
Keywords: non-small cell lung cancer, LUNX, plasma, cell-free RNA
Introduction
Lung cancer is the most common malignancy of the respiratory system, and about three-quarters are non-small cell lung cancer (NSCLC). Because no specific symptoms occurred at early stage of lung cancer, most diagnosed cases have been at advanced stage [1]. Therefore, early diagnosis and treatment is the key step to enhance the survival of lung cancer patients. Seeking for non-invasive biomarkers with high sensitivity and specificity has extremely clinical significance for early diagnosis and therapeutic evaluation of lung cancer.
Serum tumor markers, such as neuron-specific enolase (NSE), cytokeratin fragment 21-1 (CYFRA 21-1) and squamous cell carcinoma antigen (SCC-Ag), have been widely used as convenient diagnostic markers, but their lack of sufficient sensitivity and specificity for the early detection of lung cancer has limited their application [2,3]. Several studies have determined that tumor-associated circulating cell-free mRNA in plasma or serum is increased in cancer patients [4-6]. At least some of these nucleic acids are released from tumor cells and can be considered new non-invasive biomarkers for diagnosing disease or predicting prognosis [7].
The human lung-specific X protein gene (LUNX) was cloned by Iwao and colleagues using the differential-display mRNA analysis. They found that LUNX is a molecular marker for detection of micrometastasis in NSCLC [8]. Bingle et al. reported that LUNX is predominantly expressed in lung tissue but infrequent or lack of expression in tissues outside of respiratory tract [9]. In the present study, we evaluated the level of cell-free LUNX mRNA in the plasma of lung cancer patients, and compared the diagnostic values with the intracellular target mRNA in peripheral blood mononuclear cells (PBMC). We also assessed the potential utility for plasma LUNX mRNA in monitoring of response to treatment and the prediction of prognosis in NSCLC.
Materials and methods
Subjects
Primary NSCLC patients were pathologically diagnosed in Affiliated Changzhou Second Hospital of Nanjing Medical University, China during December, 2009 and April, 2013. Totally, 80 cases were recruited for the current study, including 57 males and 23 females with an average age of 65.2 years (ranging from age 31 to 87). Among those patients, 51 cases were diagnosed as squamous cell carcinoma, 24 were adenocarcinoma and 5 were large cell carcinoma. According to the TNM system to categorize pathologic stages, there were 16 cases of stage I, 27 cases of stage II and 37 cases of stage III or IV.
Eighty-six patients suffered from benign lung diseases and treated at the hospital during the same period were selected as a benign disease group, including 33 cases of lobar pneumonia, 12 pulmonary TB cases with positive sputum cultures and 41 cases of chronic bronchitis. The mean age of this group was 59.4 years (ranges: age 21 to 89). Patients with other types of primary cancer were chosen as a special control group, including 20 cases of liver cancer, 30 cases with gastric carcinoma, 20 cases of esophageal cancer, 30 colorectal cancer patients and 20 leukemia sufferers. The average age was 66.2 years (ranges: age 32 to 83). The 100 healthy controls were recruited from the subjects routinely performing physical examination at the hospital. The average age for healthy controls was 49.3 years with a range of age 21 to age 68. All healthy controls were normal for their blood test, liver/kidney functions and chest X-ray examination. All the patients and healthy controls were informed consent and the study was approved by the Institutional Review Board (IRB).
Regeants
QIAamp Viral RNA extraction Mini Kit, QIAamp RNA Blood Mini Kit and RNase-Free DNase Set were purchased from Qiagen China Co. Ltd. (Shanghai, China). PrimeScript RT-PCR quantification kit, pMD18-T vector, and plasmid extraction kit were obtained from TaKaRa, Japan. The PCR primers and Taqman-Probe were produced by Sangon Biotech Co. Ltd., Shanghai, China. They were designed to be intro-spanning as follows: forward 5’-AAGTCTGTTGAGGCTGGCTG-3’, reverse 5’-GCCAAGTCCATCAAGCAG AG-3’, probe 5’-FAM-CCTCAACAGACTTGCACCGACCA-TAMRA-3’.
RNA extraction and cDNA synthesis
Peripheral venous blood samples (3 mL) were collected in tubes containing ethylenediaminetetraacetic acid (EDTA) from untreated patients and healthy subjects. Samples were then centrifuged within 2 hours at 4000 rpm for 20 min at 4°C (Centrifuge 5810R, Eppendorf, Hamburg, Germany). A total of 800 µl plasma from each sample was subjected to RNA extraction following the manufacturer’s instruction. The total intracellular RNA was also extracted from the peripheral blood mononuclear cells (PBMC) after plasma RNA extraction by using the RNA Blood Mini Kit. The extracted RNA was treated by DNase to remove extra DNA, and was quantified by detection the absorbance at 260 nm with a spectrophotometer (Eppendorf, Hamburg, Germany). The reverse transcription was then conducted to synthesize cDNA according to the manufacture’s instruction. Each reaction comprised of 4 µl 5X PrimeScript Buffer, 4 μl Random 6 mers, 1 μl oligo dT Primer, 1 μl PrimeScript RT Enzyme Mix I, 200 ng purified RNA, and RNase Free distilled water for a volume of 20 μl. As suggested by the manufacturer, the profile was 37°C for 15 min, 85°C for 5 sec. Subsequently, the samples were briefly centrifuged and stored at -20°C.
Fluorescence real-time quantitative polymerase chain reaction
The real-time polymerase chain reaction (PCR) method was set up according to the manual of RT-PCR quantification kit. The reaction mixture consisted of 25 μL PCR buffer solution, 1 μL (0.2 μmol/L) each of the primers, 2 μL fluorescence probe (0.2 μmol/L), 5 μL cDNA template and distilled water for a total volumn of 50 μL. The reaction was carried out in the ABI PRISM 7000 thermal cycler (Applied Biosystems, CA, USA) as follows: 95°C for 30 s, then amplification for 45 cycles with denaturation at 95°C for 5 s and annealing/extension at 60°C for 31 s. In order to obtain a standard curve, the PCR products were cloned into the pMD-18 vector following the instructions. After a serial dilution, standard data was analyzed by real-time analytical software (Applied Biosystem). Both the standards and samples were processed in duplicate.
Following of patients with NSCLC
Lung cancer patients were informed by telephone to return visit every 3 months after discharge from hospital. The survival time of each person was obtained from the information of telephone call. A second blood collection for LUNX mRNA detection and computed tomography (CT) examination were consented by cancer patients 30 days after standard treatment. The efficacy of treatment was evaluated according to the following criteria of World Health Organization (WHO) for response evaluation in solid tumors: complete response (CR)—the disappearance of all target lesions; partial response (PR)—at least a 30% decrease in the sum of the longest diameter of target lesions; progressive disease (PD)—at least a 20% increase in the sum of the longest diameter of target lesions; stable disease (SD)—neither sufficient shrinkage to qualify for partial response nor sufficient increase to qualify for progressive disease [10].
Statistical analysis
Data were analyzed with SPSS 13.0 software. The quantities of LUNX mRNA in plasma or PBMC were evaluated between lung cancer patients and other groups with Kruskall-Wallis H test. The levels of plasma LUNX mRNA before and after treatment were compared using the Wilcoxon Matched-Pairs Signed-Ranks test. Chi-square test was used to assess the differences of numeration data. A P value of <0.05 was considered statistical significance. Kaplan-Meier analyses were performed to estimate the survival of NSCLC patients. The significance of survival differences was compared by the log-rank test. A P value of less than 0.05 was considered statistical significance.
Results
Levels of LUNX mRNA in plasma and PBMC before treatment
LUNX mRNA from plasma and PBMC were amplified by real-time PCR. A signal appearing within the first 40 cycles was regarded as positive for the existence of the target mRNA. The levels of LUNX mRNA from plasma and PBMC were found to be higher in lung cancer patients compared to other groups (P<0.01) (Figures 1 and 2). Figure 3 displayed the receiver operating characteristic (ROC) curve to indicate the diagnostic values. The areas under the curve (AUC) for plasma and PBMC were 0.86 and 0.63. The cut-off values of LUNX mRNA in plasma and PBMC were 11250 copies/mL and 4905 copies/mL respectively. In plasma, the sensitivity and negative predictive value of LUNX mRNA were significantly higher than in PBMC (P<0.01) (Table 1).
Figure 1.
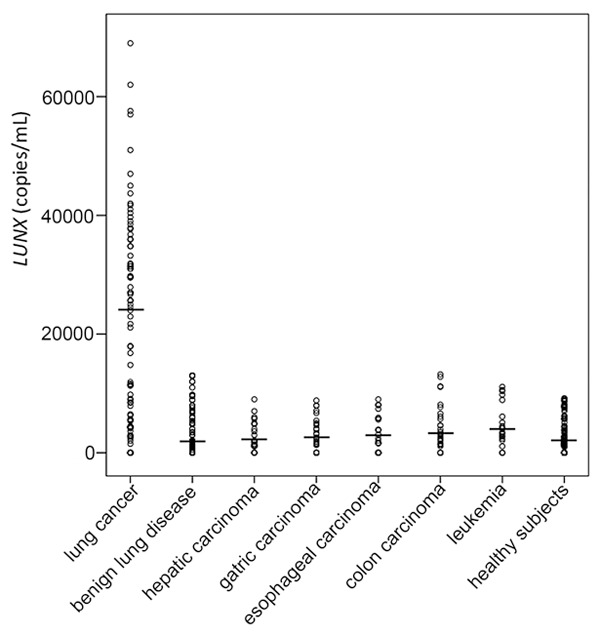
Levels of plasma LUNX mRNA in different groups. The leading dash (▬) indicates the median of each group.
Figure 2.
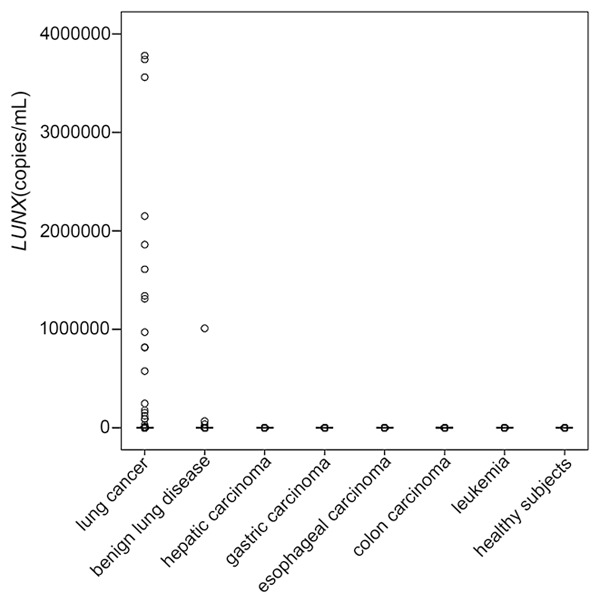
Levels of LUNX mRNA from PBMC in different groups. The leading dash (▬) indicates the median of each group.
Figure 3.
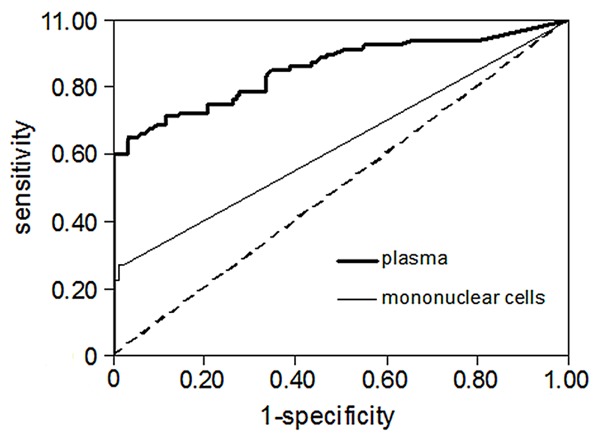
ROC curve of LUNX mRNA from plasma and PBMC.
Table 1.
Diagnostic values of LUNX mRNA from plasma and PBMC
| Source | Sensitivity | Specificity | Positive predictive value | Negative predictive value |
|---|---|---|---|---|
| Plasma | 65.0%* | 99.0% | 83.9% | 91.4%* |
| PBMC | 26.3% | 96.7% | 87.5% | 83.7% |
Compared with the source of PBMC, P<0.01.
Plasma LUNX mRNA and clinicopathologic characteristics
The possible relationship between the expression of LUNX mRNA in plasma and a series of pre-treatment tumor clinicopathologic parameters was analyzed (Table 2). A significant correlation between plasma LUNX mRNA and tumor stage was found (P<0.01) (Figure 4).
Table 2.
LUNX mRNA levels in plasma of lung cancer patients categorized by pathological characteristics
| N | LUNX [M (P 25, P 75), copies/mL] | |
|---|---|---|
| Gender | ||
| Male | 57 | 21700 (5500, 31800) |
| Female | 23 | 27000 (6301, 39700) |
| Age | ||
| <45 | 9 | 25700 (16400, 36950) |
| 45~60 | 24 | 21450 (4165, 32875) |
| >60 | 47 | 21700 (5676, 35900) |
| Histological type | ||
| Squamous carcinoma | 51 | 17900 (5517, 31900) |
| Adenocarcinoma | 24 | 27700 (5412, 40225) |
| Large cell carcinoma | 5 | 29500 (21000, 39800) |
| Stage | ||
| I | 16 | 9100 (2552, 24875) |
| II | 27 | 21700 (6500, 34800) |
| III~IV | 37 | 29600 (10100, 39100)* |
Compared with the patients of stage I, P<0.01.
Figure 4.
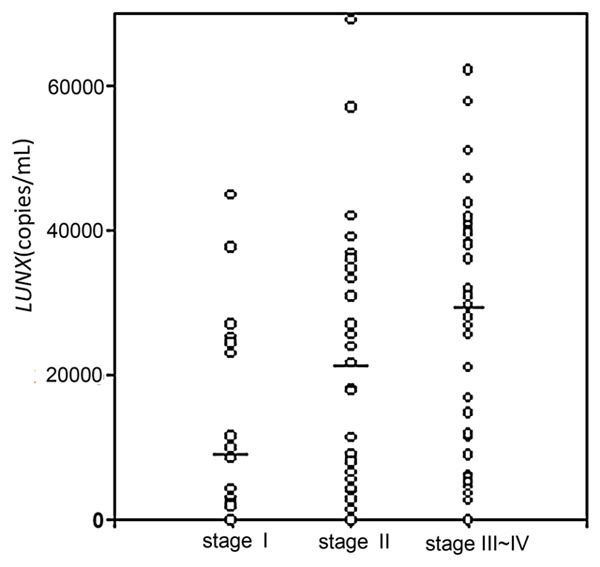
Levels of plasma LUNX mRNA in lung cancer patients under different stages. The leading dash (▬) indicates the median of each group.
Plasma LUNX mRNA and therapeutic efficacy
Comparing with the pre-treatment levels, plasma LUNX mRNA appeared significant reductions in stage I and stage II patients after therapy (P<0.01) (Figure 5). According to the data of CT, in the post-treatment patients, there were 28 cases of CR, 32 cases of PR, 9 cases of SD and 11 cases of PD. The potential correlations between plasma LUNX mRNA after treatment and the therapeutic effects were shown in Figure 6. The plasma LUNX mRNA levels in CR and PR patients after treatment were significantly decreased compared with those of prior to treatment (P<0.01). But for the PD patients, the level was increased obviously after treatment (P<0.05).
Figure 5.
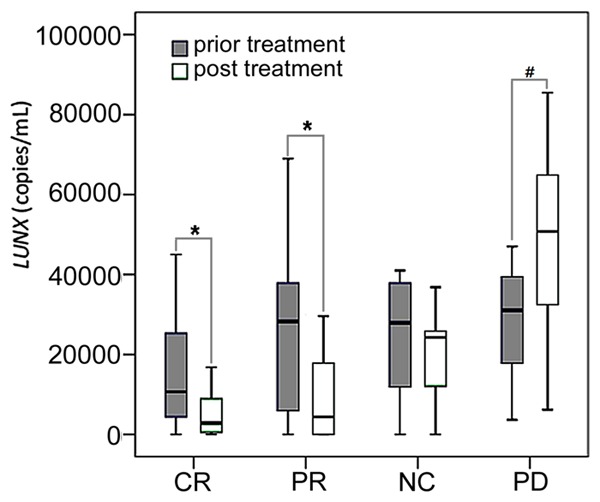
The changes of LUNX mRNA in plasma by treatment status (prior and post). In the box plots, the whiskers indicate the maximum and minimum, the boxes denote the interval between the 25th and 75th percentiles, and the lines inside the boxes mark the medians. *P<0.01; #P<0.05.
Figure 6.
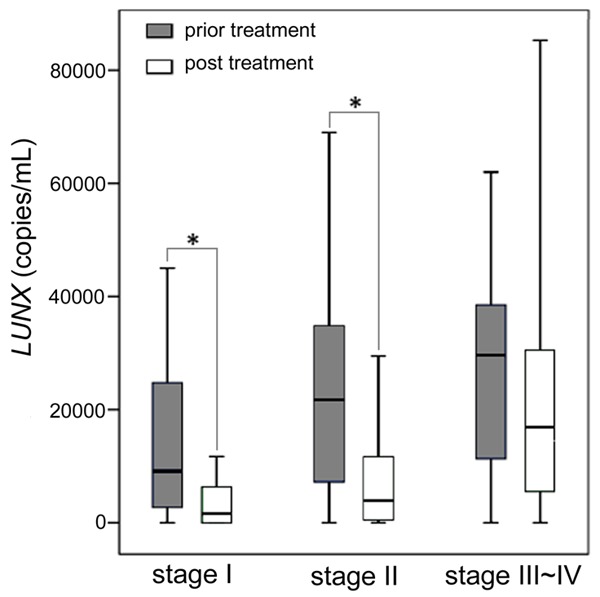
The plasma LUNX mRNA levels before and after treatment. In the box plots, the whiskers indicate the maximum and minimum, the boxes denote the interval between the 25th and 75th percentiles, and the lines inside the boxes mark the medians. *P<0.01.
Plasma LUNX mRNA and prognosis of NSCLC
Among the post-treatment lung cancer patients, 31 cases were positive for plasma LUNX mRNA (more than 11250 copies/mL), in which 19 cases of tumor metastasis or recurrence occurred within one year. On the other hand, there were 17 metastatic or recurrent lesions appeared in the patients who were negative for plasma LUNX mRNA after treatment. The incidence rate of metastasis or recurrence in patients with positive LUNX mRNA was significant higher compared to the negative group (P<0.05). The associations between plasma LUNX mRNA post therapy and survival time were revealed by Figure 7. The beginnings of patients’ survival time were calculated at the start of hospitalization. For the positive and negative LUNX mRNA patients, the median survival time were 13 and 22 months respectively. A significant difference in survival rate was observed between the two groups (P<0.01).
Figure 7.
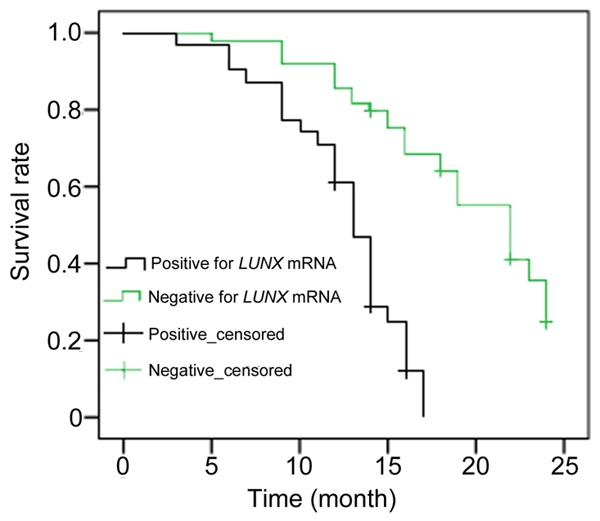
The survival curve for lung cancer patients classified by post-treatment plasma LUNX levels.
Discussion
Circulating cell-free nucleic acids mainly come from cell apoptosis or spontaneous incretion [11,12]. Most RNA combined with protein and lipid after releasing from cells to form a complex, which temporally protect RNA from nuclease degradation [13-15]. The quantities of circulating RNA were generally higher for cancer patients compared with healthy subjects because of the high proliferation and frequent apoptosis of tumor cells [16].
LUNX is a recently isolated lung tissue specific gene, which is located on chromosome 20p11.1-q12. The full-length cDNA contains 1,015 nucleotides including an open reading frame to encode a protein of 256 amino acids and a relative molecular weight of 26.7×103 [8]. It was reported that LUNX was a molecular marker for detecting micrometastasis in non-small cell lung cancer [17]. Other studies focused on the auxiliary diagnosis of lung cancer by detection of LUNX mRNA in PBMC [18,19]. To date, there has no research work focusing on the detection of circulating cell-free LUNX mRNA in patients with lung cancer. In the current study, we examined both the cellular and cell-free LUNX mRNA for NSCLC patients using fluorescent quantitative PCR. We found that the levels of LUNX mRNA in plasma or PBMC were significant higher for lung cancer patients compared with the patients of benign lung diseases, extrapulmonary tumors and healthy subjects. It suggested that both of the cell-free and intracellular LUNX mRNA may be applied in adjuvant diagnosis of lung cancer.
The diagnostic values of LUNX mRNA from plasma and PBMC were evaluated by ROC curve. We found that LUNX mRNA in both of plasma and PBMC had high specificity for lung cancer. But the sensitivity of LUNX mRNA in PBMC was quite lower than the results of previous studies [20,21]. We thought it was the different statistical methods that led to the different sensitivities of LUNX in PBMC. Because LUNX mRNA did not express in control groups in their research work, it was considered positive that the target mRNA was detectable in previous studies. However it is not applicable in current study. In our work, we found plasma LUNX mRNA was detectable not only in lung cancer patients, but also in amounts of other study populations including healthy subjects (Figure 1). So we calculated cut-off values through ROC curve to distinguish positivity and negativity for LUNX levels in plasma and PBMC, which caused part of detectable persons been regarded as negative. It was showed, in our work, that the sensitivity of LUNX mRNA detection in plasma was obviously higher than the determination in PBMC. This data may suggest that cell-free mRNA is superior to the cellular determination because of the higher sensitivity and negative predictive value.
Our data also found that plasma LUNX mRNA was significantly higher in patients with advanced lung cancer compared with those of stage I patients, which indicated the potential role of predicting tumor prognosis. The study showed that levels of plasma LUNX mRNA reflect a useful indicator of treatment effect in lung cancer patients. For the effective patients (CR and PR), LUNX levels were significantly decreased after treatment, while in the ineffective patients, it was the same or even increased. Therefore, by observing the levels of plasma LUNX mRNA before and after treatment may provide an easy method for evaluating the treatment effect.
The plasma LUNX mRNA was decreased for stage I and II lung cancer patients after treatment. We found that patients with negative post-treatment free LUNX mRNA had a favorable prognosis and survival. It suggested that measurement of post-treatment plasma LUNX mRNA may benefit patients for active monitor of chemotherapy efficacy. It may also aid patients with higher post-treatment plasma LUNX in early prevention and active treatment.
Using circulating cell-free nucleic acid as the surrogate of target tissue to detect cancer biomarkers is a convenient and non-invasive approach. It can be widely applied to cancer screening and monitoring treatment efficacy. Cancer screening is usually conducted in normal or high risk population. It requires biomarkers with sufficient sensitivity and specificity. Our data showed that the content of LUNX mRNA in plasma was quite low, and whether the sensitivity of plasma LUNX mRNA is efficient in screening of lung cancer needs further exploration. However, plasma LUNX mRNA is still a novel biomarker in auxiliary diagnosis and evaluating therapeutic efficacy of lung cancer with high specificity, especially for those at higher risk, such as smokers.
Acknowledgements
This work was supported by the College Science Foundation of Nanjing Medical University. Grant No. 09NJMUM057.
Disclosure of conflict of interest
None.
References
- 1.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 2.Karnak D, Beder S, Kayacan O, Ibis E, Oflaz G. Neuron-specific enolase and lung cancer. Am J Clin Oncol. 2005;28:586–590. doi: 10.1097/01.coc.0000177915.51805.6e. [DOI] [PubMed] [Google Scholar]
- 3.Molina R, Auge JM, Escudero JM, Marrades R, Vinolas N, Carcereny E, Ramirez J, Filella X. Mucins CA 125, CA 19.9, CA 15.3 and TAG-72.3 as tumor markers in patients with lung cancer: comparison with CYFRA 21-1, CEA, SCC and NSE. Tumour Biol. 2008;29:371–380. doi: 10.1159/000181180. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Xu W, Qian H, Yin Q, Zhu W, Yan Y. Circulating RNA as a novel tumor marker: an in vitro study of the origins and characteristics of extracellular RNA. Cancer Lett. 2008;259:50–60. doi: 10.1016/j.canlet.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Zhou H, Qian H, Bu X, Chen D, Gu H, Zhu W, Yan Y, Mao F. Combination of circulating CXCR4 and Bmi-1 mRNA in plasma: A potential novel tumor marker for gastric cancer. Mol Med Rep. 2009;2:765–771. doi: 10.3892/mmr_00000170. [DOI] [PubMed] [Google Scholar]
- 6.Milas M, Shin J, Gupta M, Novosel T, Nasr C, Brainard J, Mitchell J, Berber E, Siperstein A. Circulating thyrotropin receptor mRNA as a novel marker of thyroid cancer: clinical applications learned from 1758 samples. Ann Surg. 2010;252:643–651. doi: 10.1097/SLA.0b013e3181f5ba51. [DOI] [PubMed] [Google Scholar]
- 7.Swarup V, Rajeswari MR. Circulating (cellfree) nucleic acids--a promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 8.Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M, Tanigami A. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. Int J Cancer. 2001;91:433–437. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1059>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 9.Bingle L, Bingle CD. Distribution of human PLUNC/BPI fold-containing (BPIF) proteins. Biochem Soc Trans. 2011;39:1023–1027. doi: 10.1042/BST0391023. [DOI] [PubMed] [Google Scholar]
- 10.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 11.Stroun M, Anker P, Beljanski M, Henri J, Lederrey C, Ojha M, Maurice PA. Presence of RNA in the nucleoprotein complex spontaneously released by human lymphocytes and frog auricles in culture. Cancer Res. 1978;38:3546–3554. [PubMed] [Google Scholar]
- 12.Biggiogera M, Bottone MG, Pellicciari C. Nuclear RNA is extruded from apoptotic cells. J Histochem Cytochem. 1998;46:999–1005. doi: 10.1177/002215549804600903. [DOI] [PubMed] [Google Scholar]
- 13.Wieczorek AJ, Rhyner C, Block LH. Isolation and characterization of an RNA-proteolipid complex associated with the malignant state in humans. Proc Natl Acad Sci U S A. 1985;82:3455–3459. doi: 10.1073/pnas.82.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260:248–256. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 15.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 16.El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, Godfrey TE. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50:564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Qi H, Zhou HX, Deng CY, Zhu H, Li JF, Wang XL, Li FR. Detection of micrometastases in peripheral blood of non-small cell lung cancer with a refined immunomagnetic nanoparticle enrichment assay. Int J Nanomedicine. 2011;6:2175–2181. doi: 10.2147/IJN.S24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng M, Chen Y, Yu X, Tian Z, Wei H. Diagnostic utility of LunX mRNA in peripheral blood and pleural fluid in patients with primary nonsmall cell lung cancer. BMC Cancer. 2008;8:156. doi: 10.1186/1471-2407-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Huang X, Zhu Z, Hu Y, Ou W, Zhang L, Zhou N. Significance of combined detection of LunX mRNA and tumor markers in diagnosis of lung carcinoma. Chin J Cancer Res. 2014;26:89–94. doi: 10.3978/j.issn.1000-9604.2014.02.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitas M, Hoover L, Silvestri G, Reed C, Green M, Turrisi AT, Sherman C, Mikhitarian K, Cole DJ, Block MI, Gillanders WE. Lunx is a superior molecular marker for detection of nonsmall cell lung cancer in peripheral blood [corrected] . J Mol Diagn. 2003;5:237–242. doi: 10.1016/s1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimi S, Mohamadnia A, Nadji SA, Yadegarazari R, Khosravi A, Bahrami N, Saidijam M. Expression of two basic mRNA biomarkers in peripheral blood of patients with non-small cell lung cancer detected by real-time rt-PCR, individually and simultaneously. Iran Biomed J. 2015;19:17–22. doi: 10.6091/ibj.1397.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]


