Abstract
MicroRNAs (miRNAs) dysregulation is a common event in a variety of human diseases including breast cancer. However, clinical relevance and biological role of miR-654-5p in the progression of breast cancer remain greatly elusive. Herein, the expression levels of miR-654-5p were aberrantly downregulated in human breast cancer specimens and four breast cancer cell lines. Low expression of miR-654-5p was strongly associated with advanced TNM stage and lymph node metastasis as well as a poor survival. Functional analysis showed that miR-654-5p overexpression inhibited cell growth and invasion, and induced cell apoptosis in two aggressive breast cancer cells. Further studies demonstrated that Epithelial stromal interaction 1 (EPSTI1) was a direct target gene of miR-654-5p and showed an inverse correlation with miR-654-5p expression. Forced expression of EPSTI1 could abrogate the inhibitory effect of miR-654-5p on the growth and invasion of breast cancer cells as well as apoptosis-induced ability. In conclusion, the present study highlights that miR-654-5p acts as a tumor suppressor in breast cancer through directly targeting EPSTI1, and their functional regulation may open a novel avenue with regard to the therapeutic target for breast cancer.
Keywords: miRNA, breast cancer, miR-654-5p, EPSTI1, survival
Introduction
Breast cancer remains to be the most common cancer and a major cause of cancer death in women across the world [1]. Breast cancer is a heterogeneous disease characterized by diverse pathological features [2]. Despite the great advances in the early diagnosis and therapeutic strategies of breast cancer, the long-term survival in patients, especially patients with metastatic breast cancer, remains unsatisfactory [3]. Like that in other solid malignancies, increasing mortality of patients with breast cancer derives from tumor metastasis [4]. It has been well established that tumor metastasis is a complex, multistep and multifactor process, including tumor cells migration from the primary residence through invasiveness of blood vessel, dissemination by the circulation, and ultimately colonization at distant organs [5]. The initiator of tumor metastasis is the ability of tumor cells invasion via intravasation [6]. Although epithelial-mesenchymal transition is proposed to be critical for the development of metastatic tumor cells, mechanism with regard to acquisition of invasive capability is not fully illustrated.
MicroRNAs (miRNAs) are a class of noncoding RNAs with 18-23 nucleotides in length, which control the expression of target genes through specific binding to the 3’-untranslated region (3’-UTR) of target mRNAs [7]. MiRNAs are differentially expressed in various biological processes, including cell growth, apoptosis, and differentiation [8]. As previously described, miRNAs are commonly deregulated during carcinogenesis and tumor metastasis in a variety of tumors [9,10]. A series of miRNAs have been documented as oncogenes and/or tumor suppressor genes [11]. Due to miRNAs stability and length, a few investigations have reported that several miRNAs serve as novel diagnostic and risk prognostic biomarkers, as well as personal therapeutic targets of breast cancer. Ectopic expression of miR-1 is associated with the progression of breast cancer and is a negative prognostic factor in breast cancer patients [12]. MiR-320a functions as a tumor suppressor in breast cancer through involvement of Rab11a and Akt signaling pathway [13]. MiR-630 is involved in the regulation of metastatic potential of breast cancer through targeting metadherin [14].
Epithelial stromal interaction 1 (EPSTI1), mapped to chromosome 13q13.3, is initially reported as a stromal fibroblast-induced gene in human breast cancer and highly upregulated in tumor tissues of breast cancer patients [15,16]. Subsequent studies have been shown that abnormal expression of EPSTI1 plays an important role in breast cancer invasion, metastasis, and apoptosis [17] as well as a poor prognosis in breast cancer patients [16].
MiR-654-5p, identified from the miRNA microarrays at chromosome 14q32.31, has been verified to be deregulated and involved in prostate cancer via the androgen receptor [18]. Herein, we showed that miR-654-5p was significantly decreased in breast cancer tissues and cell lines, and its overexpression inhibited cell growth and invasion by inducing cell apoptosis. Importantly, EPSTI1 was identified to be a target gene of miR-654-5p and reversely modulated the functions of miR-654-5p. Our findings suggest a promising new insight with regard to the potential mechanism underlying tumor suppressive function of miR-654-5p in breast cancer.
Materials and methods
Tissue samples
A total of 110 patients who were diagnosed as breast cancer and had undergone surgical resection in the Zhongda Hospital of Southeast University and Yichang Central People’s Hospital from 2009 to 2010 were enrolled in this study. Written informed consent was collected from each patient, and the Ethics Committee of The First College of Clinical Medical Science, China Three Gorges University approved this procedure. All samples were histologically confirmed. None of patients received chemotherapy or radiotherapy prior to surgery. The clinical characteristics are summarized in Table 1. The tissue samples were frozen in liquid nitrogen after resection and immediately stored at -80°C until RNA extraction.
Table 1.
Association between miR-654-5p and baseline characteristics
| miR-654-5p expression | ||||
|---|---|---|---|---|
|
|
||||
| Factors | Number of cases | Low (n=57) | High (n=53) | P valuea |
| Age (years) | ||||
| <62 | 55 | 25 | 30 | 0.182 |
| ≥62 | 55 | 32 | 23 | |
| Family history | ||||
| Absent | 102 | 52 | 50 | 0.794 |
| Present | 8 | 5 | 3 | |
| Tumor grade | ||||
| I-II | 86 | 44 | 42 | 0.795 |
| III | 24 | 13 | 11 | |
| TNM stage | ||||
| I-II | 63 | 38 | 25 | 0.039 |
| III-IV | 47 | 19 | 28 | |
| Lymph node metastasis | ||||
| Absent | 53 | 22 | 31 | 0.037 |
| Present | 57 | 35 | 22 | |
| ER status | ||||
| Negative | 60 | 31 | 29 | 0.972 |
| Positive | 50 | 26 | 24 | |
| PR status | ||||
| Negative | 13 | 8 | 5 | 0.455 |
| Positive | 97 | 49 | 48 | |
| Her2/Neu status | ||||
| Negative | 32 | 16 | 16 | 0.807 |
| Positive | 78 | 41 | 37 | |
ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor receptor type 2 also referred to as Her2/Neu.
Chi-square test.
Cell lines and cell culture
Four human breast cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-468 and BT-549) and a normal breast epithelial cell line (HBL-100) were purchased from Shanghai Cell Collection of Chinese Academy of Sciences (Shanghai, China). Cells were maintained in RPMI-1640 (GIBCO, CA, USA) with 10% fetal bovine serum (FBS; GIBCO, CA, USA), and 1% of penicillin and streptomycin at 37°C with 5% CO2.
RNA extraction and quantitative real-time PCR
Total RNA and miRNA were extracted from cells and tissue samples using Trizol LS and Trizol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The cDNA was synthesized following the PrimeScript RT-polymerase assay instruction (Takara, Dalian, China) and was subsequently analyzed for quantitative real-time PCR (qPCR) using specific primers of miR-654-5p and EPSTI1 (Sangon, Shanghai, China). U6 or GAPDH was used as an internal control for miR-654-5p or EPSTI1. The qPCR was performed using Applied Biosystems 7500 Real-time PCR System (Applied Biosystems, CA, USA) and the SYBR Premix Ex TaqTM (Takara, Dalian, China). Relative expression levels of target genes were determined by the 2-∆∆Ct method [19].
Prediction of target gene and construction of recombinant vector
Target prediction of miR-654-5p was performed on the online software program from the website www.targetscan.org. To stably upregulate mature miR-654-5p, expression vectors using pCDH-CMV-MCS-EF1 (miR-654-5p) were purchased from Genechem institute (Shanghai, China). A vector with non-specific miRNA served as a control (miR-NC). As previously reported, we have also constructed a pcDNA3.1-EPSTI1 plasmid containing the gene promoter of EPSTI1 to induce human EPSTI1 overexpression [17], and an empty plasmid was used as a control.
Dual luciferase reporter assay
To construct luciferase reporter vectors, EPSTI1 3’-untranslated region (UTR) containing the predicted target binding sites or mutant sites of miR-654-5p was amplified by PCR and then cloned into pMIR-Report luciferase vector (Promega). MDA-MB-468 and BT-549 cells seeded in the 96-well plate were co-infected with miR-NC or miR-654-5p and luciferase reporter containing wild type or mutant type of EPSTI1 3’-UTR by the Lipofectamine 2000 (Invitrogen). Luciferase activity was determined through the dual luciferase reporter assay system (Promega) after 48 h infection.
Cell proliferation by CCK-8 assay
Cells were embedded in 96-well plates at a density of 5000 cells per well and cultured for 12 h, 24 h, 48 h, 72 h and 96 h, respectively. Changes of cell proliferation were determined with a Cell Counting Kit-8 (KeyGen, Nanjing, China) according to the manufacturer’s protocol. Results are shown as absorbance at 450 nm.
Cell apoptosis by flow cytometry
Cells were harvested after 48 h transfection and stained using the Annexin V-FITC and propidium iodide (BD Bioscience, CA, USA) following the manufacturer’s instructions. All apoptosis assays were conducted in triplicate.
Matrigel invasion assays
Matrigel-coated BD Falcon 8 μm pore inserts (24-well insert, BD Biosciences, CA, USA) were employed to assess the ability of cell invasion. Cells were seeded in the top chamber of Matrigel-coated inserts and cultured for 24 h at 37°C. The lower chamber was supplemented with 10% FBS. Cells that did not invade through the pores of the top chamber were removed with a cotton swab, and cells invaded to the lower chamber were stained with crystal violet and counted under a light microscope.
Western blot analysis
The MDA-MB-468 and BT-549 Cell lysates were collected following routine procedures. Protein lysates were isolated by 9% SDS-PAGE and transferred onto nitrocellulose (Axygen, CA, USA). The membranes were blocked with TBST and incubated with the following primary antibodies: rabbit polyclonal anti-EPSTI1 (1:2000, Sigma Aldrich, CA) and anti-GAPDH (1:1000, Santa Cruz, CA). Secondary antibody was added and incubated for 2 h after being washed. Binds detection was determined with an ECL system (Amersham Pharmacia, Piscataway, NJ).
Statistical analysis
Statistical analyses were performed using SPSS 20.0 software (IBM, USA). The Wilcoxon test was employed to assess miR-654-5p expression in 110 pairs of human breast cancer and corresponding normal breast tissues. The association between miR-654-5p and baseline characteristics was evaluated by chi-square test. Comparison of in vitro data was analyzed by independent t test, or one-way analysis of variance, as appropriate. The correlation between miR-654-5p and EPSTI1 mRNA was identified by Spearman’s correlation analysis. The optimal cutoff value of miR-654-5p was determined by the receiver operating characteristic curve (ROC) analysis. Survival curves were plotted by the Kaplan-Meier method and the significance was assessed by the log-rank test. The influence of miR-654-5p and clinical characteristics on overall survival was further evaluated by univariate and multivariate Cox proportional hazards model. P value less than 0.05 was considered as statistical significance.
Results
MiR-654-5p is aberrantly attenuated in human breast cancer and associated with worse outcome
The expression levels of miR-654-5p were assessed by qPCR in 110 pairs of human breast cancer and corresponding normal breast tissues. We observed that the expression of miR-654-5p was markedly attenuated in breast cancer tissues compared to corresponding normal tissues (P<0.001, Figure 1A). To further validate this result, we examined the expression of miR-654-5p in breast cancer cell lines MCF-7, MDA-MB-231, MDA-MB-468, BT-549, and breast epithelial cell line HBL-100 by qPCR. The results showed that breast cancer cell lines had lower miR-654-5p expression levels than normal breast epithelial cells (P<0.01, Figure 1B). Additionally, ROC analysis indicated that the optimal cutoff value of miR-654-5p expression was 0.35-fold with the largest Youden’s index (0.48, sensitivity 0.70, specificity 0.78) based on overall survival (Figure 1C). Patients with breast cancer were then divided into low (<0.35-fold) and high (≥0.35-fold) groups. Clinical characteristics analyses that the low expression of miR-654-5p was associated with advanced TNM stage (P=0.039) and lymph node metastasis (P=0.037); whereas, there was no significant association between miR-654-5p and other clinical factors such as age, family history, tumor grade, estrogen receptor status, progesterone status and Her2/Neu status (P>0.05, Table 1).
Figure 1.
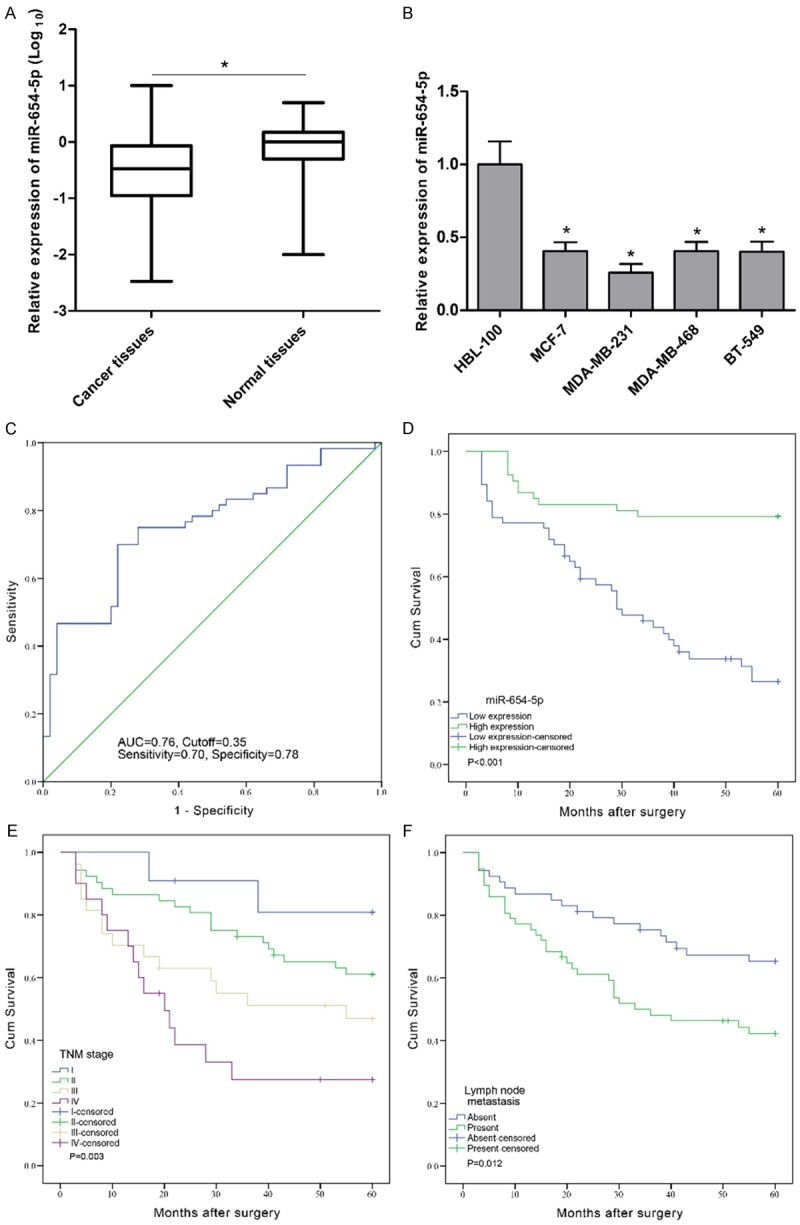
Relative expression of miR-654-5p in breast cancer tissues and cell lines as well as its correlation with overall survival of breast cancer patients. (A) MiR-654-5p expression was measured by qPCR and normalized to U6 expression in 110 paired breast cancer specimens, *P<0.001. (B) qPCR of miR-654-5p expression in HBL-100, MCF-7, MDA-MB-231, MDA-MB-468, and BT-549 cells and U6 acted as reference, *P<0.01. (C) The optimal cutoff value of miR-654-5p was determined by ROC analysis according to overall survival. Kaplan-Meier survival curve and log-rank test were employed to evaluate the associations between miR-654-5p expression (D), TNM stage (E), lymph node metastasis (F), and overall survival in breast cancer patients.
To further understand the prognostic significance of miR-654-5p in breast cancer, Kaplan-Meier survival analysis was performed according to postoperative 5-year survival for breast cancer patients. Our results demonstrated that low expression of miR-654-5p was significantly correlated with worse survival (P<0.001, Figure 1D), as well as advanced TNM stage (P=0.003, Figure 1E) and lymph node metastasis (P=0.012, Figure 1F). Furthermore, univariate and multivariate analyses showed miR-654-5p and TNM stage were identified to be independent prognostic indicators for overall survival in patients with breast cancer (Table 2). Taken together, these results suggest that the dynamic changes of miR-654-5p expression may be common events during the progression and prognosis of human breast cancer.
Table 2.
Influence of miR-654-5p expression and clinical risk factors on overall survival in breast cancer patients
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
||||
| Factors | HR (95% CI) | P value | HR (95% CI) | P value |
| Age (≥62/<62) | 1.47 (0.84-2.56) | 0.180 | ||
| Family history (Present/Absent) | 1.38 (0.55-3.48) | 0.495 | ||
| Tumor grade (III/I-II) | 1.91 (1.05-3.46) | 0.033 | 1.72 (0.95-3.13) | 0.073 |
| TNM stage (III-IV/I-II) | 2.36 (1.35-4.14) | 0.003 | 2.25 (1.28-3.96) | 0.005 |
| Lymph node metastasis (Present/Absent) | 2.05 (1.15-3.66) | 0.015 | 0.82 (0.27-2.46) | 0.717 |
| ER status (Positive/Negative) | 0.70 (0.40-1.23) | 0.218 | ||
| PR status (Positive/Negative) | 0.76 (0.34-1.70) | 0.506 | ||
| Her2/Neu status (Positive/Negative) | 0.79 (0.44-1.41) | 0.422 | ||
| miR-654-5p (Low/High) | 4.86 (2.47-9.56) | <0.001 | 4.74 (2.41-9.32) | <0.001 |
ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor receptor type 2 also referred to as Her2/Neu. HR, hazard ratio; CI, confidence interval.
Overexpression of miR-654-5p inhibits cell growth and invasion as well as induces cell apoptosis in breast cancer cell lines
To evaluate the effect of miR-654-5p on breast cancer progression, we constructed an overexpression vector carrying miR-654-5p. The aggressive breast cancer cell lines MDA-MB-468 and BT-549 were stably transfected with overexpression vectors (miR-654-5p) or negative control (miR-NC). As shown in Figure 2A and 2B, miR-654-5p but not miR-NC significantly inhibited the growth property of MDA-MB-468 and BT-549 cells in a time-dependent manner. Analysis of apoptosis suggested that ectopic expression of miR-654-5p induced cell apoptosis in MDA-MB-468 and BT-549 cells (Figure 2C and 2D). Matrigel invasion assay also showed that miR-654-5p dramatically suppressed the invasion ability of MDA-MB-468 and BT-549 cells (Figure 2E and 2F).
Figure 2.
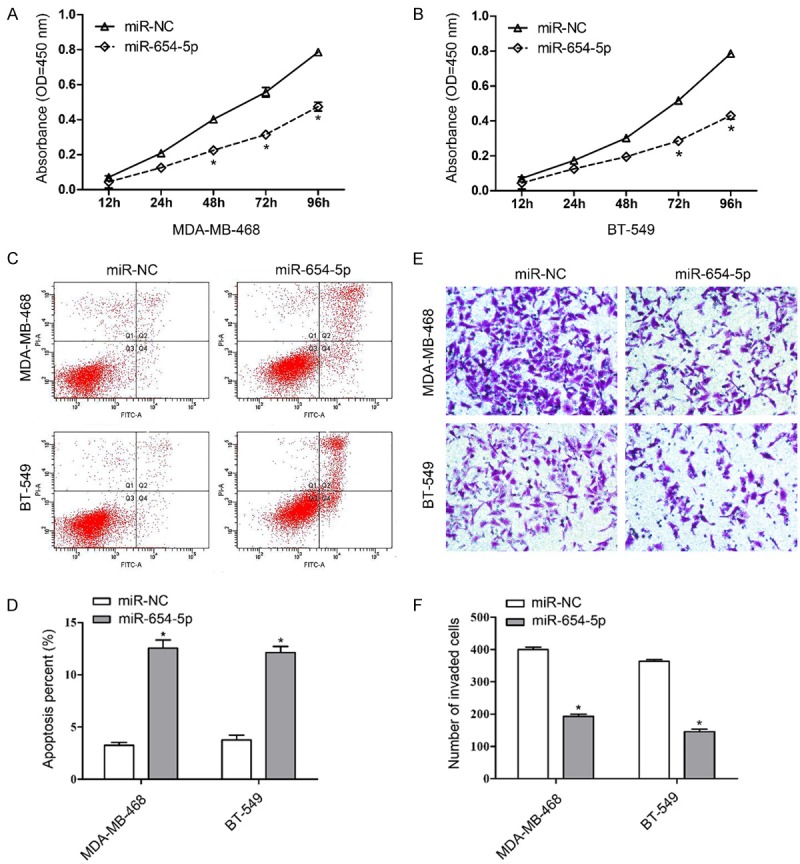
Ectopic expression of miR-654-5p inhibits cell proliferation, invasion and induces cell apoptosis of breast cancer cells. Cell proliferation was performed in MDA-MB-468 (A) and BT-549 (B) cells stably transfected with expression vectors carrying miR-654-5p or mi-NC. (C) Cell apoptosis was examined by flow cytometry with Annexin V-FITC and propidium iodide staining. (D) Cell apoptotic rate was counted according to summation of the second quadrant and fourth quadrant. (E) Matrigel invasion assays were conducted to determine breast cancer cell invasive ability. (F) The number of invaded cells was calculated, mean ± SD, *P<0.01.
MiR-654-5p directly targets EPSTI1 in human breast cancer
To systematically decipher the molecular me-chanisms underlying biological effect of miR-654-5p in breast cancer progression, bioinformatics was used to predict target sites of miRNA through the online software TargetScan [20]. Recently, it has been well established that EPSTI1, a stromal fibroblast-induced gene, highly elevated in tumor tissues of breast cancer patients [15,16]. Through bioinformatics algorithms, we observed that EPSTI1 may serve as a direct target of miR-654-5p (Figure 3A). According to these findings, we proposed a hypothesis that EPSTI1 may be involved in the biological role of miR-654-5p in breast cancer. To confirm this hypothesis, we firstly performed a dual luciferase assay and observed that miR-654-5p overexpression significantly decreased the luciferase activity of Wt-3’UTR of EPSTI1 but not that of the vector and Mt-3’UTR of EPSTI1 in these breast cancer cells (Figure 3B). Next, we explored the effect of miR-654-5p on inhibition of EPSTI1 protein and mRNA. The results showed that the expression of EPSTI1 protein and mRNA was dramatically reduced in expression vectors carrying miR-654-5p compared to miR-NC in MDA-MB-468 and BT-549 cells (Figure 3C and 3D). To further support this assertion, we randomly detected EPSTI1 expression levels of 70 paired tissue samples from 110 patients with breast cancer by qPCR. Correlation analysis of expression of miR-654-5p and EPSTI1 in 70 paired tumor tissues exhibited a significant inverse correlation between miR-654-5p and EPSTI1 (P<0.001, Figure 3E). Collectively, these results suggest that EPSTI1 may be a potential target gene of miR-654-5p.
Figure 3.
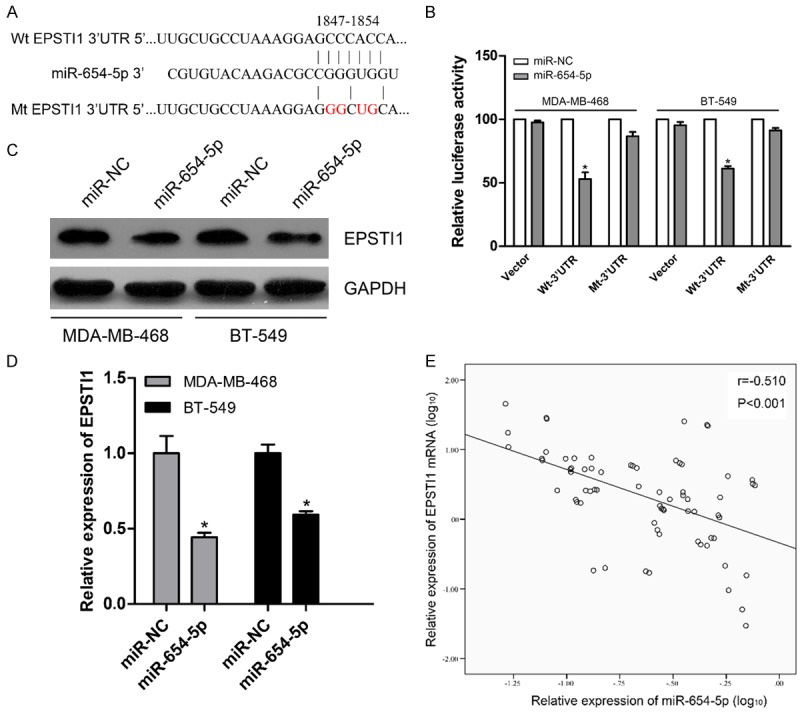
MiR-654-5p targets EPSTI1 in breast cancer cell lines. A. Putative miR-654-5p binding sites in the 3’-UTR of EPSTI1 mRNA. B. The wild type and mutant of 3’-UTR reporter vectors and control group were co-transfected into breast cancer cells with miR-654-5p or miR-NC. The relative luciferase activities were evaluated. C. Cellular lysates from breast cancer cells were used to measure EPSTI1 expression by western blot. D. The expression of EPSTI1 mRNA was examined by qPCR in the transfected breast cancer cells. E. The correlation analysis of miR-654-5p and EPSTI1 mRNA expression levels in 70 breast cancer specimens. All experiments were performed in triplicate, mean ± SD, *P<0.01.
EPSTI1 is involved in miR-654-5p-mediated cell growth, invasion and apoptosis of breast cancer cells
To determine whether miR-654-5p exerts its biological function through its target gene EPSTI1, expression vectors carrying miR-654-5p or miR-NC were co-transfected with EPSTI1-induced expression vectors or negative control respectively into MDA-MB-468 and BT-549 cells. The results of western blot showed that ectopic expression of EPSTI1 partially restored its levels inhibited by miR-654-5p (Figure 4A). Further analyses of CCK-8 and invasion assays suggested that EPSTI1 overexpression markedly reversed cell growth and invasion suppressed by miR-654-5p in MDA-MB-468 and BT-549 cells (Figure 4B and 4D). EPSTI1 also exhibited a similar reverse effect of cell apoptosis induced by miR-654-5p in breast cancer cells (Figure 4C). These results further suggest that the function of miR-654-5p in breast cancer cells is mediated through its modulation of EPSTI1 expression.
Figure 4.
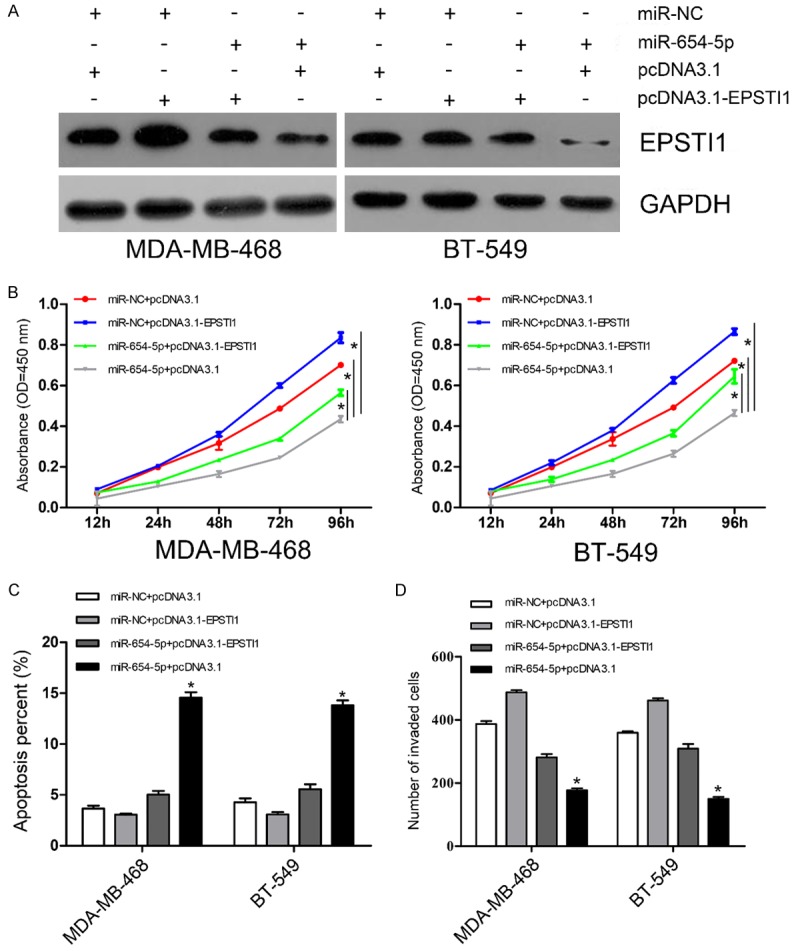
EPSTI1 attenuates the suppressive effect of miR-654-5p on cell proliferation and invasion as well as apoptosis-induced ability. A. Western blot analysis of EPSTI1 protein expression in breast cancer cells carrying miR-NC or miR-654-5p transfected with either pcDNA3.1 or pcDNA3.1-EPSTI1, GAPDH was used as a loading control. B. CCK-8 assay detecting the proliferation of breast cancer cells carrying miR-NC or miR-654-5p transfected with either pcDNA3.1 or pcDNA3.1-EPSTI1. C. Cell apoptosis was examined in breast cancer cells carrying miR-NC or miR-654-5p transfected with either pcDNA3.1 or pcDNA3.1-EPSTI1 by flow cytometry. D. Matrigel invasion assay evaluating invasion of breast cancer cells carrying miR-NC or miR-654-5p transfected with either pcDNA3.1 or pcDNA3.1-EPSTI1. Each assay was repeated three times. *P<0.05.
Discussion
It has been well established that miRNAs participate in tumorigenesis and tumor progression through the modulation of various target genes. Cancer metastasis is a major cause of all cancer related deaths [21]. Blockade of cancer development or metastasis would therefore be an approbatory approach for cancer treatment. It is very important to find potential biomarkers for inhibition of cancer metastasis, risk prognostication and personal therapy screening of breast cancer patients. In the present study, we have demonstrated that the expression of miR-654-5p was decreased in the majority of breast cancer tissues and in breast cancer cell lines. Meanwhile, analyses of clinical characteristics revealed that low expression of miR-654-5p was strongly correlated with advanced TNM stage and lymph node metastasis as well as worse overall survival. Furthermore, miR-654-5p may be an independent prognostic factor for overall survival in breast cancer patients. Further functional analysis suggested the involvement of miR-654-5p in the progression of breast cancer, and forced expression of miR-654-5p significantly inhibited proliferation and invasion as well as induced cell apoptosis in two aggressive breast cancer cell lines MDA-MB-468 and BT-549 cells. This is the first report to verify the functional performance of miR-654-5p expression in human breast cancer, and our results reveal that miR-654-5p acts as a tumor suppressor to participate in the progression of breast cancer. Accordingly, miR-654-5p provides a promising new insight with regard to the diagnostic and prognostic biomarker and therapeutic target for breast cancer.
Mounting studies focus on miRNAs as biomarkers and miRNA-mediated precise medicine for malignancies. Several differentially expressed miRNAs have been confirmed in breast cancer and associated with poor survival. For example, miR-21, miR-210, miR-373, let-7a, etc. are recently identified as diagnostic or prognostic indicators for breast cancer [22-24]. To date, miR-654-5p, mapped to the 14q32.31 locus, has been only detected in prostate cancer and exerts stronger inhibitors of tumor growth than blockade of androgen receptor or treatment with Casodex [18]. In this study, we demonstrated that downregulation of miR-654-5p was observed in majority cases of 110 breast cancer specimens. Additional correlation analysis suggested that low expression of miR-654-5p had little correlation with clinical characteristics such as age, family history, tumor grade, estrogen receptor status, progesterone status and Her2/Neu status (Table 1). Survival analysis revealed that patients with low expression of miR-654-5p had a shorter survival than those with high expression of miR-654-5p (Figure 1D). Taken together, these results suggest that aberrant expression of miR-654-5p is a frequent event in breast cancer and may be involved in breast cancer progressive process.
Furthermore, we identified EPSTI1 as a direct target of miR-654-5p in breast cancer cell lines and showed that tumor inhibitory property of miR-654-5p was fulfilled by the modulation of EPSTI1 expression. EPSTI1, an interferon response gene mapped to chromosome 13q13.3, is initially identified to be highly overexpressed in invasive breast cancer characterized by extensive epithelial-stromal interaction [15]. The subsequent experiment showed that EPSTI1 regulates tumor cell properties and epithelial-mesenchymal transition as well as substitutes for peritumoral fibroblasts [16]. Further evidence reveals an involvement of EPSTI1 in cell proliferation, invasion and apoptosis of breast cancer [17,25]. These findings suggest that EPSTI1 participates in the carcinogenesis and progression of breast cancer. Herein, we showed that EPSTI1 expression could be downregulated by forced expression of miR-654-5p in breast cancer cells and observed an inverse correlation between the expression levels of miR-654-5p (Figure 3C and 3D) and EPSTI1 mRNA in 70 paired tissues of breast cancer (Figure 3E). The dual luciferase assay further confirmed that EPSTI1 was a direct target of miR-654-5p (Figure 3A and 3B), and EPSTI1 could partially abolish the miR-654-5p-induced inhibitory property on the proliferation and invasion of breast cancer cells and protein expression of EPSTI1 as well as apoptosis-induced ability (Figure 4A-D). These phenomena suggest that the functional interactions of EPSTI1-miR-654-5p provide a potential therapeutic target in breast cancer treatment.
In summary, the present work, for the first time, identified a function for miR-654-5p as a tumor suppressor miRNA in breast cancer, implicating cell proliferation, invasion and apoptosis by direct modulation of its target EPSTI1. The results elucidate a potential molecular mechanism underlying the tumor inhibitory effect of miR-654-5p, and reveal that miR-654-5p may serve as a potential prognostic marker and therapeutic target in breast cancer management.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SE. Metastatic breast cancer: the treatment challenge. Clin Breast Cancer. 2008;8:224–233. doi: 10.3816/CBC.2008.n.025. [DOI] [PubMed] [Google Scholar]
- 4.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Hanieh H. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol Cancer. 2015;14:172. doi: 10.1186/s12943-015-0443-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu S, McDonnell K, Choi H, Gao D, Hahn M, Joshi N, Park SM, Catena R, Do Y, Brazin J, Vahdat LT, Silver RB, Mittal V. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell. 2013;23:63–76. doi: 10.1016/j.ccr.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Minemura H, Takagi K, Miki Y, Shibahara Y, Nakagawa S, Ebata A, Watanabe M, Ishida T, Sasano H, Suzuki T. Abnormal expression of miR-1 in breast carcinoma as a potent prognostic factor. Cancer Sci. 2015;106:1642–50. doi: 10.1111/cas.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Yang Z, Wang H, Cao Z, Zhao Y, Gong C, Ma L, Wang X, Hu X, Chen S. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am J Cancer Res. 2015;5:2719–2729. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou CX, Wang CL, Yu AL, Wang QY, Zhan MN, Tang J, Gong XF, Yin QQ, He M, He JR, Chen GQ, Zhao Q. MiR-630 suppresses breast cancer progression by targeting metadherin. Oncotarget. 2016;7:1288–99. doi: 10.18632/oncotarget.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen HL, Ronnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002;79:703–710. doi: 10.1006/geno.2002.6755. [DOI] [PubMed] [Google Scholar]
- 16.de Neergaard M, Kim J, Villadsen R, Fridriksdottir AJ, Rank F, Timmermans-Wielenga V, Langerod A, Borresen-Dale AL, Petersen OW, Ronnov-Jessen L. Epithelial-stromal interaction 1 (EPSTI1) substitutes for peritumoral fibroblasts in the tumor microenvironment. Am J Pathol. 2010;176:1229–1240. doi: 10.2353/ajpath.2010.090648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Lu H, Shen C, Lahiri SK, Wason MS, Mukherjee D, Yu L, Zhao J. Identification of epithelial stromal interaction 1 as a novel effector downstream of Kruppel-like factor 8 in breast cancer invasion and metastasis. Oncogene. 2014;33:4746–4755. doi: 10.1038/onc.2013.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjo A, Kangaspeska S, Edgren H, Nicorici D, Bjartell A, Ceder Y, Perala M, Kallioniemi O. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 19.Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X, Chen J, Wang S. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. doi: 10.1371/journal.pone.0103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usmani A, Shoro AA, Memon Z, Hussain M, Rehman R. Diagnostic, prognostic and predictive value of MicroRNA-21 in breast cancer patients, their daughters and healthy individuals. Am J Cancer Res. 2015;5:2484–2490. [PMC free article] [PubMed] [Google Scholar]
- 23.Muller V, Gade S, Steinbach B, Loibl S, von Minckwitz G, Untch M, Schwedler K, Lubbe K, Schem C, Fasching PA, Mau C, Pantel K, Schwarzenbach H. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Res Treat. 2014;147:61–68. doi: 10.1007/s10549-014-3079-3. [DOI] [PubMed] [Google Scholar]
- 24.Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014;60:197–205. doi: 10.1373/clinchem.2013.210542. [DOI] [PubMed] [Google Scholar]
- 25.Capdevila-Busquets E, Badiola N, Arroyo R, Alcalde V, Soler-Lopez M, Aloy P. Breast cancer genes PSMC3IP and EPSTI1 play a role in apoptosis regulation. PLoS One. 2015;10:e0115352. doi: 10.1371/journal.pone.0115352. [DOI] [PMC free article] [PubMed] [Google Scholar]


