Abstract
Recent advances in fluorescence microscopy allow three-dimensional analysis of HIV-1 preintegration complexes in the nuclei of infected cells. To extend this investigation to gammaretroviruses, we engineered a fluorescent Moloney murine leukemia virus (MLV) system consisting of MLV-integrase fused to enhanced green fluorescent protein (MLV-IN-EGFP). A comparative analysis of lentiviral (HIV-1) and gammaretroviral (MLV) fluorescent complexes in the nuclei of infected cells revealed their different spatial distributions. This research tool has the potential to achieve new insight into the nuclear biology of these retroviruses.
TEXT
To complete its replication cycle, HIV-1 has to reach the nucleus of the infected cell, where the viral cDNA integrates itself into the host chromatin (1). Although the integration reaction and the targeted chromatin regions have been well characterized, very little is known about the virus-cell interactions that take place within the nuclear compartment.
The development of fluorescence imaging approaches to study HIV-1 early replication steps has significantly contributed to our knowledge of the HIV-1 life cycle. Indeed, fluorescence imaging techniques allow the observation at the single-particle level of HIV-1 interactions with target cell structures, preserving their main three-dimensional (3D) structural properties. Imaging techniques have revealed that HIV-1 particles (2–5) and integrated proviruses (6), preferentially localize in the nuclear periphery, which is consistent with the observation that peripheral chromatin, especially that near the nuclear pore complexes, is favored for HIV-1 integration (7, 8).
Here we further analyzed the 3D nuclear distribution of HIV-1 preintegration complexes (PICs) in relation to the expression of HIV-1 integrase (IN) cofactor LEDGF/p75 (for a recent review, see reference 9). We also developed a new tool for the detection of fluorescent murine leukemia virus (MLV) that allowed us to compare the 3D nuclear distributions of gammaretroviral and lentiviral particles.
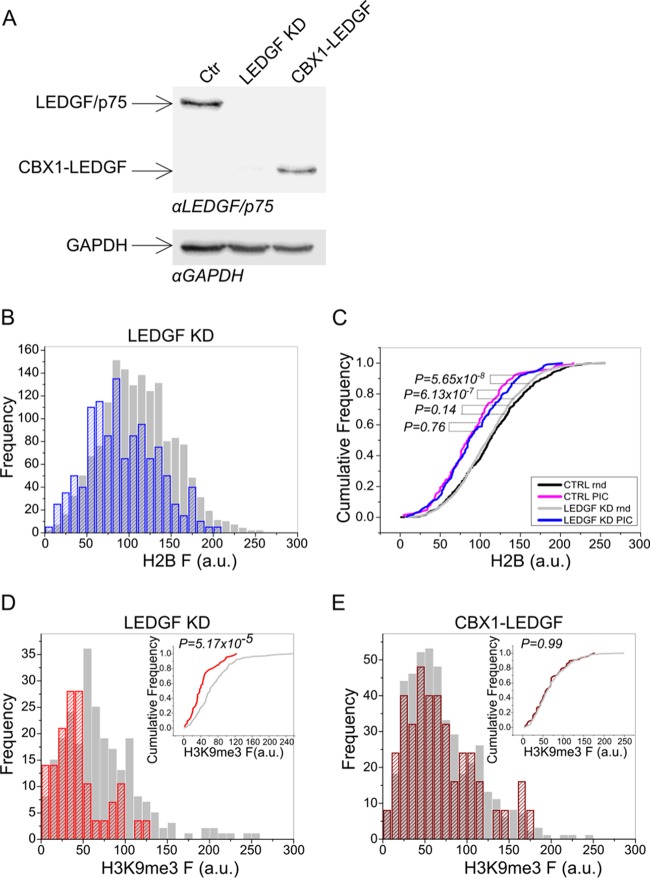
We set out to explore the role played by LEDGF/p75 in the 3D nuclear distribution of HIV-1 in the nucleus during the early phases of infection. Using a technique that allows us to produce and track fluorescent HIV-IN-enhanced green fluorescent protein (EGFP) PICs (2–4), we analyzed virus localization with respect to the levels of chromatin condensation as demarcated by the ectopic expression of histone H2B fused to red fluorescent protein (RFP) (2). Using the same procedures we previously established (2–4), we analyzed the localization of HIV-IN-EGFP PICs in HeLa-H2B-RFP cells stably silenced for LEDGF/p75 (10) (Fig. 1A). Figure 1B shows the preferential localization of HIV-IN-EGFP PICs toward less condensed euchromatic regions characterized by low H2B fluorescence. Statistical analysis using the nonparametric two-tailed Kolmogorov-Smirnov test revealed no significant difference between the distributions of HIV-1 PICs in LEDGF/p75 knockdown and control cells (P = 0.76; blue and pink curves). Conversely, the HIV-PIC distribution to euchromatin in both cell lines significantly differs from the random region of interest (ROI) distribution in the same cells (gray and black curves from control and LEDGF/p75 knockdown cells, respectively) (Fig. 1C). Analyses performed with a heterochromatin-specific marker (H3K9me3) confirmed the preferential localization of HIV-1 PICs in chromatin regions with low H3K9me3 signal intensity (Fig. 1D), which is similar to what we previously observed in wild-type cells (2). Therefore, even though LEDGF/p75 is the main factor directing HIV-1 to specific gene-rich regions (9, 11), it does not play a crucial role in the macrolocalization of HIV-1 viral complexes in the nucleus.
FIG 1.
LEDGF/p75 is dispensable for the localization of HIV-1 complexes in nuclear regions occupied by euchromatin. (A) Western blot analysis of HeLa-H2B-RFP LEDGF/p75 knockdown cells or LEDGF/p75 knockdown cells transcomplemented with CBX-LEDGF(325-530). At the top is a Western blot developed with anti-LEDGF/p75 antibodies (Bethyl Laboratories, Inc.), and at the bottom is a Western blot developed with an anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase) antibody to control protein loading. Ctr, control. (B) Frequency distribution of H2B-RFP fluorescence intensity (F) in HIV-IN-EGFP PIC ROIs (blue striped bars) and in random ROIs (gray bars) in LEDGF/p75 knockdown cells. (C) Distribution of cumulative probabilities plotted for HIV-IN-EGFP PIC ROIs or random (Rnd) ROIs toward H2B-RFP fluorescence in LEDGF/p75 knockdown (LEDGF KD) or control (CTRL) cells. Reported P values were obtained by the Kolmogorov-Smirnov test (n = 100). (D) Frequency distribution of H3K9me3 fluorescence intensity (F) in HIV-IN-EGFP PIC ROIs (red striped bars) and in random ROIs (gray bars) in LEDGF/p75 knockdown cells. In the inset, the distributions of cumulative probabilities in PIC (red) and random (gray) ROIs are plotted. The P value was obtained by the Kolmogorov-Smirnov test (n = 100). (E) Frequency distribution of H3K9me3 fluorescence intensity (F) in PIC ROIs (red striped bars) and in random ROIs (gray bars) in LEDGF/p75 knockdown cells expressing CBX-LEDGF(325-530). In the inset, the distributions of cumulative probabilities in PIC (red) and random (gray) ROIs are plotted. The P value was obtained by the Kolmogorov-Smirnov test (n = 100). a.u., arbitrary units.
To confirm that the absence of an HIV-1 PIC relocalization phenotype in LEDGF/p75 knockdown cells was not due to intrinsic limitations of the assay, the analysis was repeated with LEDGF/p75 knockdown cells stably expressing CBX-LEDGF(325-530) (12) (Fig. 1A). CBX-LEDGF(325-530) is a chimeric LEDGF/p75 molecule engineered to contain an alternative chromatin-binding domain, CBX1, and is reported to strongly relocalize HIV-1 integration toward heterochromatin (12–14). As expected, we observed that HIV-IN-EGFP PICs were randomly distributed in these cells (Fig. 1E), confirming that the HIV-1 imaging tool correctly detects the 3D macrolocalization of HIV-1.
Therefore, even though LEDGF/p75 knockdown generates a more random distribution of integrated proviruses (9, 15–17), in the absence of this IN cofactor, HIV-1 PICs preserve their localization toward subnuclear regions occupied by euchromatin.
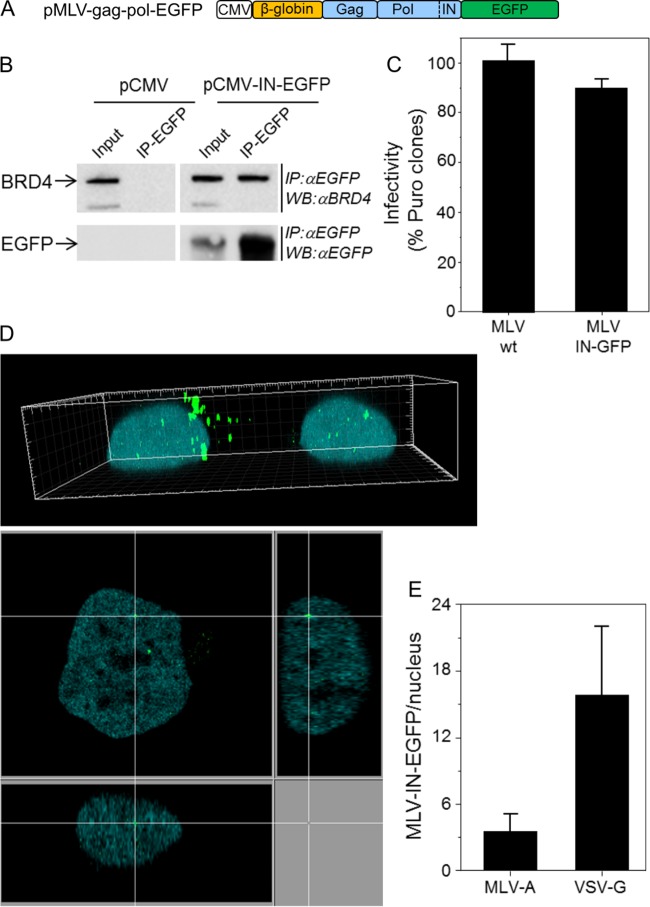
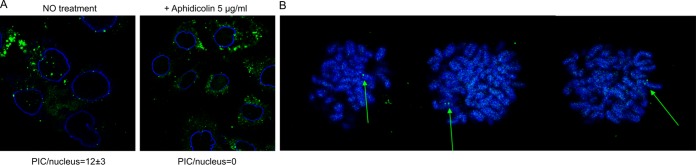
To compare the nuclear localizations of lentiviruses and gammaretroviruses, we developed MLV-IN-EGFP, a fluorescently labeled MLV. To produce fluorescently labeled viral particles, a Moloney MLV retroviral packaging vector, pMLV-gag-pol-EGFP, was generated (Fig. 2A). Fusion of EGFP at the C terminus of MLV Pol does not impair the association between IN and BRD4 (18–20), as demonstrated by a coimmunoprecipitation experiment performed with 293T cells transfected with the pMLV-gag-pol-EGFP construct (Fig. 2B). To further characterize MLV-IN-EGFP, we tested the infectivity of the retroviral transfer vector pBabe-Puro (21) packaged with pMLV-gag-pol-EGFP (Fig. 2C) and pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G). These experiments showed that the infectivity of MLV-IN-EGFP is similar to that of wild-type MLV (pBabe-Puro vector packaged with parental phCMV-intron-gag-pol) (Fig. 2C). We next analyzed the MLV-IN-EGFP viral complexes after infection of HeLa-H2B-CFP (cyan fluorescent protein) and found them to be localized both in the cytoplasm (Fig. 2D, top) and in the nuclear compartment, as indicated by three-axis nuclear analysis (Fig. 2D, bottom). Twenty-four hours postinfection, an average of three MLV-IN-EGFP complexes per nucleus were detected in viral preparations made with an amphotropic MLV envelope (strain 4070A), while with VSV-G-pseudotyped virions, we detected an average of 16 complexes per nucleus (Fig. 2E). Gammaretroviruses, such as MLV, need the target cell to go through mitosis in order to complete infection (22). This has been attributed to their inability to actively cross the nuclear envelope (22). We thus tested the cellular localization of MLV-EGFP complexes in cells blocked in G1 of the cell cycle by aphidicolin treatment. As expected, MLV complexes were restricted to the cytoplasmic compartment in aphidicolin-treated cells and nuclear complexes could not be detected (Fig. 3A). As reported earlier (23), also in this case, MLV-IN-EGFP particles associated with condensed chromatids in cells going through mitosis were detected (Fig. 3B).
FIG 2.
MLV-IN-EGFP visualization system. (A) Schematic representation of the plasmid used to produce MLV-IN-EGFP particles. The construct was prepared as follows. The 3′-terminal end of MLV pol was amplified with primers MLV pol 5′ (GATCCTCGAGCTATAGAAAATTCATCACCC) and MLV pol 3′ (GATCAGATCTCCGGGGGCCTCGCGGGTTAAC) and with the phCMV-intron-gag-pol plasmid (a gift from François-Loïc Cosset) as the template DNA and cloned into the BglII site of plasmid pEGFP-N1 (Clontech). Next, a SnaBI-XmaI fragment encompassing the β-globin intron and MLV gag-pol from phCMV-intron-gag-pol was cloned into the SnaBI and XmaI sites of pEGFP-N1 containing the 3′ end of MLV pol. (B) HEK 293T cells transfected with plasmid pMLV-gag-pol-EGFP or control plasmid pCMV were immunoprecipitated (IP) with anti-EGFP antibodies (IP-EGFP) and analyzed by Western blotting with anti-BRD4 antibodies (Bethyl Laboratories, Inc.) (top) or anti-EGFP antibodies (bottom). Crude lysates (input) were loaded as controls. (C) MLV-IN-EGFP viral supernatants were produced by transfecting 5 × 106 HEK 293T cells with 20 μg of pBabe-Puro, 10 μg of pMLV-gag-pol-EGFP (or phCMV-intron-gag-pol), and 5 μg of pVSV-G with the polyethylenimine reagent. Supernatants were collected after 48 h and filtered through a 0.45-μm-pore-size filter. The infectivity of MLV-IN-EGFP was measured by counting the HeLa colonies resistant to puromycin 2 weeks postinfection with 2.87 reverse transcriptase units (RTU; measured by PCR-enhanced reverse transcriptase (PERT) assay [3]) and approximately corresponding to a multiplicity of infection of 1. wt, wild type. (D) HeLa cells expressing H2B-CFP and infected with MLV-IN-EGFP (2.87 RTU) in a 3D image (top) and a derived confocal section (bottom). (E) Numbers of MLV-IN-EGFP complexes in the nuclei of HeLa cells at 24 h postinfection. MLV-A and VSV-G indicate the envelopes used to produce MLV-IN-EGFP viral particles.
FIG 3.
Nuclear detection of MLV complexes. (A) Confocal section obtained from HeLa cells infected with MLV-IN-EGFP particles (green spots) and immunostained with lamin A/C (blue) (left image). Confocal section obtained from HeLa cells treated with 5 μM aphidicolin, infected with MLV-IN-EGFP particles (green spots), and immunostained with lamin A/C (blue) (right image). Image analysis of fluorescent nuclear PICs is described in references 2 to 4. PIC/nucleus values indicate the average number of nuclear MLV-IN-EGFP complexes detected in 90 cells under both conditions. (B) Visualization of MLV-EGFP complexes attached to chromatids (arrows).
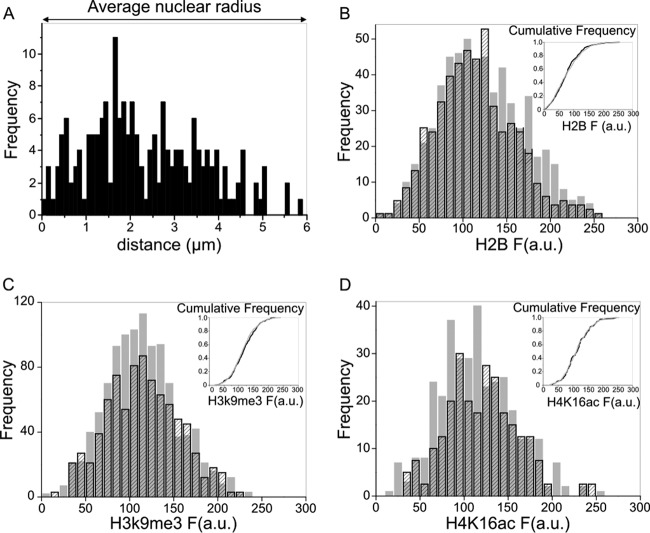
To compare the nuclear distribution of MLV complexes to that of HIV-1, the localization of MLV was assessed with respect to the nuclear periphery and chromatin condensation. Unlike HIV-1, the MLV complexes were homogeneously distributed in the volume spanning from the periphery to the center of the nuclei (Fig. 4A). Moreover, in cells marked with fluorescent H2B histones, the MLV complexes did not show any preferential localization in either more or less condensed regions of the chromatin (Fig. 4B). To better analyze this aspect, MLV distribution was analyzed in cells immunostained with an epigenetic marker of transcriptional repression (H3K9Me3) or activation (H4K16Ac) (24). Also in this case, we could not observe any preferential localization of MLV with respect to either of these two markers (Fig. 4C and D). These results demonstrate that while HIV-1 PICs are found mainly in subnuclear compartments occupied by euchromatin and near the nuclear periphery (2, 4), MLV PICs do not show any preferential localization.
FIG 4.
3D nuclear distribution of MLV complexes. (A) Distance of intranuclear MLV complexes from lamin A/C staining. (B) Frequency distribution of H2B-CFP fluorescence intensity (F) in MLV ROIs (black striped bars) and in random ROIs (gray bars). In the inset, the distributions of cumulative probabilities in MLV (black) and random (gray) ROIs are plotted. P = 0.35 (Kolmogorov-Smirnov test; n = 400). (C) Frequency distribution of H3K9me3 fluorescence intensity (F) in MLV ROIs (black striped bars) and in random ROIs (gray bars). In the inset, the distributions of cumulative probabilities in MLV (black) and random (gray) ROIs are plotted. P = 0.33 (Kolmogorov-Smirnov test; n = 270). (D) Frequency distribution of H4K16ac fluorescence intensity (F) in MLV ROIs (black striped bars) and in random ROIs (gray bars). In the inset, the distributions of cumulative probabilities in MLV (black) and random (gray) ROIs are plotted. P = >0.96 (Kolmogorov-Smirnov test; n = 100). a.u., arbitrary units.
Therefore, even though both HIV-1 and MLV prefer to integrate within transcription units (25) associated with less condensed euchromatic regions, the subnuclear 3D macrodistributions of their PICs differ significantly. This difference can probably be attributed to the different mechanisms by which HIV-1 and MLV enter the nuclei of infected cells, i.e., either through nuclear pore complexes (HIV-1) or during mitosis and nuclear envelope disassembly (MLV). On the basis of this hypothesis, nucleoporins or other factors might be relevant for 3D microdistribution. Alternatively, nonrandom distribution assumed by HIV-1 PICs might simply result from their position immediately after transition through the nuclear pore complexes. In this scenario, PIC distribution would be determined by physical events rather than by any specific cellular factor.
The microscopy tool reported here, by allowing the analysis of viral PIC nuclear distribution during infection, provides a set of information complementary to that obtained by linear sequencing, thereby shedding light on new aspects of retroviral biology.
ACKNOWLEDGMENTS
This work was supported by grants from the EU FP7 (THINC, Health-2008-201032) and the Provincia Autonoma di Trento (COFUND Project, Team 2009-Incoming). L.C.F.M. is supported by NIH-NIGMS grant R01 GM113886.
phCMV-intron-gag-pol was a gift from François-Loïc Cosset, LVRTG, ENS de Lyon, U412 INSERM, Lyon, France.
REFERENCES
- 1.Craigie R, Bushman FD. 2012. HIV DNA integration. Cold Spring Harb Perspect Med 2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanese A, Arosio D, Terreni M, Cereseto A. 2008. HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS One 3:e2413. doi: 10.1371/journal.pone.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francis AC, Di Primio C, Quercioli V, Valentini P, Boll A, Girelli G, Demichelis F, Arosio D, Cereseto A. 2014. Second generation imaging of nuclear/cytoplasmic HIV-1 complexes. AIDS Res Hum Retroviruses 30:717–726. doi: 10.1089/aid.2013.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cereseto A, Giacca M. 2014. Imaging HIV-1 nuclear pre-integration complexes. Methods Mol Biol 1087:47–54. doi: 10.1007/978-1-62703-670-2_5. [DOI] [PubMed] [Google Scholar]
- 5.Burdick RC, Hu W-S, Pathak VK. 2013. Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proc Natl Acad Sci U S A 110:E4780–4789. doi: 10.1073/pnas.1315996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Primio C, Quercioli V, Allouch A, Gijsbers R, Christ F, Debyser Z, Arosio D, Cereseto A. 2013. Single-cell imaging of HIV-1 provirus (SCIP). Proc Natl Acad Sci U S A 110:5636–5641. doi: 10.1073/pnas.1216254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, Giacca M, Lusic M. 2015. Nuclear architecture dictates HIV-1 integration site selection. Nature 521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 8.Lelek M, Casartelli N, Pellin D, Rizzi E, Souque P, Severgnini M, Di Serio C, Fricke T, Diaz-Griffero F, Zimmer C, Charneau P, Di Nunzio F. 2015. Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debyser Z, Christ F, De Rijck J, Gijsbers R. 2015. Host factors for retroviral integration site selection. Trends Biochem Sci 40:108–116. doi: 10.1016/j.tibs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, Christ F, Bushman FD, Debyser Z, Gijsbers R. 2012. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog 8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. 2006. Cellular co-factors of HIV-1 integration. Trends Biochem Sci 31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther 18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. 2010. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther 21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun M-C, Allis CD, Engelman A, Hughes SH. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci U S A 107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 16.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shun M-C, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, Gijsbers R, Debyser Z. 2013. The BET family of proteins targets Moloney murine leukemia virus integration near transcription start sites. Cell Rep 5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF. 2013. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol 87:12721–12736. doi: 10.1128/JVI.01942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res 18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roe T, Reynolds TC, Yu G, Brown PO. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elis E, Ehrlich M, Prizan-Ravid A, Laham-Karam N, Bacharach E. 2012. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog 8:e1003103. doi: 10.1371/journal.ppat.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cattoglio C, Pellin D, Rizzi E, Maruggi G, Corti G, Miselli F, Sartori D, Guffanti A, Di Serio C, Ambrosi A, De Bellis G, Mavilio F. 2010. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell RS, Beitzel BF, Schroder ARW, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]