ABSTRACT
The herpes simplex virus 1 (HSV-1) capsid is a huge assembly, ∼1,250 Å in diameter, and is composed of thousands of protein subunits with a combined mass of ∼200 MDa, housing a 100-MDa genome. First, a procapsid is formed through coassembly of the surface shell with an inner scaffolding shell; then the procapsid matures via a major structural transformation, triggered by limited proteolysis of the scaffolding proteins. Three mature capsids are found in the nuclei of infected cells. A capsids are empty, B capsids retain a shrunken scaffolding shell, and C capsids—which develop into infectious virions—are filled with DNA and ostensibly have expelled the scaffolding shell. The possible presence of other internal proteins in C capsids has been moot as, in cryo-electron microscopy (cryo-EM), they would be camouflaged by the surrounding DNA. We have used bubblegram imaging to map internal proteins in all four capsids, aided by the discovery that the scaffolding protein is exceptionally prone to radiation-induced bubbling. We confirmed that this protein forms thick-walled inner shells in the procapsid and the B capsid. C capsids generate two classes of bubbles: one occupies positions beneath the vertices of the icosahedral surface shell, and the other is distributed throughout its interior. A likely candidate is the viral protease. A subpopulation of C capsids bubbles particularly profusely and may represent particles in which expulsion of scaffold and DNA packaging are incomplete. Based on the procapsid structure, we propose that the axial channels of hexameric capsomers afford the pathway via which the scaffolding protein is expelled.
IMPORTANCE In addition to DNA, capsids of tailed bacteriophages and their distant relatives, herpesviruses, contain internal proteins. These proteins are often essential for infectivity but are difficult to locate within the virion. A novel adaptation of cryo-EM based on detecting gas bubbles generated by radiation damage was used to localize internal proteins of HSV-1, yielding insights into how capsid maturation is regulated. The scaffolding protein, which forms inner shells in the procapsid and B capsid, is exceptionally bubbling-prone. In the mature DNA-filled C capsid, a previously undetected protein was found to underlie the icosahedral vertices: this is tentatively assigned as a storage form of the viral protease. We also observed a capsid species that appears to contain substantial amounts of scaffolding protein as well as DNA, suggesting that DNA packaging and expulsion of the scaffolding protein are coupled processes.
INTRODUCTION
It has long been known that viral capsids serve as protective shells in which viral genomes are packed at high density. However, it is becoming increasingly apparent that many capsids also house substantial amounts of proteins. These internal proteins have diverse roles: for example, they can serve as scaffolding components that help specify the viral architecture (1, 2), as DNA-condensing factors (3), as proteases that guide the maturation process (4, 5), and as RNA polymerases that replicate and/or transcribe viral genomes (6, 7), or they may have a role in delivering the genome into a host cell (8). In general, the incidence of internal proteins appears to correlate with the size and complexity of the virion.
While cryo-electron microscopy (cryo-EM) and X-ray crystallography have been used successfully to determine detailed molecular structures for many kinds of capsids, information concerning the locations of internal proteins and, in some cases, their very existence has remained relatively sparse. Visualization of internal proteins has been hampered by difficulty in detecting them against a dense background of nucleic acid, and their distribution—as with that of genomic material—may not conform to the icosahedral symmetry that has facilitated structural analysis of capsids. Moreover, their internal locations have rendered them inaccessible to antibodies and other labeling agents. In this context, the recently introduced technique of bubblegram imaging (9) offers a possible approach. This technique is an adaptation of cryo-electron microscopy in which a liability—radiation-induced degradation of native structure—is turned into an asset—localization of internal proteins. Usually, cryo-EM is performed under “low dose” conditions, i.e., up to about 25 electrons/Å2, to optimize preservation of the native structure. With increasing doses, the images blur progressively until bubbles of hydrogen gas at high pressure appear at sites occupied by buried proteins (10–12). On account of their low density, even very small bubbles (∼3 nm in diameter, say) are readily visible in the resulting micrographs (“bubblegrams”). Proteins that are embedded in DNA bubble relatively early because the DNA impedes the diffusion of radiation products—in particular, hydrogen gas—away from its site(s) of origin, allowing it to build up more readily to a critical concentration at which bubbles nucleate and then grow. This property was helpful for the present application.
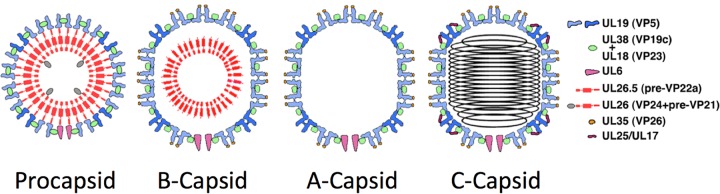
In this study, we have used bubblegram imaging to explore the internal contents of capsids of herpes simplex virus 1 (HSV-1). HSV-1 belongs to an extensive family of large and structurally elaborate double-stranded DNA (dsDNA) viruses that infect animals, but their capsids share many properties (including, in all likelihood, evolutionary origins) with those of tailed bacteriophages (reviewed in references 13 and 14). Their notably diverse protein compositions have been analyzed by mass spectrometry (15). Assembly and maturation of the HSV-1 nucleocapsid take place in the nuclei of infected cells, and their principal features have been recapitulated in vitro with purified components (16). Procapsid (the earliest precursor) and three end-product capsids are illustrated schematically in Fig. 1. The surface shell conforms to T=16 icosahedral symmetry. At one of its 12 5-fold vertices is the portal protein, a 12-fold ring. Once the procapsid is complete, the viral protease is activated and processes the scaffolding protein, excising its C-terminal 25 amino acids, which decouples the two layers. The surface shell (capsid) then undergoes a major structural transformation, while the inner layer is dismantled and expelled. In vivo, these transitions take place concomitantly with DNA packaging via the portal.
FIG 1.
Schematic diagram of the molecular anatomy of four HSV-1 capsids. UL19, the major capsid protein, forms hexamers and pentamers of the icosahedral surface lattice. The “triplexes” (green ovals) are α2β heterotrimers of UL18 and UL38. UL6 is the portal. UL25 and UL17 form heterodimers that bind to the outer surface of C capsids but only in much smaller amounts to A capsids and B capsids (31). VP24 (yellow ovals) is the protease; its location in mature capsids has been unclear and is one of the questions addressed in this study.
In this study, we investigated the internal structure of four kinds of capsid (Fig. 1): the procapsid; the C capsid, which has a fully mature surface shell and contains the 158-kbp genome arranged in a coaxial coil (17); and two mature capsids, neither of which contains DNA. These are the A capsid, which retains little, if any, scaffolding protein and is thought to be produced when DNA packaging miscarries in such a way that the genome is lost, and the B capsid, which also has an inner shell consisting of processed scaffolding protein and appears to have failed to initiate packaging. We analyzed each of these capsids by bubblegram imaging, paying particular attention to the issues of where the protease is located in mature capsids, whether any capsid proteins are particularly susceptible to bubbling, the uniformity of bubbling for a given type of capsid, and whether any unexpected sources of bubbling, i.e., caches of internal proteins, would be detected.
MATERIALS AND METHODS
Cell growth and capsid preparation.
All experiments were carried out with the KOS strain of HSV-1, which was grown at 37°C on monolayers of Vero cells as previously described (18). A, B, and C capsids were isolated from infected cells essentially according to the methods outlined in reference 17, and procapsids were prepared using methods described in reference 19.
Preparation of scaffolding protein aggregates.
G capsids, R capsids, and reconstituted aggregates of mature scaffolding protein were prepared as previously described (18). In brief, 200 μl of purified B capsids at a concentration of 1.5 mg/ml in TNE buffer (20 mM Tris-HCl, 500 mM NaCl, 1 mM EDTA [pH 7.4]) were mixed with 170 μl of TNE, 30 μl of 1 M dithiothreitol (DTT), and 200 μl of 6 M GuHCl (in TNE). The final concentration of GuHCl was 2 M, and that of DTT was 50 mM. This solution, after dialysis overnight against 1 liter of phosphate-buffered saline (PBS) at 4°C, was confirmed by negative staining EM to contain spheroidal particles ∼35 nm in diameter (scaffolding protein aggregates) and R capsids (see Results). For experiments comparing the bubbling of native scaffold inside B capsids and that of reconstituted scaffold aggregates, the latter material was mixed in approximately equal parts with a suspension of purified B capsids.
Cryo-electron microscopy and collection of bubblegram data.
In a typical experiment, a 3.5-μl drop of sample was applied to an EM grid bearing a glow-discharged continuous thin carbon film and after 1 min was blotted to a thin film and then vitrified using a Leica EM GP cryostation. The grid was transferred into a Gatan model 626 cryoholder, and micrographs were recorded on a CM200-FEG transmission electron microscope (FEI), operating at 120 keV and a magnification of ×38,000, with a nominal defocus (Δf) of ∼−1.9 μm. Data were recorded on film (Kodak SO-163). In most experiments, unless otherwise noted, the following conditions applied: each exposure typically corresponded to an electron dose of 15 to 16 electrons/Å2, there were 10-s intervals between exposures, and each dose series consisted of 16 exposures. As we have noticed some dependence of bubbling thresholds on the thickness of the ice layer (N. Cheng, unpublished data), for experiments comparing different kinds of particles, the particles involved were mixed prior to preparation of the EM grids and therefore experienced identical irradiation.
Three-dimensional image reconstruction and image analysis.
In all, 18 sets of dose series images were collected. The micrographs were digitized on a Nikon Super Coolscan 9000 scanner with a 6.35-μm step size and binned 2-fold, giving a sampling rate of 3.34 Å/pixel. EMAN (20) and EMAN2 (21) were used for image processing. To align the images in a given series, the centers of three corresponding particles on each image were marked. These three points define a triangle whose circumcenter was used to define a reference point on each image, while the vector from the circumcenter to one of the three particles was employed to calculate the rotation matrix needed to bring the images into alignment. The resulting translation and rotation parameters were used to align the particles from multiple exposures with those of the 1st exposure. e2boxer.py was employed to pick all the C capsids in the 1st-exposure images, and the coordination of the particles was recorded in a .box file and copied as the coordination for the same particles in high-exposure images. This procedure can further confirm the alignment among the images in a dose series. batchboxer was used to pick 240 1st-exposure particles for further processing. The zeroes of the contrast transfer function (CTF) were determined from the 1st-exposure images as described in reference 22 and used to perform the same phase-flipping correction on all the images in that dose series. The particles' centers (origins), initially estimated as described above, were refined using cenalignInt in EMAN1. To calculate reconstructions, capsid orientations were determined by projection matching, focusing on the 1st-exposure images and applying the same parameters throughout the dose series. Icosahedral symmetry was applied in calculating the reconstructions.
Molecular modeling.
The cryo-EM map of the procapsid used was the earliest precursor described in reference 19. The crystal structure of the HSV-2 protease (PDB code 1AT3), oriented so as to minimize its cross section, was placed manually in the channel entrances of the UL19 penton and the peripentonal hexon. UCSF Chimera (23) was used to make this simulation.
Reconstruction accession numbers.
Reconstructions have been deposited in the EMDB database with the following accession numbers: EMDB-3288 for the low dose (1st exposure), EMDB-3358 for the 7th exposure, EMDB-3359 for the 9th exposure, EMDB-3360 for the 13th exposure, and EMDB-3361 for the 15th exposure.
RESULTS
In a dose series, the first-exposure micrograph yields a relatively sharp image of the capsids. Subsequent exposures cause progressive blurring until the bubbling threshold is reached, beyond which point bubbles grow and eventually merge.
The scaffolding protein is exceptionally prone to bubbling.
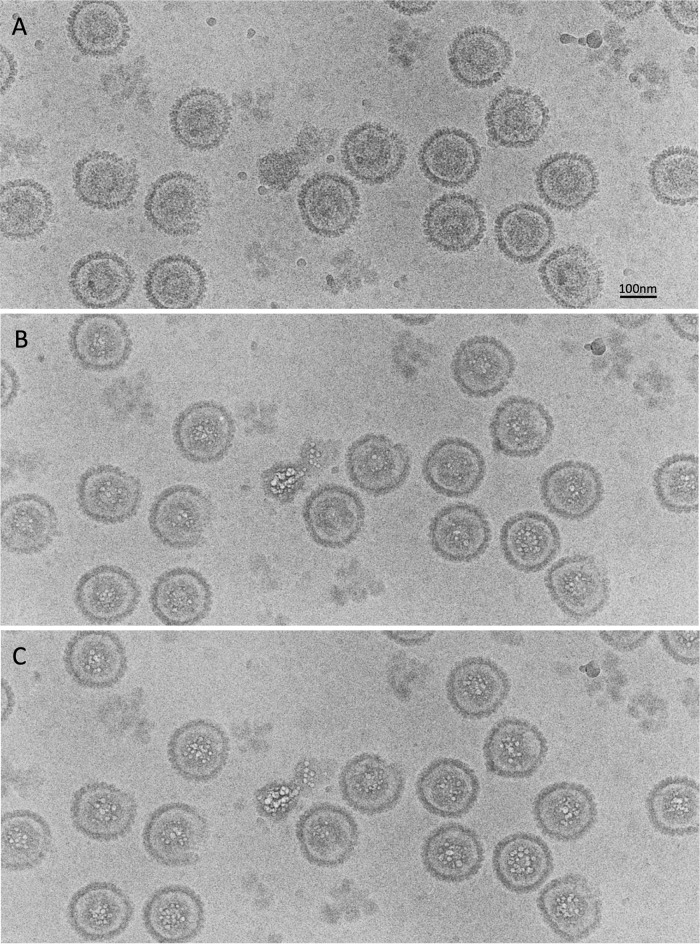
HSV-1 procapsids, which are extremely labile, were produced and purified as previously described (see Materials and Methods). Maturation was blocked by using a viral strain with a defective protease. A typical dose series is shown in Fig. 2. The first-exposure image (low dose [Fig. 2A]) can be compared with the seventh exposure (incipient bubbling [Fig. 2B]) and the eighth exposure (more developed bubbling [Fig. 2C]). In both of the last two images, the bubbling is confined to the inner (scaffolding) shell. Later in the dose series, the bubbles became larger but remained confined to the scaffolding shell (data not shown). We conclude, therefore, that the scaffolding protein, preUL26.5, is markedly more bubbling prone than the proteins of the surface shell. Eventually, upon more aggressive imaging with doses higher than the range covered in this series, small bubbles start to appear in the surface shell (Fig. 3).
FIG 2.
(A) Field of procapsids in a low-dose image (first exposure); (B) incipient bubbling (7th exposure); (C) more advanced bubbling (8th exposure). Procapsids are labile particles and exhibit various distortions. Each exposure imparted a dose of ∼12 electrons/Å2. The first small bubbles appeared in exposure 6 (data not shown).
FIG 3.
Bubbling of the procapsid scaffolding shell and surface shell at high doses. In the left-hand panel there is extensive bubbling of the scaffold but none yet in the surface shell. With additional irradiation (right-hand panel), bubbling of the scaffold is even more advanced and is accompanied by incipient bubbling in the surface shell. This dose series was performed at a high magnification (×115,000), and the left-hand and right-hand panels are from exposures 4 (∼500 electrons/Å2) and 6 (cumulative dose of ∼800 electrons/Å2), respectively.
An alternative interpretation of these images showing bubbling of the scaffolding shell would be that the surface shell impedes the outward diffusion of radiation products (primarily, H gas) generated in the procapsid interior and thus promotes bubbling of the scaffold. In principle, this “diffusion barrier” mechanism would be similar to that of densely packed DNA surrounding protein (24). A priori, this eventuality appeared to be unlikely because the procapsid surface shell is highly porous (25, 26). Nevertheless, we tested it with an experiment in which capsid-free aggregates of scaffolding protein were compared with the inner shell of B capsids (Fig. 4). The aggregates were produced by first treating purified B capsids with 2 M guanidine hydrochloride, which extracts the scaffold and the small outer capsid protein VP26 (18). We call these particles G capsids. On dialyzing out the denaturant, the solubilized scaffolding protein reassociates into aggregates of about the same size as the B-capsid cores (18); concomitantly, VP26 binds to the G capsids, giving R capsids. In relation to the design of this experiment, we note the following. (i) The propensity of a given protein to bubble is enhanced in larger aggregates which promote the local buildup of hydrogen gas to a critical concentration (27); hence, the similar sizes of the two assemblies of scaffolding protein vindicate direct comparison. (ii) The B-capsid surface shell is less porous than that of the procapsid (19); hence, any barrier effect is likely to be greater with the B capsid. (iii) There is a difference in primary structure between UL26.5, the mature scaffolding protein found in B capsids, and its precursor, preUL26.5, found in the procapsid, as the C-terminal 25 of the precursor's 329 residues are excised by the viral protease (28).
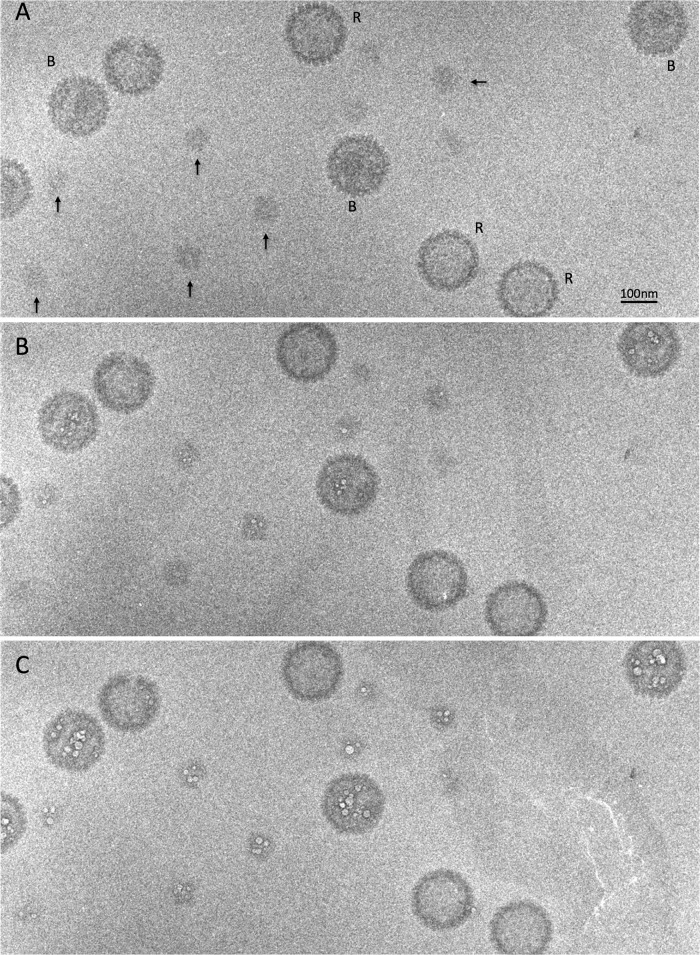
FIG 4.
A dose series of cryo-electron micrographs of a field containing B capsids, R capsids, and (arrowed) reconstituted aggregates of scaffolding protein. (A) First exposure; (B) 11th exposure; (C) 16th exposure.
Aggregates were mixed with B capsids and R capsids (included as a negative control) and prepared for cryo-EM, and dose series were recorded. The three kinds of particles are readily distinguishable in the micrographs (e.g., Fig. 4A). The aggregates and the B-capsid scaffolds started to bubble in the 11th exposure (Fig. 4B), with prominent bubbling recorded in the 16th exposure (Fig. 4C); in contrast, R capsids showed no visible bubbling over this dose range. This outcome, with the scaffolding protein bubbling at the same dose threshold in both contexts, confirms that the scaffolding protein is more prone to bubbling than the proteins of the capsid shell.
A capsids are bubbling-reluctant, while C capsids bubble in multiple locations.
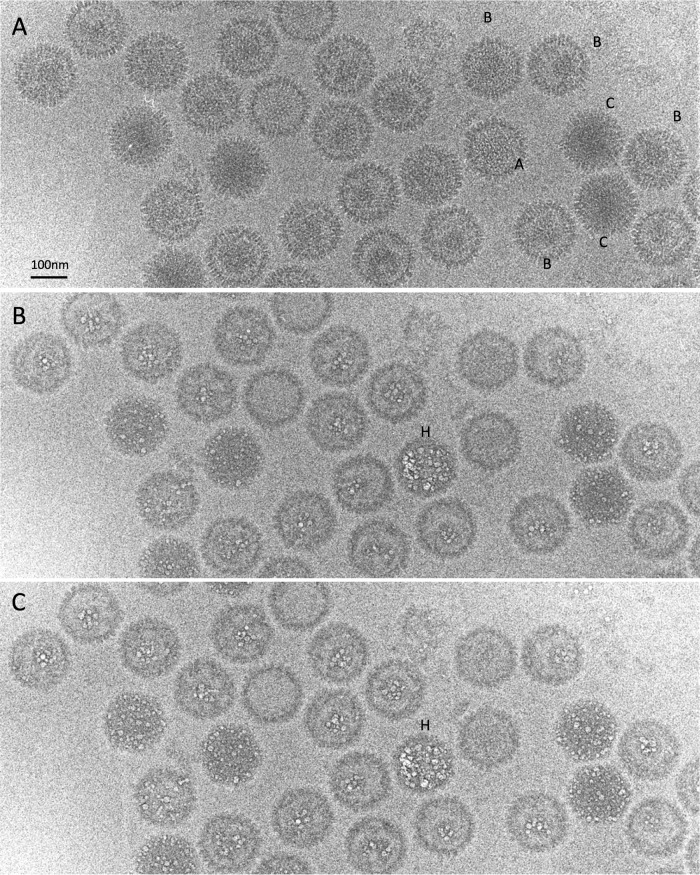
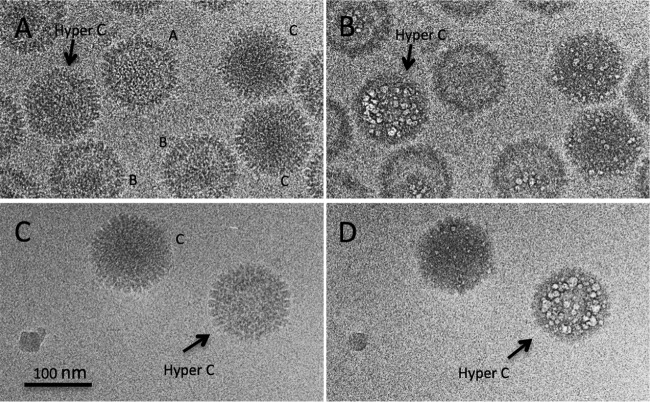
Next we addressed the respective propensities of A, B, and C capsids to bubble. A mixed population with approximately equal proportions of the three capsids was isolated, grids were prepared for cryo-EM, and dose series were recorded. A field of these capsids (first exposure) is shown in Fig. 5A; the three kinds of capsids are readily distinguishable. As all capsids in a given field received the same irradiation and are in an ice layer of approximately uniform thickness, any systematic differences between their bubblegrams must reflect intrinsic properties of the respective particles. As in the procapsid experiments (see above), successive exposures brought about a progressive blurring of the capsids. In the 10th exposure, the first small bubbles appeared in B capsids and C capsids but not in A capsids. Shown in Fig. 5B and C are the 10th and 11th exposures from this series. A capsids showed no perceptible bubbling throughout the dose series (13 exposures). In B capsids, bubbling became copious but was confined to their inner shell of scaffolding protein (as already observed in Fig. 4). C capsids, on the other hand, exhibited constellations of mostly smallish bubbles distributed throughout the particles, with a noteworthy proportion located around the capsid periphery (Fig. 5B and C). As we have observed with other specimens (24; unpublished data), there is significant variability, apparently stochastic, in the amount of bubbling observed in capsids of a given kind. However, a small minority (∼3% [5/235]) of C capsids bubble prolifically, well beyond the range of variability exhibited by the rest of that population (see the particle labeled H in Fig. 5B and C); we call them “hyperbubblers.”
FIG 5.
A dose series of cryo-electron micrographs of a mixed field of A capsids, B capsids, and C capsids. (A) First exposure; (B) 11th exposure; (C) 12th exposure. A few C capsids bubbled earlier and much more copiously than the majority; one of these hyperbubblers is labeled (H).
3D reconstruction of C-capsid bubblegrams.
To determine whether any locations within a C capsid are particularly favored for bubble formation, we calculated three-dimensional (3D) reconstructions from successive members of a dose series. If a location is consistently occupied by bubbles in symmetry-related sites in most (even, many) C capsids, it will show up in the reconstruction as a low-density region that we call a “gas cloud.” Such a cloud may be expected to grow as the dose series progresses, as existing bubbles grow and new bubbles nucleate. If, on the other hard, the bubbles are randomly distributed and account for only a minor fraction of the experimental volume, they will smear out to give an approximately uniform internal density, slightly lower than that of the first-exposure reconstruction, i.e., no gas cloud. A similar outcome will be realized if there are just a few bubbles per C capsid and icosahedral symmetry (invoking 60 symmetry operations) is applied. In general, bubbling registers earlier in individual images (projections) than in reconstructions (24).
In Fig. 6, one can compare central sections of a dose series of C-capsid reconstructions (Fig. 6B to F) with an earlier C-capsid reconstruction (Fig. 6A) calculated from many more particles and consequently at a higher resolution (for instance, the 25-Å layering of the DNA is evident). Despite the relative paucity of data, the major features of the capsid shell are already apparent in the first-exposure reconstruction from this dose series (Fig. 6B). Figure 6C shows the 7th exposure (late in the prebubbling phase). Figure 6D shows the 9th exposure, with early but significant bubbling, and Fig. 6E and F show the 13th and 15th exposures, in which bubbling was well advanced and sufficiently consistent among symmetry-related sites to register significant gas clouds in the reconstructions. Degradation of the capsid structure is already evident in Fig. 6C, for instance, in the capsomer protrusions (compare with Fig. 6A and B), but no significant gas clouds have yet developed. In Fig. 6E and F (and with the 14th-exposure reconstruction [data not shown]), well-defined gas clouds are evident underlying the pentamers at the vertex sites (two are marked by black arrows in Fig. 6A). There is no indication of similar low-density zones under the hexamers (some hexamers are marked with white arrows in Fig. 6A). There may also be a gas cloud right at the center of the C capsid, but this is hard to interpret with confidence, as it involves the part of the map most affected by noise. We conclude that there is a significant amount of protein underneath the vertices that gives rise to the observed gas clouds.
FIG 6.
Central sections of 3D reconstructions from a dose series of C capsids. For reference, an icosahedrally symmetrized reconstruction at a resolution of 19 Å (31) is shown in panel A. (B to E) Reconstructions from the dose series. The designation E#M refers to the Mth exposure. The sections are from reconstructions oriented so as to present a 2-fold view. Black arrows in panel A mark the positions of vertices (pentamers). The subvertex gas clouds have developed in panels E and F, although less copious bubbling was observed as early as the 10th exposure (cf. Fig. 5B).
An additional set of randomly distributed bubbles in C capsids.
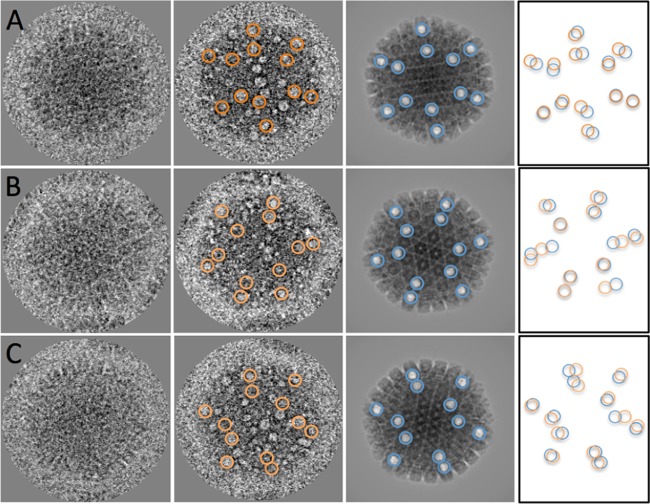
The foregoing analysis has shown that subvertex sites are favored for bubbling. However, there are only 12 vertices—1 portal and 11 nonportal—and more than 12 bubbles are generated in some C capsids, particularly late in a dose series (e.g., Fig. 5C). (The question of whether the portal vertex has a bubble is moot, as there is only one such vertex per capsid and its contribution to the reconstruction is outweighed by those of the nonportal vertices). Thus, we infer that some of these C-capsid bubbles originate in proteins that are randomly distributed through the condensed DNA. Given a 3D reconstruction and the viewing geometry (defined by a set of Euler angles) of a contributing C capsid, its vertex-associated bubbles may be identified by comparing the corresponding reprojection of the reconstruction with the original image. Some examples of such bubble-mappings are shown in Fig. 7; by elimination, it may be concluded that the other bubbles are not vertex associated.
FIG 7.
(A) Distinguishing vertex-associated bubbles from other bubbles in C capsids, illustrated for three particles (A to C). The first column shows first-exposure images. The second column shows the corresponding bubblegrams, with vertex-associated bubbles in orange rings. The third column shows corresponding reprojections of a C-capsid reconstruction with modeled bubbles at all 12 vertices in blue rings. The last column shows that the matching of the theoretical positions (blue) and the observed positions (orange) is good but not perfect, implying that bubbles tend to be slightly offset from the 5-fold axes. Other bubbles presumably derive from proteins present at other internal sites. Capsids are 125 nm in diameter.
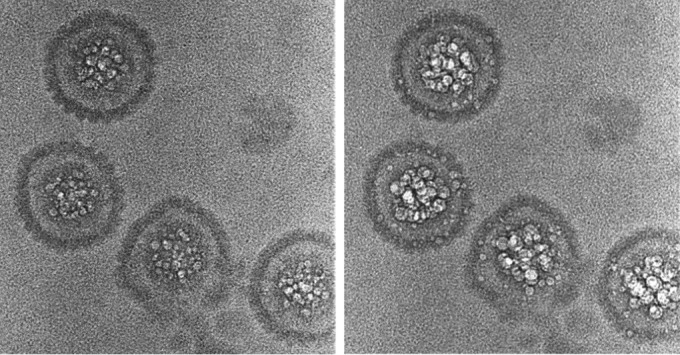
Having identified hyperbubblers in bubblegrams, we then inquired whether they exhibit any visibly different features from the majority of C capsids in low-dose micrographs. Two examples are shown in Fig. 8, alongside A, B, and regular C capsids. The hyperbubblers have less internal material than regular C capsids, but they are readily distinguished from B capsids because they lack the latter particle's conspicuous gap between its scaffold and surface shell and from DNA-filled C capsids because they lack the “fingerprint” or “punctate array” motifs (17). Although bubbles are copious in these two H particles (Fig. 8B and D), they are not seen in the outer rim of density, 10 to 15 nm thick (projection of the surface shell), suggesting that an internal component (most likely the scaffolding protein, as discussed below) is responsible. We suggest that these particles represent capsids in which DNA packaging has arrested prematurely and from which the exit of scaffolding protein is incomplete.
FIG 8.
Cryo-micrographs and bubblegrams of two hyperbubblers compared with A capsids, B capsids, and regular C capsids. In addition to the distinction between hyperbubblers and regular C capsids in terms of their respective bubbling, they are also distinguished in first-exposure micrographs because hyperbubblers tend to have less internal material than regular C capsids and from B capsids because they lack the conspicuous gap between the B-capsid scaffold and its surface shell. Although bubbles are plentiful throughout the interiors of these two hyperbubblers (B and D), they do not appear in their surface shells.
DISCUSSION
Internal proteins of herpesvirus capsids.
The HSV-1 procapsid contains two variants of the scaffolding protein: one is the core domain, preUL26.5; in the other, preUL26, the viral protease VP24 is connected via a linker at the N-terminal end of preUL26.5. In total, there are ∼1,900 scaffolding protein subunits per procapsid, of which ∼10% are the fusion protein (19). The core domains form elongated dimers, ∼225 Å long, that pack together side-by-side in assembling the scaffolding shell, with the protease domains on the inside (29). This arrangement provides an elegant mechanism for incorporating the protease into the capsid. In maturation, both variants undergo proteolytic cleavage, freeing the protease (how the first cleavage is accomplished remains unclear [30]) and detaching the scaffolding shell from the surface shell. In a B capsid, ∼50% of the scaffold (now consisting of UL26.5 and the core domain-linker protein, VP21) remains inside as a somewhat shrunken and less ordered inner shell. With A capsids or C capsids, the scaffold is mostly expelled. In contrast, the protease remains associated with all three mature capsids (17). These changes are beneficial in two ways: expulsion of the scaffolding protein frees up space for DNA, and confinement of the protease prevents it from degrading nuclear proteins that may be needed to sustain a productive infection. These considerations raise the following questions. (i) Does the protease occupy any particular position inside mature C capsids? (ii) What is the exit pathway of the scaffolding proteins? Before addressing them, we review some aspects of bubblegram interpretation.
Multiple factors affect the bubbling propensities of different proteins.
Bubbles of hydrogen gas generated by protracted electron irradiation of vitrified specimens mark the locations of buried proteins. Our experience to date suggests that most, if not all, proteins will eventually bubble (see, for example, late bubbling in the procapsid surface shell in Fig. 3), but they vary markedly in their bubbling thresholds. These may depend on several factors. One is the protein itself: the most bubbling-prone proteins that we have encountered to date are the nucleocapsid protein of HIV (27) and the scaffolding protein of HSV-1 (this study). Both are elongated molecules with high surface-to-volume ratios and may have disordered regions. It will be interesting to see if this trend is maintained as more data become available, as it may eventually become possible to draw inferences as to a protein's overall structure or its amino acid composition from its bubbling threshold. Another factor is the amount of protein present; having a substantial amount of closely packed protein is conducive to bubbling. A third factor is location: deeply buried sites are bubbling-friendly. A fourth factor concerns the imaging conditions, with (for a given dose) a short exposure with an intense beam or shorter intervals between multiple exposures favoring bubbling. All of these factors affect the local concentration of hydrogen gas (and perhaps other radiation products) and the rate at which it builds up to a critical value at which bubbles nucleate.
Proteins responsible for bubbling of immature and mature HSV-1 capsids.
The four capsids compared in this study produce widely differing patterns of bubbles, reflecting the profound structural rearrangements that take place during maturation. Which protein(s) contribute to the observed bubbling? As described above (Results and Fig. 3), the scaffolding protein is responsible for the bubbling of the inner shells of the procapsid and the B capsid. However, it is less clear which protein(s) is responsible for the bubbles seen in C capsids, which produce almost the same amount of bubbles as the B capsids, despite loss of their scaffold (Fig. 5B and C). According to SDS-PAGE of purified C capsids, an evident candidate is the protease VP24 (17, 31, 32), a 24-kDa protein of which there are about 150 copies per capsid (33). In principle, another candidate would be residual (nonexpelled) scaffolding protein, except that SDS-PAGE of C capsids has shown no significant density at the position where UL26.5 would run (17, 31; W. W. Newcomb, unpublished results), indicating that the average copy number should be very small.
To explain these observations, we suggest that as DNA packaging proceeds, protease molecules are pushed out toward the vertices by growing pressure from packaged DNA (34), but they cannot exit through the axial channel of the major capsid protein (UL19) pentamer because it is too narrow (Fig. 9). This can explain the subvertex bubbles. As for the non-vertex-associated bubbles, we entertain two possibilities that are not mutually exclusive. One invokes a subset of protease molecules that do not end up in vertex-proximal sites. The other envisages host proteins that were fortuitously incorporated into capsids during assembly. If such stowaways are diverse and their quantities small, they would not be detected by SDS-PAGE of purified capsids. Finally, noting that A capsids also retain the protease (17) but do not bubble at these electron doses, we infer that the protease is dispersed throughout the interior of the A capsid, thus not reaching a local concentration that supports bubbling. This model (random dispersal) also explains why no VP24-related density is seen at vertex-adjacent sites in cryo-EM reconstructions of A capsids (31).
FIG 9.
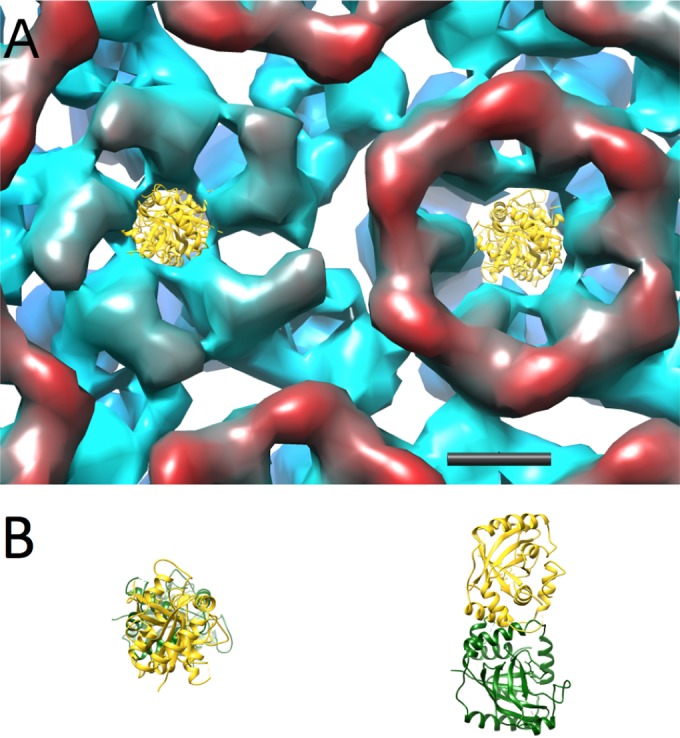
Model of a portion of the HSV-1 procapsid surface viewed from inside and colored radially from red (inside) to blue (outside), with the crystal structure of the HSV-2 protease (PDB 1AT3) placed in the channel vestibules of the UL19 penton and a peripentonal hexon. The dimeric protease is shown in side view and top view in panel B. The penton channel is clearly too narrow to admit passage of the protease. Considered on steric grounds alone, the hexon channel is marginally navigable, but because the protease remains inside mature capsids (17), other factors must contribute to blocking its passage. Scale bar = 40 Å.
Turning now to expulsion of the scaffolding protein, we posit that these elongated dimers reptate out through the axial channels of UL19 hexamers, perhaps boosted by outward pressure from packaged DNA (34). It appears unlikely that UL19 pentamers can be used for this purpose, as their axial channel is markedly narrower than that of hexamers (Fig. 9). Furthermore, a buildup of protease molecules in the vertex-proximal regions (see above) would impede access to the pentamer channel. Finally, it is noteworthy that a cluster of rods—putatively α-helices—has been visualized underlying the pentamers but not the hexamers in a high-resolution reconstruction of the HSV-1 virion (J. F. Conway, personal communication). No such feature was seen underlying the hexamers. This cluster, if also present in C capsids, would further impede the exiting of scaffold via the pentamers.
Finally, the bubblegram images provide the first evidence that isolated C capsids do not constitute a homogeneous population: the hyperbubblers apparently contain additional protein that is not present in the majority of C capsids. We suggest that this protein is scaffold and it is not detected by SDS-PAGE of purified C capsids because the hyperbubblers—although unmistakably recognizable in bubblegram images—account for such a small fraction of these isolates. We speculate that these particles represent capsids fortuitously captured at a stage of incomplete DNA packaging, when a significant proportion of scaffold molecules are still inside the surface shell. In this scenario, DNA packaging and the dismantling and exit of scaffold are coupled, not separate processes.
ACKNOWLEDGMENTS
We thank Bernard Heymann for support with resources for computational analysis and Juan Fontana for helpful discussions.
This work was supported by the Intramural Research Program of NIAMS, with additional support from an NIGMS PRAT fellowship (to A.A.).
Funding Statement
The funder had no role in the study design, data collection and interpretation, or the decision to submit the work for publication
REFERENCES
- 1.Dokland T. 1999. Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci 56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevelige PE, Fane BA. 2012. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol 726:325–350. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- 3.Saper G, Kler S, Asor R, Oppenheim A, Raviv U, Harries D. 2013. Effect of capsid confinement on the chromatin organization of the SV40 minichromosome. Nucleic Acids Res 41:1569–1580. doi: 10.1093/nar/gks1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong L. 2002. Viral proteases. Chem Rev 102:4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 5.Lee SK, Potempa M, Swanstrom R. 2012. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem 287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. 2003. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol 10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mindich L. 2012. Packaging in dsRNA viruses. Adv Exp Med Biol 726:601–608. doi: 10.1007/978-1-4614-0980-9_26. [DOI] [PubMed] [Google Scholar]
- 8.Chang CY, Kemp P, Molineux IJ. 2010. gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology 398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu W, Thomas JA, Cheng N, Black LW, Steven AC. 2012. Bubblegrams reveal the inner body of bacteriophage phiKZ. Science 335:182. doi: 10.1126/science.1214120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol 111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 11.Sun S, Shi S, Leapman R. 1993. Water distributions of hydrated biological specimens by valence electron energy loss spectroscopy. Ultramicroscopy 50:127–139. doi: 10.1016/0304-3991(93)90003-G. [DOI] [PubMed] [Google Scholar]
- 12.Meents A, Gutmann S, Wagner A, Schulze-Briese C. 2010. Origin and temperature dependence of radiation damage in biological samples at cryogenic temperatures. Proc Natl Acad Sci U S A 107:1094–1099. doi: 10.1073/pnas.0905481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardone G, Heymann JB, Cheng N, Trus BL, Steven AC. 2011. Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions, p 423–439. In Rossmann MG, Rao V (ed), Viral molecular machines. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rixon FJ, Schmid MF. 2014. Structural similarities in DNA packaging and delivery apparatuses in herpesvirus and dsDNA bacteriophages. Curr Opin Virol 5:105–110. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Engel EA, Song R, Koyuncu OO, Enquist LW. 2015. Investigating the biology of alpha herpesviruses with MS-based proteomics. Proteomics 15:1943–1956. doi: 10.1002/pmic.201400604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown JC, Newcomb WW. 2011. Herpesvirus capsid assembly: insights from structural analysis. Curr Opin Virol 1:142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. 1991. Liquid-crystalline, phage-like, packing of encapsidated DNA in herpes simplex virus. Cell 64:1007–1015. doi: 10.1016/0092-8674(91)90324-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb WW, Brown JC. 1991. Structure of the herpes simplex virus capsid: Effects of extraction with guanidine-HCl and partial reconstitution of extracted capsids. J Virol 65:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aksyuk AA, Newcomb WW, Cheng N, Winkler DC, Fontana J, Heymann JB, Steven AC. 2015. Subassemblies and asymmetry in assembly of herpes simplex virus procapsid. mBio 6(5):e01525-15. doi: 10.1128/mBio.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 21.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Wu W, Chen Z, Cheng N, Watts NR, Stahl SJ, Farci P, Purcell RH, Wingfield PT, Steven AC. 2013. Specificity of an anti-capsid antibody associated with hepatitis B virus-related acute liver failure. J Struct Biol 181:53–60. doi: 10.1016/j.jsb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Cheng N, Wu W, Watts NR, Steven AC. 2014. Exploiting radiation damage to map proteins in nucleoprotein complexes: the internal structure of bacteriophage T7. J Struct Biol 185:250–256. doi: 10.1016/j.jsb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol 263:447–462. doi: 10.1016/S0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 26.Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol 10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 27.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 89:9765–9780. doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelletier A, Do F, Brisebois JJ, Lagace L, Cordingley MG. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol 71:5197–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol 74:1663–1673. doi: 10.1128/JVI.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang K, Wills EG, Baines JD. 2012. Release of the herpes simplex virus 1 protease by self cleavage is required for proper conformation of the portal vertex. Virology 429:63–73. doi: 10.1016/j.virol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol 425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and triplexes. J Mol Biol 232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 34.Bauer DW, Li D, Huffman J, Homa FL, Wilson K, Leavitt JC, Casjens SR, Baines J, Evilevitch A. 2015. Exploring the balance between DNA pressure and capsid stability in herpesviruses and phages. J Virol 89:9288–9298. doi: 10.1128/JVI.01172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]