ABSTRACT
Chronic immune activation/inflammation driven by factors like microbial translocation is a key determinant of human immunodeficiency virus/simian immunodeficiency virus (HIV/SIV) disease progression. Although extensive research on inflammation has focused on studying protein regulators, increasing evidence suggests a critical role for microRNAs (miRNAs) in regulating several aspects of the immune/inflammatory response and immune cell proliferation, differentiation, and activation. To understand their immunoregulatory role, we profiled miRNA expression sequentially in intestinal lamina propria leukocytes (LPLs) of eight macaques before and at 21, 90, and 180 days postinfection (dpi). At 21 dpi, ∼20 and 9 miRNAs were up- and downregulated, respectively. However, at 90 dpi (n = 60) and 180 dpi (n = 44), ≥75% of miRNAs showed decreased expression. Notably, the T-cell activation-associated miR-15b, miR-142-3p, miR-142-5p, and miR-150 expression was significantly downregulated at 90 and 180 dpi. Out of ∼10 downregulated miRNAs predicted to regulate CD69, we confirmed miR-92a to directly target CD69. Interestingly, the SIV-induced miR-190b expression was elevated at all time points. Additionally, elevated lipopolysaccharide (LPS)-responsive miR-146b-5p expression at 180 dpi was confirmed in primary intestinal macrophages following LPS treatment in vitro. Further, reporter and overexpression assays validated IRAK1 (interleukin-1 receptor 1 kinase) as a direct miR-150 target. Furthermore, IRAK1 protein levels were markedly elevated in intestinal LPLs and epithelium. Finally, blockade of CD8+ T-cell activation/proliferation with delta-9 tetrahydrocannabinol (Δ9-THC) significantly prevented miR-150 downregulation and IRAK1 upregulation. Our findings suggest that miR-150 downregulation during T-cell activation disrupts the translational control of IRAK1, facilitating persistent gastrointestinal (GI) inflammation. Finally, the ability of Δ9-THC to block the miR-150-IRAK1 regulatory cascade highlights the potential of cannabinoids to inhibit persistent inflammation/immune activation in HIV/SIV infection.
IMPORTANCE Persistent GI tract disease/inflammation is a cardinal feature of HIV/SIV infection. Increasing evidence points to a critical role for miRNAs in controlling several aspects of the immune/inflammatory response. Here, we show significant dysregulation of miRNA expression exclusively in the intestinal lamina propria cellular compartment through the course of SIV infection. Specifically, the study identified miRNA signatures associated with key pathogenic events, such as viral replication, T-cell activation, and microbial translocation. The T-cell-enriched miR-150 showed significant downregulation throughout SIV infection and was confirmed to target IRAK1, a critical signal-transducing component of the IL-1 receptor and TLR signaling pathways. Reduced miR-150 expression was associated with markedly elevated IRAK1 expression in the intestines of chronically SIV-infected macaques. Finally, Δ9-THC-mediated blockade of CD8+ T-cell activation in vitro significantly inhibited miR-150 downregulation and IRAK1 upregulation, suggesting its potential for targeted immune modulation in HIV infection.
INTRODUCTION
A major hallmark of untreated human immunodeficiency virus/simian immunodeficiency virus (HIV/SIV) infection is chronic generalized immune activation and inflammation that is linked to viral replication, loss of CD4+ T cells, immunological dysfunction, and disease progression (1–3). Further, chronic immune activation has been reported to persist even in HIV-infected individuals successfully treated with combination antiretroviral therapy (cART), suggesting that proinflammatory signaling does not completely subside in treated individuals (4). While the exact mechanisms driving chronic immune activation in HIV/SIV infection remain to be determined, several independent studies have implicated HIV persistence, coinfections with hepatitis C virus/hepatitis B virus/cytomegalovirus (HCV/HBV/CMV), compensatory homeostatic mechanisms, and chronic stimulation by translocated intestinal microbial products to drive this phenomenon. Among these, microbial translocation from a structurally and functionally damaged gastrointestinal (GI) tract has received considerable attention and is considered to significantly contribute to persistent systemic inflammation/immune activation and HIV/SIV disease progression (2, 3). Recent evidence has identified a central role for noncoding RNAs, particularly miRNAs, in the pathogenesis of chronic inflammatory diseases of the GI tract (5). While it is known that increased inflammatory gene transcription sustains persistent inflammation/immune activation in the intestine during HIV/SIV infection, the posttranscriptional mechanisms that fine-tune inflammatory gene activation/expression remain unknown and unaddressed.
Numerous studies have shown that microRNAs (miRNAs), a class of small noncoding RNAs, can posttranscriptionally regulate immune cell proliferation/differentiation/migration, cytokine signaling/production, and the inflammatory response (6–8). miRNA dysregulation has been associated with several chronic inflammatory diseases, such as inflammatory bowel disease (9), experimental autoimmune encephalomyelitis (10), multiple sclerosis (11), and rheumatoid arthritis (12). Lately, the enhanced expression of miR-21 (13) and miR-142 (14) was correlated with macrophage activation and encephalitis in the brain of SIV-infected rhesus macaques. Focusing on the GI tract, an important site of CD4+ T-cell depletion and HIV/SIV replication, we (15, 16) and others (17) reported significant miRNA dysregulation in intact colon during SIV infection. Unlike our two previous studies that utilized intact intestinal tissue (15, 16), in the present study, we focused exclusively on the intestinal lamina propria compartment to obtain a deeper understanding of the link between aberrant miRNA expression and chronic immune activation/inflammation in the intestine. In tune with this objective, we collected serial intestinal resection segments from the same animals before and at 21, 90, and 180 dpi and separated the different mucosal compartments (epithelium, intraepithelial lymphocytes, lamina propria leukocytes [LPLs], and fibrovascular stroma). This strategy enabled the identification of distinct miRNA profiles related to T-cell proliferation/expansion, activation, viral replication, and microbial translocation. Specifically, we found the expression of 4 miRNAs (miR-15b, miR-142-3p, miR-142-5p, and miR-150) previously linked to T-cell activation to be consistently downregulated at 90 and 180 dpi. Similar to the LPLs, miR-150 showed markedly reduced expression in jejunal and colonic epithelium. Further, we showed that miR-150 can directly regulate protein expression of IRAK1 (interleukin-1 receptor-associated kinase 1), an important serine/threonine protein kinase involved in Toll-like receptor (TLR) and interleukin-1R (IL-1R) signaling pathways. Furthermore, decreased miR-150 expression was associated with elevated protein expression of IRAK1 in both colonic epithelium and LPLs that may contribute to persistent inflammatory signaling in the intestine during chronic SIV infection. Finally, and more importantly, we demonstrate that delta-9 tetrahydrocannabinol (Δ9-THC)-mediated inhibition of CD8+ T-cell activation and proliferation successfully prevented miR-150 downregulation and IRAK1 upregulation, an important finding with strong translational significance.
MATERIALS AND METHODS
Animal care, ethics, and experimental procedures.
All experiments using rhesus macaques were approved by the Tulane Institutional Animal Care and Use Committee (protocol 3574). The Tulane National Primate Research Center (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility (AAALAC number 000594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071-01. All clinical procedures, including the administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Animals were anesthetized with ketamine hydrochloride for blood collection procedures. Intestinal pinch and resection biopsy specimens were performed by laboratory animal veterinarians. Animals were preanesthetized with ketamine hydrochloride, acepromazine, and glycopyrolate, intubated, and maintained on a mixture of isoflurane and oxygen. All possible measures were taken to minimize discomfort of all animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state, and local laws
Animals and experimental design.
Jejunum and colon tissues were collected from 32 Indian-origin rhesus macaques, including 16 animals infected with SIVmac251 or SIVmac239 and 16 uninfected macaques. Animal identifiers (IDs), SIV inoculum, duration of infection, plasma and intestinal viral loads, and intestinal histopathology in all SIV-infected and uninfected macaques with diarrhea and colitis are provided in Tables 1 and 2. Serial resection segments (∼6 to 8 cm long) of jejunum were collected from eight macaques (Table 1) before infection and again at 21 and 90 dpi and necropsy (180 dpi) and were used for the initial TaqMan low-density array (TLDA) microRNA profiling. An additional eight chronic SIV-infected and 12 control macaques (Table 2) were included for quantitative reverse transcription-PCR (qRT-PCR) confirmation studies. Colon resection segments (∼5 cm long) were collected from four macaques (HD08, HH07, HF54, and HR42) to be used as controls (Table 2). Among the 16 uninfected animals we included four that were necropsied for chronic diarrhea unresponsive to treatment (non-SIV infected, with diarrhea and colitis) (15, 18, 19) (Table 2). These animals were included to determine whether miR-150 downregulation was in response to immune activation or a consequence of CD4+ T-cell depletion that generally occurs in HIV/SIV infection. Peripheral blood (EDTA) was collected from 13 additional uninfected macaques to determine the effect of Δ9-THC on CD8+ T-cell activation and proliferation and on miR-150 and IRAK1 protein expression.
TABLE 1.
Viral loads and intestinal histopathology in SIV-infected rhesus macaques used for TLDA miRNA profilinga
| Animal ID | Viral inoculum | Viral load in: |
Intestinal histopathology |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma (106/ml) |
Jejunum (106/mg RNA) |
Colon (106/mg RNA) |
||||||||||
| 21 dpi | 90 dpi | 180 dpi | 21 dpi | 90 dpi | 180 dpi | 21 dpi | 90 dpi | 180 dpi | Jejunum | Colon | ||
| HC36 | SIVmac251 | 20 | 0.1 | 1 | 2 | ND | 100 | 1 | 0.2 | NA | ND | ND |
| HB31 | SIVmac251 | 10 | 70 | 3,000 | 1 | 0.97 | 10,000 | 2 | 7 | 200 | Lymphoid hyperplasia | Lymphoid hyperplasia |
| HF27 | SIVmac251 | 9 | 10 | 40 | 5 | 2 | 30 | 0.06 | 40 | 0.04 | Lymphoid hyperplasia | Lymphoid hyperplasia |
| HB48 | SIVmac251 | 40 | 20 | 20 | 6 | 100 | 10 | 300 | 0.1 | 0.8 | Mild enteritis | ND |
| GK31 | SIVmac251 | 2 | 30 | 30 | 0.04 | 0.05 | 2,000 | 2 | 1 | 3 | ND | ND |
| HR57 | SIVmac251 | 6 | 80 | NA | 1 | 4 | NA | 0.4 | 5 | NA | NA | NA |
| GA19 | SIVmac251 | 2 | 100 | 100 | 0.5 | 0.04 | 800 | 10 | 2 | 600 | ND | Lymphoid hyperplasia |
| HV95 | SIVmac251 | 2 | 7 | 10 | 0.5 | 0.08 | 4 | 0.2 | 0.5 | 0.1 | ND | Moderate colitis |
All 8 macaques were infected intravenously with 100 TCID50 SIVmac251. NA, not available. ND, none detected.
TABLE 2.
Viral loads and colon histopathology in chronic SIV-infected and uninfected rhesus macaques with diarrhea and colitisa
| Animal ID and type | SIV inoculum | Duration of infection | Viral load |
Colon histopathology | Opportunistic infection | |
|---|---|---|---|---|---|---|
| Plasma (106/ml) | Colon (106/mg RNA) | |||||
| Chronic SIV infected | ||||||
| FT11 | SIVmac251 | 145 | 500 | 2075 | Mild colitis | ND |
| HL01 | SIVmac251 | 180 | 7 | 0.8 | None | ND |
| HN15 | SIVmac251 | 177 | NA | 0.3 | Granulomatous colitis | Mycobacterium avium intracellulare |
| FE24 | SIVmac251 | 271 | 3.1 | 0.3 | Mild colitis | ND |
| EI15 | SIVmac251 | 300 | 0.4 | 0.9 | Mild enteritis | ND |
| FE53 | SIVmac251 | 140 | 0.3 | 5 | ND | ND |
| GH25 | SIVmac239 | 148 | NA | 300 | Mild suppurative colitis | ND |
| HB31 | SIVmac251 | 180 | 3000 | 200 | Lymphoid hyperplasia | ND |
| HF27 | SIVmac251 | 180 | 40 | 0.04 | Lymphoid hyperplasia | ND |
| HB48 | SIVmac251 | 180 | 20 | 0.8 | ND | ND |
| GA19 | SIVmac251 | 180 | 100 | 600 | Lymphoid hyperplasia | ND |
| HD08 | SIVmac251 | 90 | NA | 300 | Moderate colitis/cryptitis | ND |
| Non-SIV infected, with diarrhea and colitis | ||||||
| FL94 | NA | NA | NA | NA | Moderate colitis/cryptitis | NA |
| JA58 | NA | NA | NA | NA | Moderate hemorrhagic colitis | NA |
| IP74 | NA | NA | NA | NA | Moderate colitis/cryptitis | NA |
| IP87 | NA | NA | NA | NA | Moderate colitis | NA |
| Uninfected controls | ||||||
| HT74 | NA | NA | NA | NA | NA | NA |
| IT62 | NA | NA | NA | NA | NA | NA |
| IT16 | NA | NA | NA | NA | NA | NA |
| IT05 | NA | NA | NA | NA | NA | NA |
| IH95 | NA | NA | NA | NA | NA | NA |
| HT73 | NA | NA | NA | NA | NA | NA |
| EJ34 | NA | NA | NA | NA | NA | NA |
| IT13 | NA | NA | NA | NA | NA | NA |
| HD08 | NA | NA | NA | NA | NA | NA |
| HH07 | NA | NA | NA | NA | NA | NA |
| HF54 | NA | NA | NA | NA | NA | NA |
| HR42 | NA | NA | NA | NA | NA | NA |
NA, not applicable; ND, none detected.
At necropsy, all tissues were collected in RNAlater (Ambion, TX) for total RNA extraction and qRT-PCR. For histopathologic evaluation, intestinal tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 7 μm, and stained with hematoxylin and eosin for analysis.
Intestinal epithelial and lamina propria cell isolation.
The protocol and purity of epithelial leukocytes and LPLs isolated from surgical resection segments collected before and at 21, 90, and 180 dpi were described previously (20, 21). Briefly, surgical resection segments (6 to 8 cm long) for miRNA profiling studies first were incubated with vigorous shaking in Ca2+ Mg2+-free Hanks' balanced salt solution (HBSS) containing 1 mM EDTA for two 30-min incubations at 37°C to separate the intestinal epithelial cells. Following incubation, the epithelial cells in the supernatant were harvested by centrifugation at 500 × g for 10 min, followed by subjecting the cells to Percoll density gradient centrifugation to separate intraepithelial lymphocytes. After dislodging the epithelial cells the tissue segments were incubated twice for a 30-min duration in RPMI 1640 containing 20 U of collagenase per ml while rapidly shaking at 37°C to separate LPLs from the fibrovascular stroma. The tissue homogenates were subjected again to Percoll density gradient centrifugation to separate LPLs from the fibrovascular stroma. The purified LPLs then were used for flow cytometry and miRNA profiling as indicated below.
Intestinal resections were collected from SIV-infected macaques in our previous studies (15, 16, 20, 21); the animals tolerate this procedure extremely well. The three resections (preinfection and 21 and 90 days post-SIV infection) were spaced wide apart from one another (∼2.5 to 3 months), and complete healing occurs within 2 to 3 weeks. The animals were treated with antibiotics only on the day of surgery; therefore, neither surgery nor antibiotic treatment is expected to have any marked effects on the reported changes in miRNA expression profiles. LPLs from three preinfection (GK31, GA19, and HV95), one 90-dpi (GK31), and three 180-dpi (HC36, HR57, and HV95) samples did not yield the required concentration of total RNA per microliter (120 ng/μl) for TLDA microRNA profiling and, as a result, were excluded.
Flow cytometry to quantify T-cell dynamics in intestine and peripheral blood.
LPLs from duodenal pinch biopsies were isolated as described in the previous section and adjusted to a concentration of 107/ml. For in vivo T-cell and in vitro CD8+ T-cell immunophenotyping, ∼100-μl aliquots were stained with appropriately diluted, directly conjugated monoclonal antibodies to CD3 (Pacific blue, SP34-2), CD4 (SK3, peridinin chlorophyll protein-Cy5.5), and CD8 (phycoerythrin-Texas red, 3B5), CD69 (FN50 and V450), and Ki67 (556026, fluorescein isothiocyanate) (BD Biosciences, San Jose, CA). For surface markers, samples were stained for 30 min in the dark at room temperature. The samples then were permeabilized with a BD Cytofix/Cytoperm fixation/permeabilization solution kit (554714; BD Biosciences) and stained for the intracellular marker Ki67 for 25 min in the dark at room temperature. Once staining was complete, the samples were fixed in BD stabilizing fixative buffer (338036; BD Biosciences) and stored in the dark at 4°C overnight for acquisition the next day. Samples were acquired on LSR II flow cytometry equipment (BD Biosciences) and analyzed with Flow Jo software (Tree Star Inc., Ashland, OR). The cells first were gated on singlets followed by lymphocytes, CD3+ T cells, and then CD3+ CD4+ and CD3+ CD8+ T-cell subsets. For in vitro CD8+ T-cell immunophenotyping, cells first were gated on singlets followed by lymphocytes, CD3+ T cells, and then CD3+ CD4+, CD3+ CD8+, CD3+ CD8+ CD69+, and CD3+ CD8+ Ki67+ T-cell subsets.
Global microRNA profiling using TLDA.
Approximately 350 ng of total RNA from LPLs first was reverse transcribed, preamplified, and loaded onto TLDA plates and processed as described previously (15, 16). Total RNA was extracted from LPL samples using the miRNeasy total RNA isolation kit (Qiagen Inc., CA). RNA integrity was assessed by running an aliquot on a denaturing agarose gel followed by staining with ethidium bromide to visualize intact 28S and 18S rRNA bands. For TLDA miRNA profiling, ∼350 ng of total RNA first was reverse transcribed by following the ABI microRNA TLDA reverse transcription reaction protocol. Briefly, two master mixes were prepared for each RNA sample representing either TLDA panel (panel A or panel B) and consisted of the following reaction mixture components: 0.80 μl MegaPlex RT primers (10×), 0.20 μl deoxynucleoside triphosphate (dNTP) with dTTP (100 mM), 1.50 μl MultiScribe reverse transcriptase (50 U/μl), 0.80 μl 10× RT buffer, 0.90 μl MgCl2 (25 mM), 0.10 μl RNase inhibitor, and 0.20 μl nuclease-free water (20 U/μl). Three microliters of total RNA (350 ng) was loaded into appropriate wells of a 96-well plate containing 4.5 μl of the RT reaction mix and after a brief 5-min incubation on ice was subjected to the following thermal cycling conditions on the ABI 7900 HT Fast PCR system: standard or max ramp speed, 16°C for 2 min, 42°C for 1 min, 50°C for 1 s (40 cycles), and 85°C for 5 min (hold).
Approximately 2.5 μl of the resulting cDNA from each sample was mixed with a total of 22.5 μl of preamplification reaction mix consisting of 12.5 μl TaqMan PreAmp master mix (2×), 2.5 μl Megaplex PreAmp primers (10×), 7.5 μl nuclease-free water and was preamplified on the ABI 7900 HT Fast PCR system according to the TLDA miRNA preamplification protocol outlined by the manufacturer (Life Technologies). The preamplification thermal cycling conditions were the following: hold at 95°C for 10 min, hold at 55°C for 2 min, hold at 72°C for 2 min, and 12 cycles at 95°C for 15 s and 60°C for 4 min.
The preamplified product first was diluted 4-fold with 75 μl of 0.1× Tris-EDTA, pH 8.0, and mixed, following which 9 μl of the diluted PreAmp product was mixed with 450 μl of 2× TaqMan universal PCR master mix with no UNG (AmpErase) and 441 μl of nuclease-free water to bring the final volume to 1 ml. After proper mixing and centrifuging, 100 μl of the PCR mix was loaded into each port of the TaqMan array human microRNA A+B card set, v3.0. The TLDA cards then were centrifuged, sealed, and processed on the ABI 7900 HT sequence detection system using 384-well TLDA default thermal cycling conditions.
Quantitative real-time TaqMan stem-loop microRNA RT-PCR.
The expression of a select list of differentially expressed (DE) miRNAs at 21, 90, and 180 dpi was further confirmed in both intestinal LPLs (∼100 ng total RNA) and epithelium (∼250 ng total RNA) using the TaqMan microRNA predesigned and preoptimized assays (Life Technologies) as described previously (15, 16). Briefly, total RNA was reverse transcribed using the stem-loop primers provided in the predesigned kit in a total volume of 15 μl. Approximately 4 μl of cDNA was subjected to 40 cycles of PCR in a total volume of 20 μl on the ABI 7900 HT fast PCR system (Life Technologies) using the following thermal cycling conditions: 95°C for 10 min, followed by 40 repetitive cycles of 95°C for 15 s and 60°C for 1 min. As a normalization control for RNA loading, parallel reactions in duplicate wells to amplify snoU6 were run in the same or a different multiwell plate. Comparative real-time PCR was performed in duplicate wells, including no-template controls, and relative changes in gene expression were calculated using the comparative threshold cycle (ΔΔCT) method. PCR efficiency analysis was performed using serial 10-fold RNA dilutions (500, 50, 5, and 0.5 ng for all target miRNAs) (40, 4, 0.4, and 0.04 ng for RNU48 and snoU6). The amplification curves for all assays were linear and based on slope values (−3.09 to −3.12). All assays had 103 to 107% efficiency.
Immunofluorescence for cellular localization of IRAK1 in colon.
Immunofluorescence studies for the detection of IRAK1 (9F3 at 1:750) (Abcam) was done as described earlier (18, 19). Formalin-fixed, paraffin-embedded tissues were sectioned at 7 μm and stained with the appropriate primary and secondary antibody as well as hematoxylin and eosin to microscopically assess the histological status of the tissue. Briefly, slides were blocked with 100 to 200 μl of blocking buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 3% bovine serum albumin [BSA], 10% normal goat serum, and 0.1% Triton X-100) for 45 min to 1 h followed by 1 h of incubation at 37°C with rabbit polyclonal IRAK1 primary antibody (9F3 at 1:750) (Abcam). The slides were washed three times in buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton X-100) followed by the addition of goat anti-rabbit secondary antibody conjugated to Alexa Fluor 488 (1:1,000) (Invitrogen Corp., CA). This was followed by mouse anti-CD8 (IgG1 at 1:10) (Dako Laboratories) at room temperature for 1 h. The slides were washed three times and incubated for 1 h with goat anti-mouse secondary antibody conjugated with Alexa Fluor 568 (1:1,000). Confocal microscopy was performed using a Leica TCS SP2 confocal microscope equipped with three lasers (Leica Microsystems, Exton, PA). Individual optical slices represent 0.2 μm, and 32 to 62 optical slices were collected at 512- by 512-pixel resolution. NIH Image (version 1.62) and Adobe Photoshop (version 7.0) were used to assign colors to the three channels collected: Alexa 568 (Invitrogen Corp) is red, Alexa 488 (Invitrogen Corp) is green, and the differential interference contrast (DIC) image is in grayscale. The three channels were collected simultaneously. The colocalization of antigens is demonstrated by the addition of colors as indicated in the figure legends.
Immunophenotyping of IRAK1-positive cells was done using CD8 (1:10) (Dako, Denmark) and appropriate Alexa Fluor-conjugated secondary antibodies (Lifetech Corp). Images from all macaques were captured using the same laser (red and green) strength.
Quantitative image analysis.
Quantitation of cells and regions of interest (ROI) labeled by IRAK1 was performed using Volocity 5.5 software (PerkinElmer Inc., MA, USA) after capturing images on a Leica confocal microscope. Several ROI were hand drawn on the epithelial and LPL regions in the images captured from colon of chronic SIV-infected and uninfected macaques and processed utilizing the same brightness, density, and black level settings. The data first were graphed and then analyzed using Mann-Whitney U test employing the Prism v5 software (GraphPad software). P < 0.05 was considered statistically significant.
Cloning of 3′ UTRs of CD69, IRAK1 mRNA, and Dual-Glo luciferase reporter gene assay.
The 3′ untranslated region (UTR) of the rhesus CD69 mRNA contains three highly conserved predicted binding sites for miR-92a (TargetScan 6.2) (22) (see Table S1 in the supplemental material). Similarly, miR-150 has up to 4 predicted binding sites on the 3′ UTR of IRAK1 mRNA that are conserved in the human, chimpanzee, and rhesus macaque (see Table S2) (22). Oligonucleotide design (containing wild-type [WT] and deleted miRNA binding sites), synthesis, processing, pmirGLO vector cloning, and Dual Glo luciferase reporter assay were performed as described previously (15, 16). Briefly, a short 42- to 44-nucleotide-long sequence representing the 3′ UTR containing the predicted miRNA binding site was synthesized (IDT DNA Technologies Inc., IA) for cloning into the pmirGLO dual-luciferase vector (Promega Corp., Madison, WI). A second oligonucleotide with the binding site deleted also was synthesized to serve as a negative control. Both oligonucleotide sequences were synthesized with a PmeI site on the 5′ end and an XbaI site on the 3′ end for directional cloning. The pmirGLO vector first was cut with PmeI and XbaI restriction enzymes, gel purified, and ligated with either wild-type (WT) sequence containing the miRNA binding site or deleted (Del) sequence. HEK293 cells were plated at a density of 5 × 104 cells per well of a 96-well plate. At 50% confluence, cells were cotransfected with ∼100 ng WT or Del UTR miRNA luciferase reporter vector and 100 nM miRNA mimic using the DharmaFECT duo transfection reagent (ThermoFisher Scientific). In separate wells, cells also were transfected with pmirGLO vector (Promega Corp) as a normalization control. After 48 h, the Dual-Glo luciferase assay was performed according to the manufacturer's recommended protocol using the BioTek H4 Synergy plate reader (BioTek, Winooski, VT). The normalized Firefly/Renilla ratio was calculated to determine the relative reporter activity. Experiments were performed in 6 replicates and repeated thrice.
Peripheral blood CD8+ T-cell isolation and culture.
Peripheral blood mononuclear cells (PBMCs) were isolated from EDTA blood by density gradient centrifugation using lymphocyte separation medium (LSM) as previously described (15). Briefly, after the removal of plasma, reconstituted blood was layered onto LSM for the collection of lymphocytes. Red blood cell contamination was removed by lysis with ACK buffer, and cells were washed prior to resuspension in complete RPMI 1640 containing 5% heat-inactivated fetal bovine serum, l-glutamine, and penicillin-streptomycin. CD8+ T cells were purified from PBMCs (3.9 × 107 to 14.2 × 107 cells/animal) using a magnetic activated cell sorting CD8+ T-cell isolation kit, nonhuman primate version (Miltenyi Biotec), by following the manufacturer's protocol. CD8+ T-cell yields ranged from 4 × 106 to 14 × 106/ml. Δ9-THC was provided by the National Institute on Drug Abuse (NIDA; Rockville, MD), supplied as a solution of 200 mg/ml in absolute ethanol, and stored at −20°C. Δ9-THC was concentrated with a Vacufuge (Eppendorf, Hamburg, Germany) to remove ethanol and resuspended in dimethyl sulfoxide (DMSO). Purified CD8+ T cells (∼4 × 106 to 14 × 106 cells) from each animal were resuspended in complete RPMI 1640 and preincubated with either 15 μM Δ9-THC or an equal volume of vehicle (DMSO) for 30 min, followed by activation with 40 nM PMA (phorbol 12-myristate 13-acetate)–0.5 μM ionomycin as described previously by Kaplan et al. (24). For each animal, isolated CD8+ T cells were split equally between the two treatments. Δ9-THC or vehicle was present throughout the 24-h incubation period.
Transient overexpression of miR-150.
HEK293 cells were cultured in 6-well plates in 2 ml of complete Eagle's minimum essential medium containing 10% fetal bovine serum (FBS). At 80% confluence, cells were transfected with 2 μg of pEP-miR-150 plasmid (Cell Biolabs, San Diego, CA, USA) for primary miR-150 overexpression or pEP-Null vector (control plasmid) in duplicate wells using Dharmfect Duo transfection reagent as described previously (15) and cultured for 96 h.
Immunoprecipitation and Western blotting.
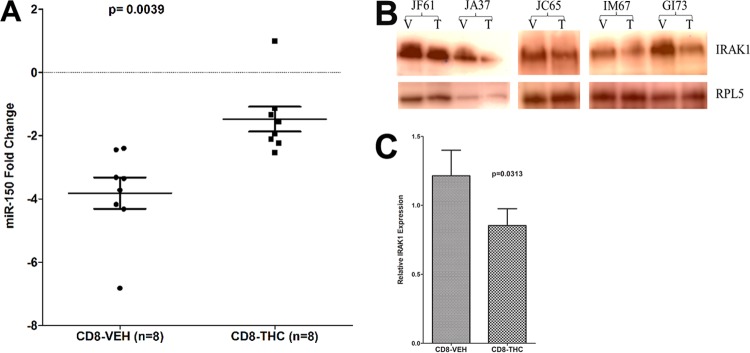
Total protein from HEK293 cells (pEP-Null or pEP-miR-150 transfected) and CD8+ T cells (vehicle or Δ9-THC treated) was extracted using a lysis buffer (Cell Signaling Technology, Inc., Beverly, MA) containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, protease inhibitor cocktail, and phosphatase inhibitor cocktail (Sigma Chemical Company, MO). Total protein was quantified using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA). Approximately 150 to 300 μg of total protein extract was immunoprecipitated with ∼5 μl of a rabbit polyclonal antibody against IRAK1 (Santa Cruz Biotechnologies, CA) overnight at 4°C, followed by incubation with 25 μl (50%, wt/vol) of protein G agarose beads (Invitrogen Corp., CA) at 4°C for 4 to 5 h. Identical protein concentrations (CD8+ T cells) representing both treatments (vehicle versus THC) from the same animal were used for immunoprecipitation. The supernatant was removed and transferred to a separate 1.5-ml microcentrifuge tube and immunoprecipitated using goat polyclonal antibody (∼5 μl) (Santa Cruz Biotechnologies, CA) to either β-actin or ribosomal protein L5 (RPL5) at 4°C overnight on a shaker. Immunoprecipitated IRAK1, β-actin, and RPL5 proteins were heat denatured for 5 min at 100°C in sample loading buffer containing 62.5 mM Tris-HCl, 5% 2-mercaptoethanol, 10% glycerol, 2% SDS, and bromophenol blue, resolved on 8 to 12% SDS-PAGE gels, and transferred to 0.2-μm polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, CA). The membranes were probed with a rabbit polyclonal primary antibody against IRAK1 (Cell Signaling Technologies, MA) and rabbit polyclonal antibody to β-actin (Santa Cruz) or ribosomal protein L5 (RPL5) (Cell Signal, MA), followed by the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz, CA). Membranes were treated with West-Dura chemiluminescent substrate (Pierce Biotechnology Inc., Rockford, IL) for 5 min, and the signal was captured using the FluorChem-R imaging system (ProteinSimple) and quantified using ImageJ software (NIH). Since CD8+ T cells were harvested on different days, immunoprecipitation/Western blotting was performed separately when the cells were available. Hence, Fig. 9B shows three separate images.
FIG 9.
THC treatment prevented miR-150 downregulation (A) and IRAK1 upregulation (B) in CD8+ T cells 24 h after in vitro activation with PMA-ionomycin compared to those treated with vehicle (VEH). (C) The quantification of Western blotting revealed significant THC-mediated reduction of IRAK1 protein levels in CD8+ T cells compared to those treated with vehicle. miR-150 expression was normalized to snoU6. The error bars represent standard errors of the means of fold changes within each group. T, THC; V, vehicle (DMSO).
Quantitation of plasma and mucosal viral loads.
Total RNA samples (plasma and intestine) from all SIV-infected animals were subjected to a quantitative real-time TaqMan two-step RT-PCR analysis to determine the viral load as described earlier (15, 16). Total RNA samples from all SIV-infected animals were subjected to a quantitative real-time TaqMan two-step RT-PCR analysis to determine the viral load in SIV-infected colon and jejunum samples. Briefly, primers and probes specific to the SIV long terminal repeat sequence were designed and used in the real-time TaqMan PCR assay. Probes were conjugated with a fluorescent reporter dye (6-carboxyfluorescein) at the 5′ end and a quencher dye at the 3′ end. Fluorescence signal was detected with an ABI 7900 HT fast PCR system (Life Technologies). Data were captured and analyzed with Sequence Detector software (PE Applied Biosystems). The viral copy number was determined by plotting CT values against a standard curve (y = −3.351x + 40.377) (r2 = 0.998) generated with in vitro-transcribed RNA representing known viral copy numbers.
Data analysis.
TLDA and individual miRNA qRT-PCR data analyses were performed as described before (15, 16). miRNA expression data were normalized to a combination of two endogenous controls (snoU6 and RNU48). Given the exploratory nature of the study with a small sample size, we decided not to apply multiple-comparison correction (Benjamini-Hochberg method for false discovery rate) mainly to avoid type-II errors (false negatives). However, to avoid type I errors (false positives), we confirmed the expression of several miRNAs using qRT-PCR at 21, 90, and 180 dpi.
For LPL qRT-PCR confirmation studies, miRNA fold changes were calculated using an average of all control animal ΔCT values. For the quantification of miR-150 in LPLs of SIV-uninfected macaques with diarrhea and colitis (see Fig. S1 in the supplemental material) and vehicle- or THC-treated CD8+ T cells (Fig. 9A), one uninfected control macaque or THC-treated CD8+ T-cell sample with the lowest ΔCT value served as the calibrator/reference and was assigned a value of 1. miR-150 fold changes in all other macaques, including THC- and vehicle-treated CD8+ T-cell samples, are shown as an n-fold difference relative to the sample. The stepwise calculation of fold change using this approach is shown in Table S3. We used this approach as it facilitated graphing all samples so that the variation within both groups can be displayed. Individual LPL miRNA qRT-PCR data were analyzed using Wilcoxon's rank-sum test using the RealTime StatMiner package (Integromics). Intestinal CD4+ and CD8+ T-cell data at 21, 90, and 180 dpi were analyzed using Kruskal-Wallis test, and post hoc analysis was done using Dunn's multiple-group comparison (Prism v5 software; GraphPad Software, La Jolla, CA). P values of <0.05 were considered significant. Quantitative image analysis data were analyzed using Mann-Whitney U test (Prism v5 software). Firefly/Renilla ratios were analyzed using an unpaired t test. Data related to the effect of THC on CD8+ T-cell activation, proliferation, miR-150 expression, and IRAK1 protein quantification were analyzed using the Wilcoxon matched-pairs signed-rank test (Prism v5 software).
Microarray data accession number.
TLDA miRNA data were deposited in the GEO database under accession number GSE77134.
RESULTS
Mucosal immunophenotyping, viral loads, and intestinal histopathology.
All SIV-infected macaques used for TLDA miRNA profiling had substantial plasma and intestinal viral loads at 21, 90, and 180 dpi (Table 1). As shown in Fig. 1A, a statistically significant reduction in intestinal CD4+ T-cell percentages was observed at 180 dpi. The CD4+ T-cell data were included in a previous publication (15), as these animals were used in that study. We have included these data once again for the convenience of the reader. In contrast to CD4+, intestinal CD8+ T-cell percentages increased at all three time points (Fig. 1B). Similar to CD4+, CD8+ T-cell increases only at the 180-dpi time point showed statistical significance (Fig. 1B).
FIG 1.
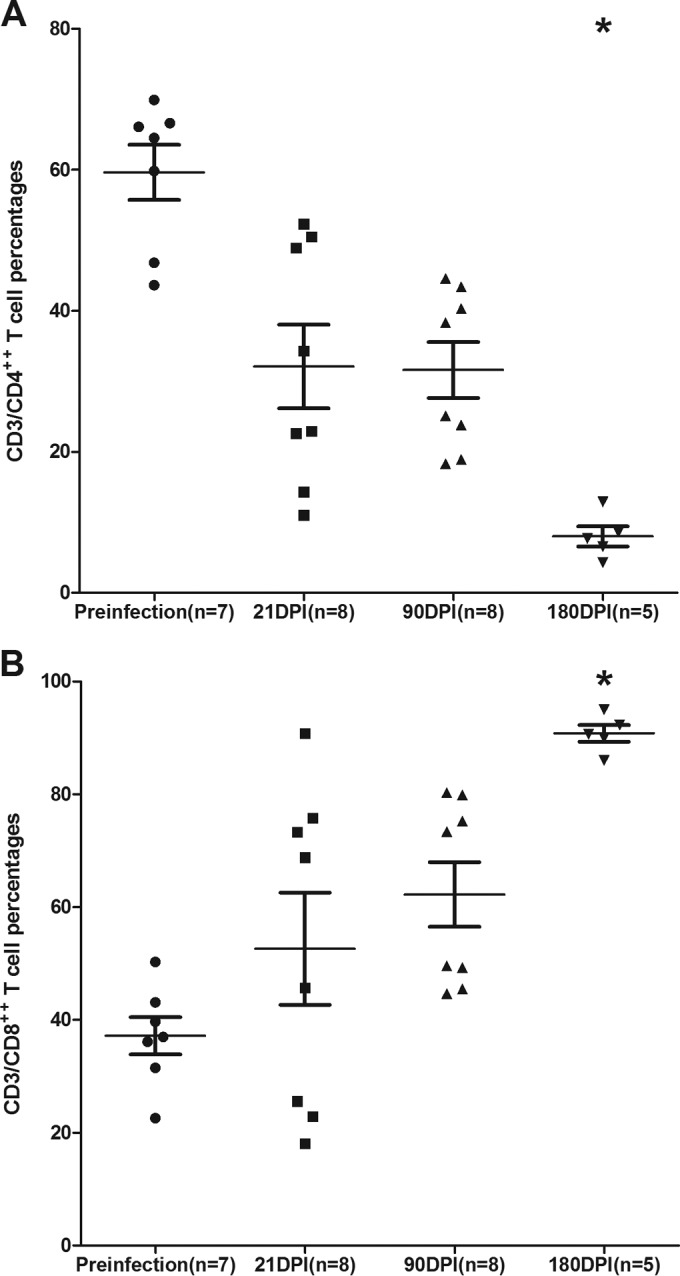
Percentage of CD4+ (A) and CD8+ (B) T cells among lamina propria leukocytes (LPLs) isolated at different time points after SIV infection. The cells were first gated on singlets followed by lymphocytes and CD3, and then they were gated on CD3+ CD4+ and CD3+ CD8+ T-cell subsets. Data analysis using Kruskal-Wallis test identified significant differences among the different groups for both CD4 (P = 0.0003) and CD8 (P = 0.0020) T cells. Post hoc analysis using Dunn's multiple-group comparison identified both CD4+ and CD8+ T cells at 180 dpi to be significantly different from the preinfection group. *, P < 0.05. The error bars represent standard errors of means for CD4+ or CD8+ T-cell percentages within each group.
Histologic evaluation of colon sections (Table 2) from all but three chronic SIV-infected macaques revealed the presence of mild to severe colitis, including lesions such as crypt abscess/cryptitis (Table 2). Opportunistic pathogens such as Mycobacterium avium intracellulare were detected in only 1 out of the 12 chronic SIV-infected macaques (Table 2).
miRNAs associated with CD8+ T-cell expansion, macrophage activation, and viral replication showed marked alterations at 21 dpi.
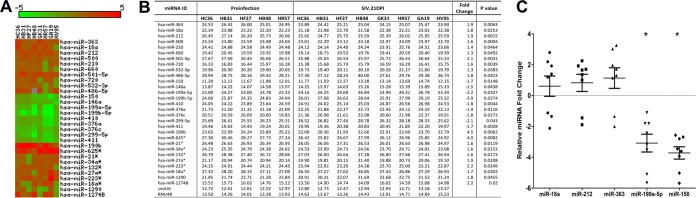
To identify differentially expressed (DE) miRNAs in LPLs during the course of SIV infection, we obtained serial jejunal resections mainly due to the higher LPL yields than those from colon samples. miRNA expression profiling of LPLs at 21 dpi identified 29 DE miRNAs (Fig. 2A and B). Out of these, 20 showed increased and 9 showed decreased expression (Fig. 2A and B). The notable miRNAs that were upregulated included miR-18a, miR-363, miR-212, and miR-190b. Strikingly, miR-18a and miR-363, which belong to the miR-17-92 cluster, and its paralog, miR-106a-363, were significantly upregulated. Previous studies in mice had identified a role for the miR-17-92 cluster in early CD8+ T-cell proliferation and clonal expansion (25, 26). Although flow cytometry data (Fig. 1B) showed increased CD8+ T-cell percentages in LPLs at 21 dpi compared to that at the preinfection time point, future studies are needed to confirm whether increased expression of miRNAs belonging to the miR-17-92 cluster during acute HIV/SIV infection is due to CD8+ T-cell proliferation and expansion. Similarly, miR-212 previously was linked to macrophage activation (27), and its upregulation may be an early indication of macrophage activation during acute SIV infection. Interestingly, miR-150, an important miRNA known to be downregulated during T-cell (28–30) and B-cell (31, 32) activation, showed significantly reduced expression (Fig. 2B), providing early evidence of immune activation. The finding that miR-150 expression also decreased in LPLs isolated from non-SIV-infected macaques with diarrhea and colitis (see Fig. S1 in the supplemental material) suggests that its downregulation is not due to CD4+ T-cell depletion but likely is due to T-cell activation, as these animals had moderate colitis (Table 2) but no CD4+ T-cell depletion (15, 18, 19). Further, miR-150 is a lymphoid-enriched miRNA, and the low CT value of ∼11 to 12 (Fig. 2B) provides strong evidence that the LPLs are highly enriched for lymphocytes. As reported previously in colon tissue from irritable bowel syndrome (IBS) patients (33), miR-199a-5p and miR-199b-5p showed decreased expression in LPLs at 21 dpi. The downregulation of both miRNAs may be in response to mucosal inflammation that is common to both IBS and HIV/SIV infection. qRT-PCR studies confirmed the significantly reduced expression of miR-150 and miR-199a-5p (Fig. 2C). In contrast, miR-18a (P = 0.18), miR-363 (P = 0.219), and miR-212 (P = 0.0726) expression was increased yet did not reach statistical significance (Fig. 2C). Interestingly, the significantly enhanced expression of miR-190b at 21 dpi (Fig. 2A and B) further confirms our recent demonstration that its upregulation is driven in response to viral replication and not immune/inflammatory responses (15). Collectively, miRNA profiles at 21 dpi in LPLs show selective upregulation of miRNAs previously associated with early CD8+ T-cell proliferation (miR-17-92 cluster), macrophage activation (miR-212), and SIV replication (miR-190b), and the concurrent downregulation of miR-150 may be an early indication of T-cell and, possibly, B-cell activation.
FIG 2.
Changes in miRNA expression in intestinal LPLs of rhesus macaques at 21 days post-SIV infection compared to the preinfection time point. (A) The heat map shows all differentially expressed (P < 0.05) miRNAs in intestinal LPLs of rhesus macaques at 21 dpi. (B) Table shows raw CT, fold change, and P values for all differentially expressed miRNAs in LPLs at 21 dpi. miRNA species originating from the opposite arm of the precursor are denoted with an asterisk. (C) qRT-PCR confirmation of select differentially expressed miRNAs at 21 dpi (n = 8) relative to preinfection samples (n = 5). Data were analyzed using the nonparametric Wilcoxon's rank-sum test for independent samples. The error bars represent standard errors of means of fold changes within each group. A single asterisk indicates statistical significance (P < 0.05) compared to preinfection controls.
A four-miRNA signature linked to T-cell activation is consistently downregulated at viral set point (90 dpi) and in chronic SIV infection (180 dpi).
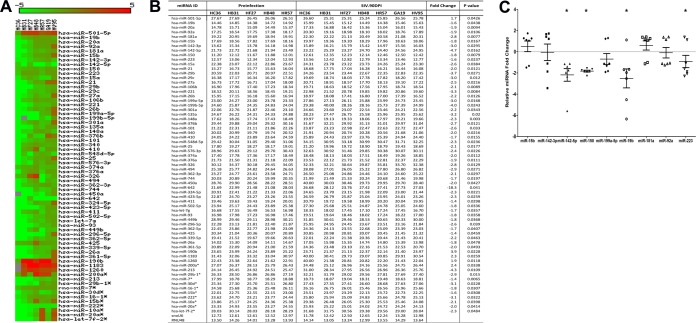
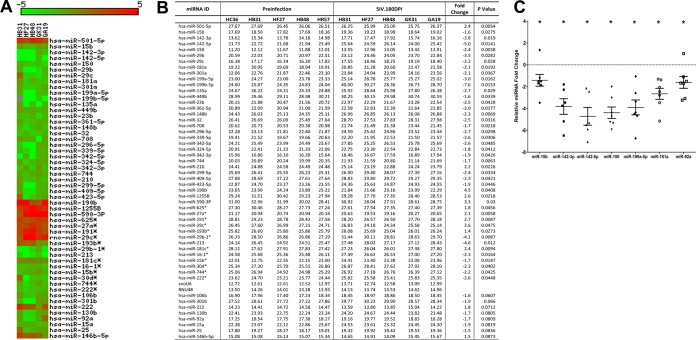
In contrast to results at 21 dpi, at the viral set point (90 dpi), 65 miRNAs showed differential expression (Fig. 3A and B). Sixty of the 65 DE miRNAs (92%) showed significantly decreased expression (Fig. 3A and B). Compared to results at 21 dpi, the number of downregulated miRNAs at 90 dpi increased ∼7-fold (n = 60). At 180 dpi, 44 DE miRNAs were identified. Similar to results at 90 dpi, the majority (∼75%) of the DE miRNAs at 180 dpi were downregulated (n = 33) and 25% (n = 11) were upregulated (Fig. 4A and B), suggesting that as disease progresses the miRNAome is downregulated.
FIG 3.
Changes in miRNA expression in intestinal LPLs of rhesus macaques at 90 days post-SIV infection compared to the preinfection time point. (A) Heat map shows all differentially expressed (P < 0.05) miRNAs in intestinal LPLs of rhesus macaques at 90 dpi. (B) Table shows raw CT, fold change, and P values for all differentially expressed miRNAs in LPLs at 90 dpi. miRNA species originating from the opposite arm of the precursor are denoted with an asterisk. (C) qRT-PCR confirmation of select differentially expressed miRNAs at 90 dpi (n = 7) relative to preinfection samples (n = 5). Data were analyzed using the nonparametric Wilcoxon's rank-sum test for independent samples. The error bars represent standard errors of means of fold changes within each group. A single asterisk indicates statistical significance (P < 0.05) compared to preinfection controls.
FIG 4.
Changes in miRNA expression in intestinal LPLs of rhesus macaques at 180 days post-SIV infection compared to the preinfection time point. (A) Heat map shows all differentially expressed (P < 0.05) miRNAs in intestinal LPLs of rhesus macaques at 180 dpi. (B) Table shows raw CT, fold change, and P values for all differentially expressed miRNAs in LPLs at 180 dpi. miRNA species originating from the opposite arm of the precursor are denoted with an asterisk. (C) qRT-PCR confirmation of select differentially expressed miRNAs at 180 dpi (n = 6) relative to preinfection samples (n = 5). Data were analyzed using the nonparametric Wilcoxon's rank-sum test for independent samples. The error bars represent standard errors of means of fold changes within each group. A single asterisk indicates statistical significance (P < 0.05) compared to preinfection controls.
At 90 and 180 dpi, several miRNAs belonging to the miR-17-92 cluster, namely, miR-19b, miR-20a, miR-17, miR-18a, miR-20b, and miR-92a, were significantly downregulated. These in vivo findings are consistent with three previous reports that showed the downregulation of the miR-17-92 cluster during CD8+ T-cell differentiation and activation in vitro (25, 26, 34, 35). Interestingly, miR-181a expression decreased at both 90 and 180 dpi. More importantly, the expression of four miRNAs (miR-15b, miR-142-3p, miR-142-5p, and miR-150), previously shown to be associated with T-cell activation (28, 29), was significantly downregulated in LPLs at 90 and 180 dpi. Further, miR-223, previously associated with NK-cell activation, also showed decreased expression at 90 dpi (36, 37). Furthermore, the expression of the Th17-enriched miR-301a (38) was significantly decreased at both time points, possibly be due to the virus-induced depletion of these cells (1, 2). Figure S2 in the supplemental material shows the expression of 12 differentially expressed miRNAs in five animals compared longitudinally to their preinfection levels.
qRT-PCR studies confirmed the statistically significant downregulation of miR-150, miR-142-5p, miR-19b, and miR-223 in LPLs at 90 dpi (Fig. 3C). In contrast, the expression of miR-142-3p (P = 0.072), miR-15b (P = 0.072), miR-92a (P = 0.29), and miR-181a (P = 0.29) did not show statistical significance (Fig. 3C). However, at 180 dpi, qRT-PCR confirmed statistically significant downregulation of miR-15b, miR-142-3p, miR-142-5p, miR-150, miR-199a-5p, and miR-181a (Fig. 4C). At 180 dpi, although miR-92a downregulation based on TLDA profiling did not show statistical significance (P = 0.0809), qRT-PCR confirmation demonstrated statistical significance (Fig. 4C). Similar to results at 21 dpi, the expression of the SIV-induced miR-190b was significantly elevated at 90 and 180 dpi (Fig. 3 and 4), confirming our previous report showing its persistent elevation throughout SIV infection (15).
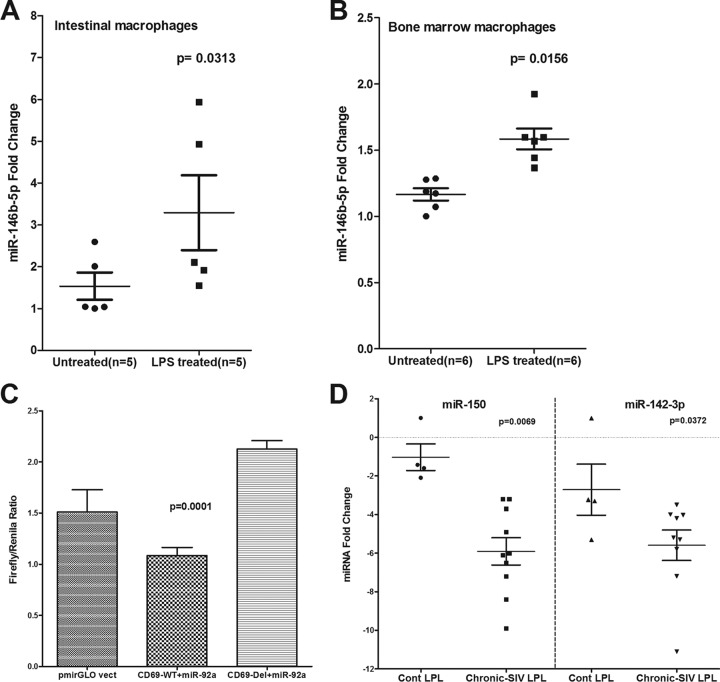
At 180 dpi, miR-146b-5p, a lipopolysaccharide (LPS)-responsive miRNA in THP-1 monocytes (39), showed a statistically nonsignificant (P = 0.0873) (Fig. 4B) increase, suggesting that its elevated expression is triggered by translocated intestinal bacteria and their products. To test this possibility, we isolated (15) and treated in vitro-cultured primary intestinal and bone marrow-derived macrophages from macaques necropsied for diarrhea and colitis with LPS. Unlike resident intestinal macrophages, recently emigrated monocytes from blood (40) and inflammatory macrophages in the intestine adhere to culture flasks (41), proliferate (15), and express high levels of the LPS coreceptor CD14 (40, 41), which makes them responsive to LPS treatment in vitro. As shown in Fig. 5, miR-146b-5p expression increased significantly in intestinal (P = 0.0313) (Fig. 5A) and bone marrow-derived macrophages (P = 0.0156) (Fig. 5B) 16 h after LPS treatment. Overall, the miRNA expression profiles in LPLs at 90 and 180 dpi were dominated by downregulated miRNAs, and the attenuated expression of members of the miR-17-92 cluster, miR-15b, miR-142-3p, miR-142-5p, and miR-150, in LPLs is suggestive of chronic persistent T-cell activation, a hallmark of untreated HIV/SIV infection.
FIG 5.
miR-146b-5p, an LPS-responsive miRNA, is significantly elevated in primary intestinal (A) and bone marrow-derived (B) macrophages in response to in vitro LPS treatment (100 ng) for 24 h. (C) miR-92a can physically bind to the 3′ UTR of rhesus macaque CD69 mRNA and downregulate its expression (P = 0.0001). HEK293 cells were cotransfected with 50 nM miR-92a mimic and 100 ng of luciferase reporter constructs containing wild-type (WT) or deleted (Del) CD69 3′ UTR sequences. Firefly and Renilla luciferase activities were detected using the Dual-Glo luciferase assay system 72 h after transfection. The ratio of luciferase activities (Firefly/Renilla) was calculated and normalized to the wells transfected with only unmanipulated pmir-GLO vector. (D) miR-150 and miR-142-3p showed statistically significant downregulation in colonic LPLs from chronically SIV-infected macaques compared to uninfected control (Cont) macaques. Data were analyzed using the nonparametric Wilcoxon's rank-sum test. The error bars represent standard errors of the means of fold changes within each group.
miR-92a can directly target the lymphocyte proliferation and activation marker CD69.
As T-cell activation is associated with enhanced gene expression, it is logical to assume that miRNA downregulation may be necessary to relax the repression of genes that control their effector functions. One such gene is CD69, a C-type lectin and an early activation marker required for lymphocyte proliferation, migration, and regulation of immune responses (42). Moreover, marked increases in the levels of CD8+ CD69+ T cells were detected in the intestine after SIV infection (43). Using the TargetScan algorithm (22), predicted binding sites for ∼10 downregulated miRNAs (miR-15a, miR-15b, miR-20a, miR-20b, miR-106b, miR-301a, miR-181a, miR-92a, miR-21, and miR-142-5p) were identified on the 3′ UTR of the rhesus macaque CD69 mRNA (see Table S1 in the supplemental material). Since miR-92a had three predicted conserved binding sites on the 3′ UTR of CD69 mRNA, we further tested its ability to bind the 3′ UTR using the luciferase reporter assay. As shown in Fig. 5C, cotransfection of HEK293 cells with CD69 WT vector and miR-92a mimic reduced Firefly/Renilla ratios by ∼50% compared to those of cells transfected with the CD69 Del vector in three independent experiments. These results demonstrate that miR-92a can posttranscriptionally regulate CD69 expression by physically interacting with its 3′ UTR during T-cell activation, and its decreased expression is consistent with the assumption that the miRNA-mediated posttranscriptional silencing needs to be shut down during T-cell activation to relieve the repression of genes (7) required to fulfill their effector functions.
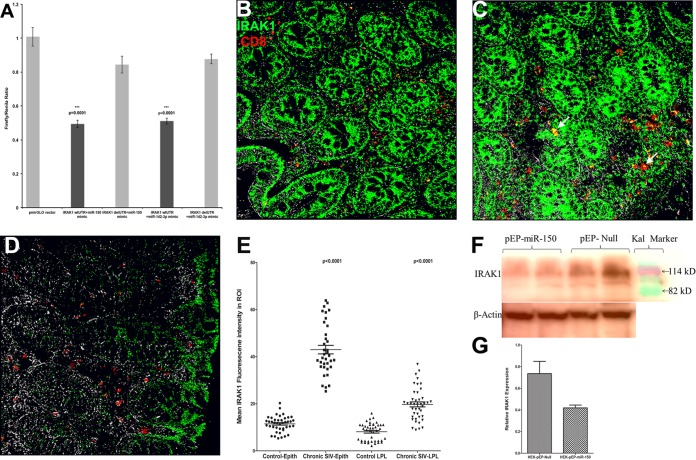
IRAK1 is a direct target of miR-150, and its protein expression is markedly elevated in colonic epithelium and LPLs during chronic SIV infection.
To understand the biological significance of miRNA downregulation associated with T-cell activation, we focused on the predicted targets of miR-150, as its expression has been confirmed by multiple studies to be rapidly and extremely downregulated during T-cell activation (28, 29). Further and more importantly, miR-150 showed reduced expression at all three time points in LPLs from jejunum (Fig. 2C, 3C, and 4C) and colon (Fig. 5D). Similar to miR-150, miR-142-3p also showed statistically significant downregulation in colonic LPLs of chronically SIV-infected macaques (Fig. 5D). Since translocating microbial products from a disrupted intestinal epithelial barrier can activate LPLs via the TLR pathway, we focused on IRAK1, a serine/threonine protein kinase and a critical component of the IL-1 receptor and TLR signaling pathways (23) predicted to be a direct target of miR-150 (22). Specifically, miR-150 has four predicted binding sites on the 3′ UTR of IRAK1 mRNA that are conserved in the human, chimpanzee, and rhesus macaque (see Table S2 in the supplemental material) (22). Interestingly, miR-142-3p also has a single predicted binding site on the 3′ UTR of IRAK1 mRNA that is highly conserved across several mammalian species (see Table S2 in the supplemental material) (22). As shown in Fig. 6A, cotransfection of pmirGLO-wtIRAK1 with miR-150 and miR-142-3p mimic resulted in significant reduction (∼40% for miR-150 and ∼51% for miR-142-3p) in Firefly/Renilla ratios (P = 0.0001). In contrast, the cotransfection of pmirGLO-delIRAK1, in which the predicted miR-150 and miR-142-3p binding sites were deleted, abolished their inhibitory effect on IRAK1 (Fig. 6A) reporter gene activity. These results provide indirect evidence that mml-miR-150 and mml-miR-142-3p can physically bind to the 3′ UTR of IRAK1 and negatively regulate its expression. Although both miRNAs showed significantly decreased expression, we decided to focus on miR-150 over miR-142-3p owing to its high relative abundance (CT of 11 for miR-150 versus 13 for miR-142-3p) (Fig. 3B and 4B) in LPLs and the presence of multiple predicted binding sites on the IRAK1 mRNA 3′ UTR (22). Accordingly, we next hypothesized that the decreased expression of miR-150 in the LPLs would relieve the translational repression exerted on IRAK1, resulting in their elevated protein expression. As shown in Fig. 6B and C, IRAK1 protein expression was significantly elevated in LPLs of chronically SIV-infected macaques. Further, immunoprecipitation/Western blotting revealed increased IRAK1 protein expression in colon and jejunum longitudinally as disease progressed (see Fig. S3 in the supplemental material). Using double labeling, CD8+ T cells were found to strongly express IRAK1 (Fig. 6B and C). Image analysis also identified a higher number of CD8+ T cells in the colonic lamina propria of chronic SIV-infected macaques that also expressed IRAK1 compared to control macaques (45.4 ± 7.8 versus 22.7 ± 2.6). In addition, elevated IRAK1 protein expression also was detected in cells other than CD8+ T cells, possibly macrophages, B cells, and dendritic cells (Fig. 6B and C). In contrast, IRAK1 protein expression was significantly weaker in the control macaque (Fig. 6D). Further, image analysis clearly demonstrated quantitative differences in protein expression (Fig. 6E). While markedly elevated IRAK1 protein expression was detected in both colon and jejunum, we have shown data from the colon, as it represents a major contributor to microbial translocation owing to its large concentration of bacterial (1011 to 1012 cells/g of intestinal contents) and other microbial flora (44). Finally, the overexpression of miR-150 significantly reduced IRAK1 protein expression (∼80 kDa), strongly underscoring the possibility that miR-150 directly targets IRAK1 through a negative feedback loop, and its downregulation in LPLs during SIV infection may facilitate the persistence of a chronic inflammatory environment in the intestine.
FIG 6.
(A) IRAK1 is a direct target of miR-150. Luciferase reporter vectors containing all four miR-150 binding sites (seed region) on the rhesus macaque 3′ UTR of IRAK1 mRNA or the corresponding construct with the binding sites deleted were cotransfected into HEK293 cells with 50 nM miRNA mimic. Firefly and Renilla luciferase activities were detected using the Dual-Glo luciferase assay system 72 h after transfection. The ratio of luciferase activities (Firefly/Renilla) was calculated and normalized to the wells transfected with only unmanipulated pmir-GLO vector. Triple asterisks indicate statistical significance (P < 0.001) compared to cells transfected with pmirGLO-delIRAK1 vector. IRAK1 protein expression is markedly elevated in the colon of chronic SIV-infected rhesus macaques (B and C) compared to that of the uninfected control macaque (D). Panels B, C, and D involve double labels with IRAK1 (green) and CD8 (red) for T cells. The gray channel represents differential interference contrast (DIC) to reveal tissue architecture. Colocalizations of green (IRAK1) and red (CD8) appear light yellow (arrows). In panels B and C, note the intense IRAK1 staining in the epithelium and lamina propria and markedly increased numbers of CD8+ IRAK1+ T cells (white arrowheads) in the colon of the SIV-infected macaques. In contrast, panel D shows that IRAK1 staining is extremely weak in the colonic epithelium and LPLs of the control macaque. All panels are ×40 magnification. Quantification of cells and regions of interest (ROI) labeled by IRAK1 was performed using Volocity 5.5 software after capturing images on a Leica confocal microscope. (E) Several ROI were hand drawn on the epithelial and LPL regions in the images from colon. Image analysis data were analyzed using nonparametric Wilcoxon's rank-sum test. (F) miR-150 can negatively regulate IRAK1 protein expression. Representative Western blot (F) and quantification (G) show the reduction in protein expression of IRAK1 (∼80 kDa) 96 h posttransfection of HEK293T cells with pre-miR-150-expressing plasmid (pEP-miR-150) (duplicate wells) compared to those transfected with pEP-Null control vector (duplicate wells). Kal is a marker (Bio-Rad kaleidoscope prestained marker).
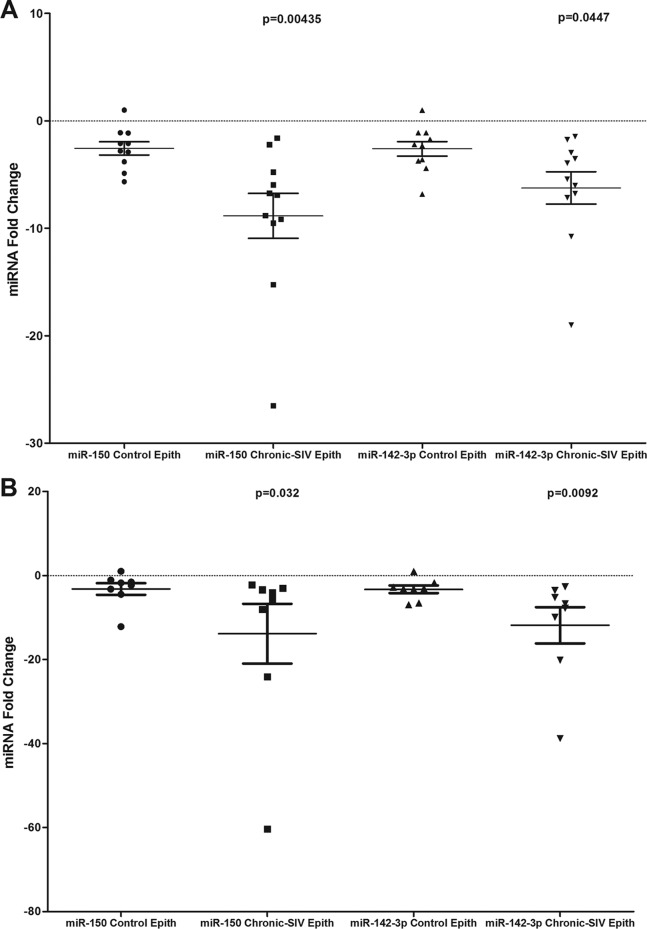
miR-150 and miR-142-3p expression is significantly decreased in both jejunal and colonic epithelium during chronic SIV infection.
In addition to LPLs, the significantly elevated IRAK1 protein expression in the colonic epithelium (Fig. 6B and C) prompted us to determine if similar reductions in miR-150 and miR-142-3p expression occurred in the intestinal epithelium. As shown in Fig. 7A and B, miR-150 and miR-142-3p expression decreased significantly in both colonic and jejunal epithelium. These findings suggest that as disease progresses, the diminished expression of miR-150 and possibly miR-142-3p reduce the constraints exerted on IRAK1 protein translation, leading to its elevated expression and in turn resulting in persistent inflammation, inflammatory cytokine production, epithelial barrier disruption, microbial translocation, and disease progression.
FIG 7.
miR-150 and miR-142-3p expression is significantly (P < 0.05) downregulated in colonic (A) and jejunal (B) epithelium (Epith) of chronically SIV-infected rhesus macaques compared to those of uninfected controls. Data were analyzed using the nonparametric Wilcoxon's rank-sum test. The error bars represent standard errors of the means of fold changes within each group.
In vitro blockade of peripheral blood CD8+ T-cell activation and proliferation with Δ9-THC significantly inhibited miR-150 downregulation and IRAK1 upregulation.
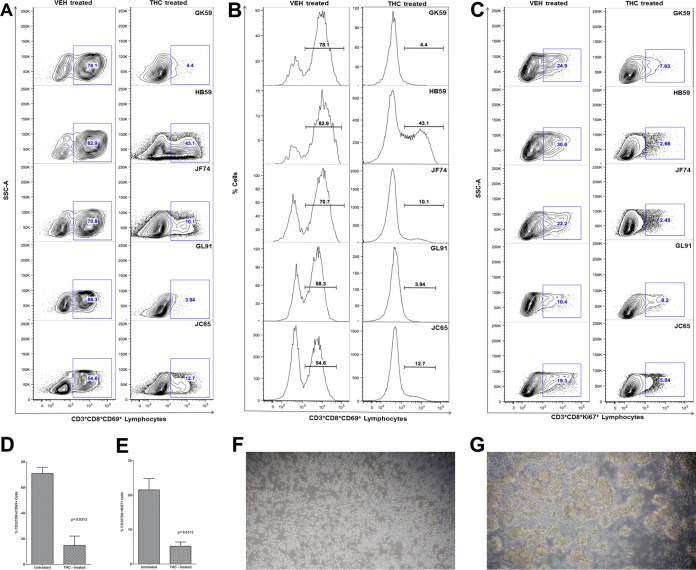
We recently demonstrated that chronic Δ9-THC administration induced the expression of several anti-inflammatory miRNAs in the intestines of SIV-infected macaques compared to those receiving vehicle (45). In the same study, THC treatment prevented the downregulation of miR-150 (45), indirectly suggesting that THC inhibits immune activation by modulating miRNA expression. Although the findings demonstrated translational relevance, it was unclear whether THC exerted these effects directly or indirectly (46). To address this question, we isolated and pretreated peripheral blood CD8+ T cells with 15 μM THC or vehicle (DMSO) for 30 min, followed by activation in vitro with 40 nM PMA–0.5 μM ionomycin and culture for 24 h (23). This concentration of THC previously has been shown to inhibit B-cell activation in vitro without reducing cell viability (46). As shown in Fig. 8, THC treatment significantly reduced the percentage of CD3+ CD8+ CD69+ (Fig. 8 A, B, and D) and CD3+ CD8+ Ki67+ (Fig. 8C and E) cells compared to the level of vehicle (DMSO)-treated controls, suggesting strong inhibition of T-cell activation and proliferation. The inhibitory effects of THC on T-cell activation and proliferation are clearly visible in Fig. 8F, as evidenced by the presence of individual cells compared to numerous cellular clumps (activation-induced adherence) in vehicle-treated controls (Fig. 8G). Interestingly, pretreatment of CD8+ T cells with THC also prevented the downregulation of miR-150 (Fig. 9A; also see Table S3 in the supplemental material), suggesting a direct effect. Note that in all 8 animals, the miR-150 CT values (see Table S3) are greater in vehicle-treated cells than in cells treated with THC, suggesting rapid downregulation in response to activation. Accordingly, we next hypothesized that THC-mediated inhibition of miR-150 downregulation in CD8+ T cells will prevent the upregulation of its predicted target, IRAK1. We chose this immunomodulatory strategy to examine the relationship between miR-150 and its target, IRAK1, because of its strong translational feasibility. As evidenced in Fig. 9B and C, the prevention of miR-150 downregulation in THC-treated cells resulted in reduced IRAK1 protein expression compared to that of vehicle-treated controls, where IRAK1 protein showed significantly elevated expression. Note that within each animal, IRAK1 expression is clearly higher in cells treated with vehicle than THC. Overall, these findings strongly confirm miR-150 downregulation in LPLs to be associated with immune cell activation and are not a consequence of CD4+ T-cell depletion. Finally and most importantly, the ability of Δ9-THC to prevent miR-150 downregulation and IRAK1 upregulation highlights the strong therapeutic potential of cannabinoids for the management of chronic immune activation/inflammation, a hallmark of HIV/SIV infection.
FIG 8.
THC inhibits CD8+ T-cell activation in vitro. THC-treated CD8+ T cells show significantly reduced expression of the lymphocyte activation marker CD69 (A and B) and proliferation marker Ki67 (C). Note the significantly reduced percentage of CD3+ CD8+ CD69+ and CD3+ CD8+ Ki67+ cells in all five animals 24 h after in vitro activation with PMA-ionomycin. Mean percentages of CD8+ CD69+ (D) and CD8+ Ki67+ (E) are significantly lower in THC-treated than in vehicle (VEH)-treated cells. THC-mediated inhibition of in vitro CD8+ T-cell activation (A and B) and proliferation (C) in response to PMA-ionomycin stimulation is evidenced by the absence of cellular clumps (F) compared to cells treated with vehicle (DMSO) (G). The error bars represent standard errors of the means of fold changes within each group.
DISCUSSION
The GI tract and intestinal immune system suffer significant structural and functional damage during the course of HIV/SIV infection. The LPL compartment, which is the effector arm of the GI immune system, is enriched for viral target cells and consequently is a major site of viral replication, CD4+ T-cell depletion, and dissemination. Additionally, active viral replication in this compartment stimulates increased proinflammatory cytokine/chemokine production, which, in addition to amplifying the inflammatory reaction, also disrupts the intestinal epithelial barrier, facilitating microbial translocation and chronic local and systemic immune activation/inflammation (1, 2). Consequently, the numerous effector cells that traffic into this compartment directly interact with cytokines and translocated microbial products, resulting in their persistent hyperactivation, thus setting up a vicious cycle involving viral replication, immune activation/inflammation, epithelial barrier disruption, and microbial translocation that eventually leads to AIDS progression. To better understand the molecular pathological networks/mechanisms underlying HIV/SIV-induced GI disease/dysfunction, we recently described differential gene expression separately in LPLs and epithelial cells at 21 and 90 dpi (20, 21). While differential gene expression is central to immune cell differentiation and activation, it is regulated in a synchronized fashion with contributions from transcription factors, histone-modifying enzymes, DNA methylation, chromatin remodeling complexes, chromatin-interacting protein kinases, and so on. Recently, miRNAs have emerged as potent posttranscriptional regulators of gene expression, and increasing evidence suggests a critical role for these small regulatory RNAs in controlling several aspects of the immune/inflammatory response (5–8). To investigate the posttranscriptional regulation of the immune/inflammatory response to SIV infection in the intestine, we collected serial jejunal resection segments and isolated and profiled miRNA expression exclusively in LPLs at 21, 90, and 180 dpi. This approach helped identify several miRNAs previously linked to innate and adaptive immune responses, including the reduced expression of at least four miRNAs previously associated with T-cell activation in the intestine at 90 and 180 dpi. Further, CD69 was validated as a direct target of miR-92a, and miR-150 and miR-142-3p expression was significantly downregulated in both LPLs and epithelium during chronic SIV infection. Decreased miR-150 expression was associated with elevated protein expression of IRAK1 in both colonic epithelium and LPLs, highlighting a very important link between perturbations in miRNA levels and the establishment of persistent inflammation/immune activation in the intestine.
Among the 22 DE miRNAs identified in LPLs at 21 dpi, miR-18a and miR-363, belonging to the miR-17-92 and miR-106a-363 cluster, showed enhanced expression and previously were linked to early CD8+ T-cell proliferation and clonal expansion (25, 26). In addition, the induction of miR-212 was shown to occur during macrophage activation (27). In contrast, at 90 and again at 180 dpi, ≥75 to 86% of the DE miRNAs showed decreased expression. At least four miRNAs, miR-15b, miR-142-3p, miR-142-5p, and miR-150, previously shown to be significantly decreased during CD4+ and CD8+ T-cell activation (28, 29), showed consistent downregulation at both 90 and 180 dpi. Further, other members of the miR-17-92 cluster and their paralogs, miR-18a, miR-19b, miR-20a, miR-20b, miR-92a, and miR-106a, showed significantly reduced expression at 90 or 180 dpi or both. Interestingly, miR-146b-5p (P = 00873), an LPS-responsive miRNA, showed statistically nonsignificant elevation at 180 dpi. Recent in vitro studies showed miR-146b-5p to be significantly upregulated following LPS treatment of THP monocytes; however, they showed delayed kinetics (39). This raised the question of whether translocated luminal bacteria and their products in the lamina propria (47) are triggering miR-146b-5p upregulation. To address this question, we isolated intestinal and bone marrow macrophages and found significantly elevated miR-146b-5p expression following a 16-h LPS treatment in vitro. At all time points, miR-501-5p and miR-190b showed elevated expression. We recently characterized miR-190b in the intestine and found its expression to be significantly upregulated throughout SIV infection (15). Further, we also demonstrated its upregulation to be driven by viral replication and not immune/inflammatory responses accompanying viral replication (15). Similarly, miR-150, miR-199a-5p, and miR-199b-5p showed differential expression at all time points but at a significantly reduced level. The downregulation of miR-150, particularly in CD4+ T-cell activation (29), also may promote viral replication, as it has been demonstrated to exert anti-HIV effects by directly targeting HIV (48) and, possibly, SIV RNA. Similar to several previous studies that have linked miR-150 downregulation to T-cell (28–30) and B-cell (31, 32) activation, a recent study in IBS patients found decreased miR-199a-5p and miR-199b-5p to result in chronic visceral pain and nociception (33). The study also suggested that the downregulation of both miRNAs is driven in response to colonic inflammation (33). Unlike colon (16), we did not detect elevated miR-34a expression in jejunum LPLs. This may be due to our previous finding that HIV/SIV impacts the colon more severely than the jejunum (18, 19). Overall, these findings are novel, as they identify a potential role for specific miRNAs in key pathogenic events during HIV/SIV infection, such as immune activation (miR-15b, miR-142-3p, miR-142-5p, and miR-150) and microbial translocation (miR-146b-5p). Finally and more importantly, these findings would not have been possible had we used intact intestinal segments.
Previous and more recent studies have demonstrated that with the exception of a few (miR-155 and miR-29b), the expression of most miRNAs is downregulated during T-cell activation (6). This global miRNA downregulation was shown to be caused by selective ubiquitylation and proteosomal degradation of AGO2 (Argonaute proteins), a critical component of the miRNA processing machinery (29). Additionally, to evade the silencing effect of a few upregulated miRNAs (miR-155 and miR-29b), activated T cells have been shown to transcribe mRNAs with shortened 3′ UTRs through alternative polyadenylation (49). A shortened 3′ UTR can protect mRNAs from miRNA-mediated silencing, as most miRNA binding sites are located on the distal regions of the 3′ UTR (49). Consistent with this hypothesis, we found predicted binding sites for at least 10 downregulated miRNAs on the 3′ UTR of the rhesus macaque CD69 mRNA (22), an immunoregulatory molecule induced following T-cell activation. Among these, miR-15a/b, miR-20a, miR-20b, miR-21, miR-106b, miR-301a, and miR-181a have one predicted binding site (22). Further, miR-142-5p and miR-92a were found to have two and three binding sites, respectively, on the 3′ UTR of CD69 mRNA (22). The results of the luciferase reporter assay confirmed the ability of miR-92a to directly bind the 3′ UTR of CD69 mRNA and potentially downregulate its expression. Overall, these findings show that the global and selective downregulation of miRNA subsets previously demonstrated in in vitro-activated T cells (28, 29) occurs in intestinal LPLs during SIV infection. Given the importance of genes such as CD69 to lymphocyte activation and proliferation, the large-scale downregulation of miRNAs targeting CD69 and possibly other immunoregulatory proteins, such as perforin and IL-2Rα (30), may be necessary to assist T cells overcome the miRNA barrier to stabilize gene expression.
Although global miRNA downregulation can stabilize gene expression to help activated cells mount an immune response, paradoxically it can disrupt the translational control of proinflammatory signaling molecules and cytokines. In the setting of HIV/SIV infection, such drastic reductions in the LPL miRNAome can enhance proinflammatory protein translation, leading to persistent proinflammatory signaling and inflammation. To explore this possibility, we focused on miR-150, as it was significantly downregulated at all three time points and shown by multiple studies to be significantly and drastically downregulated following CD4+ and CD8+ T-cell activation (28, 29). Further, bioinformatic analysis (22) revealed the presence of up to four miR-150 binding sites on the 3′ UTR of the rhesus macaque IRAK1 mRNA, a serine/threonine protein kinase, and a key signal transducer of the IL-1-receptor and TLR signaling cascades. In addition to miR-150, the IRAK1 3′ UTR also has a highly conserved binding site for miR-142-3p, suggesting the possibility of direct targeting by both miRNAs. Furthermore, mice lacking IRAK1 fail to mount an inflammatory response and produce significantly lower levels of proinflammatory cytokines, suggesting their critical requirement for activation of the NF-κB signaling pathway and the subsequent inflammatory response (23). Finally, recent studies have confirmed the central role of IRAK1 in driving intestinal inflammation (50, 51). Consistent with miR-150 downregulation, IRAK1 protein expression significantly increased in colonic LPLs of chronically SIV-infected macaques. Elevated IRAK1 protein expression also was detected in the jejunum of chronically SIV-infected macaques (data not shown). Surprisingly, quantitative image analysis revealed severalfold higher IRAK1 protein expression in the epithelium compared to that of the LPLs, raising the important question of whether decreases in miR-150 expression also occurred in the epithelium. Similar to the LPLs, qRT-PCR confirmed significantly reduced miR-150 expression in colonic and jejunal epithelium. Moreover, the results of the luciferase reporter and overexpression assays provided strong evidence that miR-150 can directly target IRAK1 and potentially regulate its protein expression in the epithelium and LPLs. Lastly and most importantly, treatment of CD8+ T cells with Δ9-THC, a potent psychotropic anti-inflammatory cannabinoid in marijuana, effectively prevented CD8+ T-cell activation and proliferation, miR-150 downregulation, and IRAK1 protein upregulation. Collectively, elevated IRAK1 protein expression in response to miR-150 downregulation may greatly facilitate the downstream transduction of proinflammatory signals, thus creating a persistent inflammatory environment in the intestine, as demonstrated recently using an IRAK1 knockout mouse model (51). In addition, our findings also suggest that cannabinoids provide a feasible strategy to decrease persistent immune activation/inflammation in HIV/SIV infection.
To summarize, unlike our previous transcriptome (mRNA) profiling studies using LPLs (20), the present study for the first time clearly identified miRNA signatures associated with key pathogenic events, such as immune activation and microbial translocation, and firmly confirmed our previously reported miR-190b induction in response to SIV replication (15). Despite the LPLs being a heterogeneous cellular population (predominantly lymphocytes with smaller numbers of macrophages, plasma cells, and dendritic cells), we successfully identified specific miRNAs associated with early CD8+ T-cell proliferation, macrophage activation (both at 21 dpi), chronic T-cell activation (90 and 180 dpi), microbial translocation (180 dpi), and SIV replication (all three time points). In addition to the well-established miR-146a (52), the present study identified miR-150, a very-high-abundance lymphocyte-enriched miRNA, to directly regulate IRAK1 expression. It is now known that inflammation and immune dysregulation directly or indirectly lead to most of the morbidity and mortality in HIV infection, even in patients on suppressive antiretroviral therapy. Accordingly, the ability of THC to inhibit immune activation/inflammation via modulating miRNA and proinflammatory protein expression suggests that cannabinoids could potentially have a significant impact on the course of HIV/SIV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ronald S. Veazey, Andrew A. Lackner, Pyone P. Aye, Xavier Alvarez, Maurice Duplantis, Yun Te Lin, Lawrance C. Chandra, Cecily C. Midkiff, Christopher Monjure, and Coty Tatum for their technical assistance in the study. We also thank the National Institute on Drug Abuse for providing Δ9-THC.
Funding Statement
This study was funded by National Institutes of Health (NIH) grants (R01DK083929 and R01DA042524 to M.M and TNPRC base grant OD011104 [formerly RR00164]).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00189-16.
REFERENCES
- 1.Brenchley JM. 2013. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol 6:657–665. doi: 10.1038/mi.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klatt NR, Funderburg NT, Brenchley JM. 2013. Microbial translocation, immune activation, and HIV disease. Trends Microbiol 21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristoff J, Haret-Richter G, Ma D, Ribeiro RM, Xu C, Cornell E, Stock JL, He T, Mobley AD, Ross S, Trichel A, Wilson C, Tracy R, Landay A, Apetrei C, Pandrea I. 2014. Early microbial translocation blockade reduces SIV-mediated inflammation and viral replication. J Clin Investig 124:2802–2806. doi: 10.1172/JCI75090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassol E, Malfeld S, Mahasha P, Bond R, Slavik T, Seebregts C, Poli G, Cassol S, van der Merwe SW, Rossouw T. 2013. Impaired CD4+ T-cell restoration in the small versus large intestine of HIV-1-positive South Africans receiving combination antiretroviral therapy. J Infect Dis 208:1113–1122. doi: 10.1093/infdis/jit249. [DOI] [PubMed] [Google Scholar]
- 5.Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. 2015. MicroRNAs: new players in IBD. Gut 64:504–517. doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Podshivalova K, Salomon DR. 2013. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit Rev Immunol 33:435–476. doi: 10.1615/CritRevImmunol.2013006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeker LT, Bluestone JA. 2013. MicroRNA regulation of T-cell differentiation and function. Immunol Rev 253:65–81. doi: 10.1111/imr.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connell RM, Rao DS, Baltimore D. 2012. MicroRNA regulation of inflammatory responses. Annu Rev Immunol 30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 9.Peck BC, Weiser M, Lee SE, Gipson GR, Iyer VB, Sartor RB, Herfarth HH, Long MD, Hansen JJ, Isaacs KL, Trembath DG, Rahbar R, Sadiq TS, Furey TS, Sethupathy P, Sheikh SZ. 2015. MicroRNAs classify different disease behavior phenotypes of Crohn's disease and may have prognostic utility. Inflamm Bowel Dis 21:2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu E, Wang X, Zheng B, Wang Q, Hao J, Chen S, Zhao Q, Zhao L, Wu Z, Yin Z. 2014. miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORγt and STAT3. J Immunol 192:5599–5609. doi: 10.4049/jimmunol.1303488. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, Singh N, Nagarkatti M, Nagarkatti P. 2014. Expression, regulation and function of microRNAs in multiple sclerosis. Int J Med Sci 11:810–818. doi: 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Huo R, Xiao L, Zhu X, Xie J, Sun S, He Y, Zhang J, Sun Y, Zhou Z, Wu P, Shen B, Li D, Li N. 2014. A novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol 66:49–59. doi: 10.1002/art.38142. [DOI] [PubMed] [Google Scholar]
- 13.Yelamanchili SV, Chaudhuri AD, Chen LN, Xiong H, Fox HS. 2010. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis 1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri AD, Yelamanchili SV, Marcondes MC, Fox HS. 2013. Up-regulation of microRNA-142 in simian immunodeficiency virus encephalitis leads to repression of sirtuin1. FASEB J 27:3720–3729. doi: 10.1096/fj.13-232678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohan M, Chandra LC, Torben W, Aye PP, Alvarez X, Lackner AA. 2014. miR-190b is markedly upregulated in the intestine in response to SIV replication and partly regulates myotubularin related protein-6 expression. J Immunol 193:1301–1313. doi: 10.4049/jimmunol.1303479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohan M, Kumar V, Lackner AA, Alvarez X. 2015. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic SIV infection of rhesus macaques. J Immunol 194:291–306. doi: 10.4049/jimmunol.1401447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaulke CA, Porter M, Han YH, Sankaran-Walters S, Grishina I, George MD, Dang AT, Ding SW, Jiang G, Korf I, Dandekar S. 2014. Intestinal epithelial barrier disruption through altered mucosal microRNA expression in human immunodeficiency virus and simian immunodeficiency virus infections. J Virol 88:6268–6280. doi: 10.1128/JVI.00097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. 2007. Gastrointestinal disease in simian immunodeficiency virus-infected rhesus macaques is characterized by proinflammatory dysregulation of the interleukin-6-janus kinase/signal transducer and activator of transcription-3 pathway. Am J Pathol 171:1952–1965. doi: 10.2353/ajpath.2007.070017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan M, Aye PP, Borda JT, Alvarez X, Lackner AA. 2008. CCAAT/enhancer binding protein beta is a major mediator of inflammation and viral replication in the gastrointestinal tract of SIV-infected rhesus macaques. Am J Pathol 173:106–118. doi: 10.2353/ajpath.2008.080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. 2012. Focused examination of the intestinal lamina propria yields greater molecular insight into mechanisms underlying SIV induced immune dysfunction. PLoS One 7:e34561. doi: 10.1371/journal.pone.0034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. 2013. Focused examination of the intestinal epithelium reveals transcriptional signatures consistent with disturbances in enterocyte maturation and differentiation during the course of SIV infection. PLoS One 8:e60122. doi: 10.1371/journal.pone.0060122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Gottipati S, Rao NL, Fung-Leung WP. 2008. IRAK1: a critical signaling mediator of innate immunity. Cell Signal 20:269–276. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan BL, Springs AE, Kaminski NE. 2008. The profile of immune modulation by cannabidiol (CBD) involves deregulation of nuclear factor of activated T cells (NFAT). Biochem Pharmacol 76:726–737. doi: 10.1016/j.bcp.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Wieland A, Araki K, Davis CW, Ye L, Hale JS, Ahmed R. 2012. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 109:9965–9970. doi: 10.1073/pnas.1207327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan AA, Penny LA, Yuzefpolskiy Y, Sarkar S, Kalia V. 2013. MicroRNA-17∼92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 121:4473–4483. doi: 10.1182/blood-2012-06-435412. [DOI] [PubMed] [Google Scholar]
- 27.Nahid MA, Yao B, Dominguez-Gutierrez PR, Kesavalu L, Satoh M, Chan EK. 2013. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J Immunol 190:1250–1263. doi: 10.4049/jimmunol.1103060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. 2007. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, Rao A, Ansel KM. 2013. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trifari S, Pipkin ME, Bandukwala HS, Äijö T, Bassein J, Chen R, Martinez GJ, Rao A. 2013. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 110:18608–18613. doi: 10.1073/pnas.1317191110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 32.de Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. 2013. Regulation of B-cell development and function by microRNAs. Immunol Rev 253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Q, Yang L, Larson S, Basra S, Merwat S, Tan A, Croce C, Verne GN. 13 February 2015. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut doi: 10.1136/gutjnl-2013-306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salaun B, Yamamoto T, Badran B, Tsunetsugu-Yokota Y, Roux A, Baitsch L, Rouas R, Fayyad-Kazan H, Baumgaertner P, Devevre E, Ramesh A, Braun M, Speiser D, Autran B, Martiat P, Appay V, Romero P. 2011. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J Transl Med 9:44. doi: 10.1186/1479-5876-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohyashiki M, Ohyashiki JH, Hirota A, Kobayashi C, Ohyashiki K. 2011. Age-related decrease of miRNA-92a levels in human CD8+ T-cells correlates with a reduction of naïve T lymphocytes. Immun Ageing 8:11. doi: 10.1186/1742-4933-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, Heinz ME, Crosby SD, Nagarajan R, Ramsingh G, Link DC, Ley TJ, Mardis ER. 2010. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res 20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenu EW, Ashton KJ, van Driel M, Staines DR, Peterson D, Atkinson GM, Marshall-Gradisnik SM. 2012. Cytotoxic lymphocyte microRNAs as prospective biomarkers for chronic fatigue syndrome/myalgic encephalomyelitis. J Affect Disord 141:261–269. doi: 10.1016/j.jad.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 38.Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. 2012. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci U S A 109:E1248–E1257. doi: 10.1073/pnas.1114325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. 2013. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A 110:11499–11504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bain CC, Mowat AM. 2014. Macrophages in intestinal homeostasis and inflammation. Immunol Rev 260:102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T. 2008. Unique CD14+ intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN gamma axis. J Clin Investig 118:2269–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancho D, Gómez M, Sánchez-Madrid F. 2005. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol 26:136–140. doi: 10.1016/j.it.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Xu H, Alvarez X, Pahar B, Moroney-Rasmussen T, Lackner AA, Veazey RS. 2011. Distinct expression patterns of CD69 in mucosal and systemic lymphoid tissues in primary SIV infection of rhesus macaques. PLoS One 6:e27207. doi: 10.1371/journal.pone.0027207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artis D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol 8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 45.Chandra LC, Kumar V, Torben W, Vande Stouwe C, Winsauer P, Amedee A, Molina PE, Mohan M. 2015. Chronic THC administration induces an intestinal anti-inflammatory microRNA expression during acute SIV infection of rhesus macaques. J Virol 89:1168–1181. doi: 10.1128/JVI.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngaotepprutaram T, Kaplan BL, Carney S, Crawford R, Kaminski NE. 2013. Suppression by Δ-tetrahydrocannabinol of the primary immunoglobulin M response by human peripheral blood B cells is associated with impaired STAT3 activation. Toxicology 310:84–91. doi: 10.1016/j.tox.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, Hirsch VM, Silvestri G, Douek DC, Miller CJ, Haase AT, Lifson J, Brenchley JM. 2010. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog 6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantri CK, Mantri JV, Pandhare J, Dash C. 2014. Methamphetamine inhibits HIV-1 replication in CD4+ T cells by modulating anti-HIV-1 miRNA expression. Am J Pathol 184:92–100. doi: 10.1016/j.ajpath.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chassin C, Hempel C, Stockinger S, Dupont A, Kübler JF, Wedemeyer J, Vandewalle A, Hornef MW. 2012. MicroRNA-146a-mediated downregulation of IRAK1 protects mouse and human small intestine against ischemia/reperfusion injury. EMBO Mol Med 4:1308–1319. doi: 10.1002/emmm.201201298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heiseke AF, Jeuk BH, Markota A, Straub T, Lehr HA, Reindl W, Krug AB. 2015. IRAK1 drives intestinal inflammation by promoting the generation of effector Th cells with optimal gut-homing capacity. J Immunol 195:5787–5794. doi: 10.4049/jimmunol.1501874. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. 2012. miR-146a controls the resolution of T cell responses in mice. J Exp Med 209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.