ABSTRACT
An altered intestinal microbiome during chronic human immunodeficiency virus (HIV) infection is associated with mucosal dysfunction, inflammation, and disease progression. We performed a preclinical evaluation of the safety and efficacy of fecal microbiota transplantation (FMT) as a potential therapeutic in HIV-infected individuals. Antiretroviral-treated, chronically simian immunodeficiency virus (SIV)-infected rhesus macaques received antibiotics followed by FMT. The greatest microbiota shift was observed after antibiotic treatment. The bacterial community composition at 2 weeks post-FMT resembled the pre-FMT community structure, although differences in the abundances of minor bacterial populations remained. Immunologically, we observed significant increases in the number of peripheral Th17 and Th22 cells and reduced CD4+ T cell activation in gastrointestinal tissues post-FMT. Importantly, the transplant was well tolerated with no negative clinical side effects. Although this pilot study did not control for the differential contributions of antibiotic treatment and FMT to the observed results, the data suggest that FMT may have beneficial effects that should be further evaluated in larger studies.
IMPORTANCE Due to the immunodeficiency and chronic inflammation that occurs during HIV infection, determination of the safety of FMT is crucial to prevent deleterious consequences if it is to be used as a treatment in the future. Here we used the macaque model of HIV infection and performed FMT on six chronically SIV-infected rhesus macaques on antiretroviral treatment. In addition to providing a preclinical demonstration of the safety of FMT in primates infected with a lentivirus, this study provided a unique opportunity to examine the relationships between alterations to the microbiome and immunological parameters. In this study, we found increased numbers of Th17 and Th22 cells as well as decreased activation of CD4+ T cells post-FMT, and these changes correlated most strongly across all sampling time points with lower-abundance taxonomic groups and other taxonomic groups in the colon. Overall, these data provide evidence that changes in the microbiome, particularly in terms of diversity and changes in minor populations, can enhance immunity and do not have adverse consequences.
INTRODUCTION
Antiretroviral treatment (ART) has substantially decreased the progression to AIDS in human immunodeficiency virus (HIV)-infected individuals. However, despite the ability to suppress viremia, HIV-infected individuals in whom the infection is suppressed by ART have increased rates of morbidity and mortality compared to those of uninfected individuals (1). One mechanism underlying the increased mortality is dysfunction of the gastrointestinal (GI) tract, which is associated with inflammation during HIV infection. Several lines of evidence support the role of GI dysfunction in HIV-related disease, including (i) a consistent association between mortality in HIV-infected individuals and markers of microbial translocation and gut epithelial dysfunction (2–4), (ii) the association of microbial products that translocate during HIV or simian immunodeficiency virus (SIV) infection and inflammation (5–7), and (iii) dysbiosis of the gut microbiome during HIV infection, which results in inflammation (8–10).
The recent awareness of the importance of the gut microbiome in human health has greatly improved our understanding of the interactions between GI bacteria and the immune system, as well as the importance of maintaining healthy microbial communities at mucosal surfaces. During HIV infection, the delicate balance of commensal bacterial communities is perturbed, resulting in microbial dysbiosis. This is typically characterized by an overall loss of diversity, with alterations to the phyla Bacteroidetes, Firmicutes, and Proteobacteria (11). Specifically, the loss of beneficial bacterial genera, such as Bacteroides, Lactobacillus, and Bifidobacterium, has been observed and associated with pathogenesis (9, 10, 12, 13). Furthermore, the levels of several pathogenic Proteobacteria have been observed to increase during HIV infection, including those within the genera Campylobacter, Escherichia, Acinetobacter, Desulfovibrio, and Pseudomonas, as have those of Prevotella species (9–11). Indeed, these bacteria have been found to be more adherent to the epithelium, to be more prone to translocate, and to be drivers of inflammation in the context of HIV and SIV infections (8–10, 14). In an effort to reverse the deleterious effects of gut dysbiosis, studies have been performed using probiotic microbes to improve health in HIV-infected individuals and using the SIV-infected macaque model. In pathogenic SIV infection, animals in which SIV infection is suppressed by ART have enhanced immunity and CD4+ T cell restoration in the GI tract after probiotic therapy or therapy with probiotics supplemented with interleukin-21 (IL-21) (15, 16). In HIV-infected individuals, probiotics have been shown to decrease inflammation and microbial translocation (17, 18). Thus, replenishing the gut microbiome with beneficial commensals may improve immunity, decrease inflammation, and improve health in individuals with HIV infection.
Fecal microbiota transplantation (FMT), wherein donor fecal material is transplanted into the intestine of a recipient, has been demonstrated to robustly alter microbial communities (19). FMT has been used to treat bacterial diseases, such as Clostridium difficile and vancomycin-resistant Enterococcus (VRE) infections, which occur when there is dysbiosis (typically after antibiotic use) and these pathogenic bacteria overtake the microbiome and cause diarrheal disease; such infections are typically resistant to treatment (20–22). Unlike antibiotic therapy, FMT has a rate of efficacy of restoring a diverse microbiota and decreasing GI symptoms of nearly 90% (20, 23, 24). Clinical trials evaluating the benefits of FMT in inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), and other gastrointestinal diseases are under way (19, 25).
Given that microbial dysbiosis occurs during HIV infection and is associated with pathogenesis, we sought to determine whether FMT would be a safe and efficacious method of modifying the bacterial community and improving health in HIV-infected individuals. Due to the immunodeficiency and chronic inflammation that occurs in HIV infection, determining the safety of FMT is crucial to prevent deleterious consequences if it is to be used as a treatment in the future. Here we used the macaque model of HIV infection and performed FMT on six chronically SIV-infected rhesus macaques on ART.
(Preliminary data from this study were previously presented at the Human Immunity and the Microbiome in Health and Disease meeting in Montreal, Canada, September 2015.)
MATERIALS AND METHODS
Study animals.
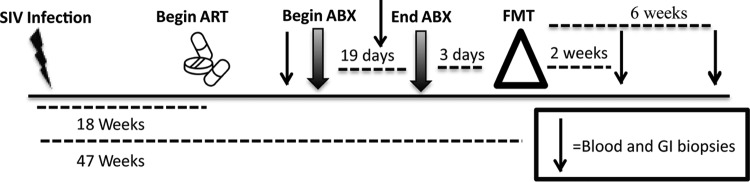
The animals were housed and cared for in Association for the Assessment and Accreditation of Laboratory Animal Care international (AAALACi)-accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Washington. Six male rhesus macaques (Macaca mulatta) were infected intrarectally with SIVMAC239x and started on ART at 130 days after infection. ART consisted of subcutaneous tenofovir (9-R-2-phosphonomethoxypropyl adenine; 20 mg/kg of body weight) and emtricitabine (FTC; 30 mg/kg) and oral raltegravir (50 mg twice a day [b.i.d.]). For longitudinal assessment of the effects of FMT, procedures and sampling were performed in two groups, with the study schedule varying by 1 day between groups. The times at which samples were obtained from the two groups are referred to collectively as the number of weeks post-FMT (an overview is presented in Fig. 1). Specifically, blood and biopsy specimens of the colon, rectum, and jejunum were taken before antibiotic treatment (pre-ABX; 3 weeks prior to FMT) and after 2 weeks on antibiotic treatment, which was 1 week prior to FMT. Post-FMT samples were taken at 2 and 6 weeks after transplantation. In addition, prior to FMT, we collected plasma and peripheral blood mononuclear cells (PBMCs) before SIV infection (−47 weeks) and pre-ART (−33 weeks).
FIG 1.
Timeline showing the times of sampling of blood and GI tract biopsy specimens from 6 SIV-infected rhesus macaques before and after antibiotic treatment and fecal microbial transplantation. The macaques were intrarectally challenged with 100,000 50% tissue culture infective doses of SIVMAC239x. ART consisted of subcutaneous tenofovir and emtricitabine and oral raltegravir. ABX consisted of vancomycin for 7 days, followed by a combination of vancomycin and enrofloxacin (Baytril) for 12 additional days. FMT consisted of administration of stool from SIV-negative rhesus macaques homogenized in PBS by gavage to the upper and lower GI tracts of the 6 SIV-infected rhesus macaques.
Antibiotic treatment and fecal microbial transplantation.
First, animals were placed on an ABX regimen for 19 total days. This regimen consisted of oral vancomycin (15 mg/kg orally b.i.d.) for 7 days, followed by a combination of vancomycin and enrofloxacin (10 mg/kg orally b.i.d.) for 12 additional days. This ABX regimen was chosen to target both Gram-positive and Gram-negative intestinal bacteria. Stool samples from healthy (SIV-negative) rhesus macaque donors also housed at the Washington National Primate Research Center (WaNPRC) were processed for transplantation as previously described in protocols for humans (26). In brief, stool was homogenized in phosphate-buffered saline (PBS), and fibrous material was filtered out using a 70-μm-pore-size filter. The solution was centrifuged, and the pellet was resuspended in PBS with 10% glycerol and frozen at −80°C until use. For transplantation, stool sample mixtures containing 50 g of fecal donor material were thawed for each recipient animal. The mixtures were centrifuged, the pellet was resuspended in 80 ml PBS, and 40 ml of the suspension was gavaged into the duodenum and 40 ml was gavaged into the upper colon via an endoscope. The transplant procedure was performed 3 or 4 days after the end of ABX, depending on the sampling group (see Fig. 1).
Blood and tissue processing.
Plasma and PBMCs were separated by density gradient centrifugation and cryopreserved for later analyses. Gut biopsy specimens were immediately processed as follows: 1 to 2 biopsy specimens per tissue were frozen in RNAlater solution (Qiagen, Valencia, CA) for subsequent DNA extraction and 16S rRNA sequencing. The remaining biopsy specimens were enzymatically digested with RPMI-1640 medium (GE Healthcare Life Sciences, Logan, UT) supplemented with Liberase (40 μg/ml; Sigma-Aldrich, St. Louis, MO) and DNase (4 μg/ml; Sigma-Aldrich) for 1 h at 37°C with vigorous stirring, ground through a 70-μm-pore-size cell strainer into a single-cell suspension, and then analyzed by flow cytometry. Complete blood counts with differential were measured on a Beckman Coulter AC*T 5diff CP hematology analyzer. Viral loads were determined by real-time reverse transcription (RT)-PCR using primers specific for SIV gag as previously detailed (27).
Genomic DNA extraction and sequencing.
Genomic DNA was extracted from colon tissue biopsy specimens using an Omni Bead Ruptor with 2.8-mm ceramic beads and homogenized in Buffer RLT (Qiagen, Valencia, CA). Genomic DNA was extracted by use of an RNA/DNA All-Prep kit (Qiagen, Valencia, CA), and 5 ng of genomic DNA per biopsy specimen was used to generate a 460-bp amplicon by targeting the V3 and V4 variable regions of the 16S rRNA gene. All genomic DNA underwent 20 cycles of PCR, and 16S rRNA amplicons were cleaned using 0.8× AMPure XP beads (Beckman Coulter, Brea, CA). Nextera XT dual index adaptors (Illumina Inc., San Diego, CA) were incorporated by performing 12 PCR cycles using a FailSafe PCR system (Epicentre, Madison, WI), cleaned using 1.1× AMPure XP beads (Beckman Coulter, Brea, CA), quantified using a Qubit DNA high-sensitivity assay kit (Life Technologies, Carlsbad, CA), and multiplexed using an equal molar ratio of DNA for each sample. Final 16S rRNA libraries were loaded on a 600-cycle MiSeq kit at 2 pM with 5% phiX phage as a control and sequenced using Nextera sequencing read and index primers (all from Illumina, San Diego, CA).
Analysis of 16S rRNA gene sequencing data.
We first combined paired-end reads using the PANDAseq assembler (28) and then analyzed 16S rRNA gene sequence data using the QIIME software package (29). We initially clustered sequences into operational taxonomic units (OTUs) at 97% similarity and assigned a taxonomy to each OTU using the Greengenes database (release 13.5) (30). We performed principal-coordinate analysis (PCoA) using the tools within QIIME software.
Antibodies and flow cytometry. (i) Surface staining.
Single cells isolated from biopsy specimens were stained immediately after isolation using the following surface antigen antibodies (clones are denoted in parentheses), all of which were from BD Biosciences, unless stated otherwise: CD45 peridinin chlorophyll protein (PerCP; clone D058-1283), CD3 phycoerythrin (PE) CF594 (clone SP34-2), CD4 Brilliant Violet 605 (clone OKT4; BioLegend), CD8 Brilliant Violet 570 (clone RPA-T8; BioLegend), CD14 Brilliant Violet 786 (clone M5E2; BioLegend), and HLA-DR Brilliant Violet 711 (clone G46-6).
(ii) Intracellular cytokine staining.
Cryopreserved PBMCs were thawed using R10 medium with 5 μg/ml DNase. Thawed PBMCs were rested for 4 h and then stimulated for 14 h at 37°C with 10 ng/ml phorbol myristate acetate (PMA; Sigma-Aldrich) and 1 μg/ml ionomycin (Life Technologies) in R10 medium with 1 mg/ml of brefeldin A (Sigma-Aldrich). The stimulated cells were then stained with the following surface antigen antibodies, all of which were from BD Biosciences, unless otherwise stated: CD45 PerCP (clone D058-1283), CD3 PE CF594 (clone SP34-2), CD4 Brilliant Violet 605 (clone OKT4; BioLegend), CD8 allophycocyanin-H7 (clone SK1), and CD14 Brilliant Violet 786 (clone M5E2; BioLegend). Following surface staining, stimulated cells were permeabilized using Cytofix/Cytoperm (BD Biosciences) and stained using the following intracellular antigen antibodies from eBioscience: IL-17 PE (catalog number ebio64CAP17) and IL-22 PerCP-eFluor710 (catalog number IL022JOP). Stained samples were fixed in 1% paraformaldehyde and collected on an LSR II flow cytometer (BD Biosciences, La Jolla, CA). Analysis was performed with FlowJo software (version 9.7.6; TreeStar Inc., Ashland, OR).
Assessment of plasma cytokines and soluble factors.
Cytokine and chemokine levels in plasma were analyzed using a Milliplex MAP (multianalyte profiling) nonhuman primate cytokine magnetic bead panel premixed 23-plex kit (Millipore, Darmstadt, Germany). The levels of the analytes were assessed on a Bio-Plex 200 system (Bio-Rad, Hercules, CA). Other soluble factors were assessed via enzyme-linked immunosorbent assay (ELISA) using the following preprepared kits: for soluble CD14 (sCD14), a Quantikine ELISA human sCD14 immunoassay (R&D Systems Inc., Minneapolis, MN); for lipopolysaccharide (LPS) binding protein (LBP), an ELISA kit from Biometic (Brixen, Italy); for monkey C-reactive protein (CRP), an ELISA kit from Life Diagnostics (West Chester, PA); and for rhesus macaque soluble IL-6R, an ELISA from RayBiotech, Inc. (Norcross, GA). ELISA results were read using an iMark microplate reader (Bio-Rad, Hercules, CA).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism statistical software (version 6; GraphPad Software, San Diego, CA). The results obtained at the post-ABX time point (week −1) and the post-FMT time points (weeks 2 and 6) were compared to those obtained at the pre-ABX time point (week −3), which was used as the baseline, and significance was evaluated using a paired t test. The statistical significance of the difference in the results between the pre-SIV infection and all post-SIV infection time points was also evaluated using a paired t test when cryopreserved samples obtained before SIV infection were available for comparison. We generated correlation matrices using the R statistical software package.
RESULTS
Microbiome composition changes induced by antibiotic treatment and fecal microbial transplantation.
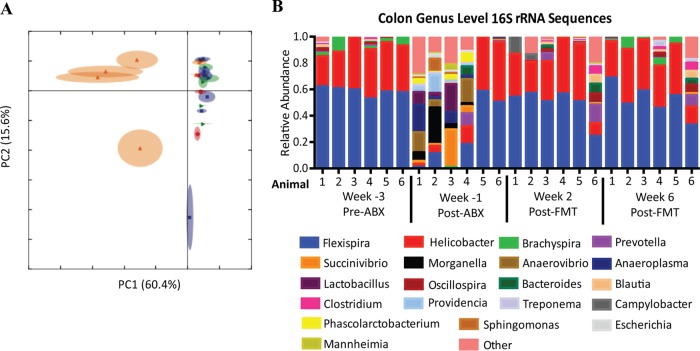
The use of antibiotics to reduce or eliminate pathogenic bacteria, such as C. difficile, is a standard prerequisite for patients receiving FMT. The first goal of our study was to determine the impact of vancomycin-enrofloxacin antibiotic treatment and FMT on the colonic microbiome. By comparing the relative abundances of bacterial genera across all time points, we found that the composition of the microbiota post-ABX clustered separately from that at the pre-ABX and post-FMT time points, with 60.4% of the variation being explained by the PC1 axis (Fig. 2A). Specifically, ABX significantly reduced the relative abundance of all Gram-positive bacteria except Lactobacillus and the majority of aerobic Gram-negative bacteria. Antibiotic treatment had the largest impact on the abundance levels of the most prevalent bacterial genera, Helicobacter and Flexispira, while it enriched the composition of the microbiome with anaerobic Gram-negative bacteria, resulting in a greater diversity (Fig. 2B). The microbiome composition quickly reverted by 2 weeks post-FMT to be dominant in Helicobacter and Flexispira, similar to the findings pretransplantation. However, differences in the abundances of the minor populations remained, with two of the six animals (animals 4 and 6) retaining increased microbial diversity.
FIG 2.
ABX and FMT treatment resulted in microbiome composition changes. (A) Principal-component analysis using the relative abundances of all genera from colon biopsy specimens across all time points. Green, pre-ABX (week −3); orange, post-ABX (week −1); blue, 2 weeks post-FMT (week 2); red, 6 weeks post-FMT (week 6). (B) Relative abundance of bacterial genera from colon biopsy specimens pre- and post-ABX and post-FMT. The 20 most abundant genera are shown. Less abundant genera were grouped into the “other” category (pink).
No negative side effects of fecal microbiota transplantation.
Due to the immunocompromised nature of HIV-infected patients, it will be critical to monitor the effects of microbial therapeutics on the overall health of the HIV-infected recipient. It will be particularly important to ensure that introduction of outside bacteria does not affect the welfare of the recipient or cause increased inflammation and immune activation or bacteremia. In this study, FMT in SIV-infected rhesus macaques was well tolerated, with no change in animal behavior and only a minor reduction in the mean animal weight being noted at 2 weeks post-FMT (Table 1). There were no significant changes in the leukocyte counts measured by use of a complete blood count, with the exception of monocytes, the levels of which were reduced at 6 weeks post-FMT to a level more closely resembling the levels pre-SIV infection (Table 1 and data not shown). Although all animals were on ART, they maintained low but measurable plasma virus levels pretransplantation ranging from 36 to 208 copies/ml (Table 1). There was a slight yet nonsignificant increase in plasma virus levels in some animals post-ABX, but these returned to pre-ABX levels by 6 weeks post-FMT. Finally, we measured the levels of cytokines and soluble markers of microbial translocation and inflammation in the plasma posttreatment to ensure that bacteremia or inflammation did not occur. ABX resulted in increases in the levels of the cytokines IL-2 and IL-15, but the levels of both returned to pre-ABX levels by 2 weeks post-FMT (see Fig. S1 in the supplemental material). Interestingly, the levels of vascular endothelial growth factor (VEGF), a cytokine whose levels are increased in patients with HIV-associated central nervous system diseases (31), transiently decreased at 2 weeks post-FMT. We observed no significant changes in the levels of the other cytokines measured. The level of soluble CD14 (sCD14), a marker of microbial translocation, began increasing after ABX and remained significantly elevated at 2 and 6 weeks post-FMT. On the contrary, we observed no changes in the levels of LPS binding protein (LBP), a marker of LPS stimulation, or the systemic inflammation markers C reactive protein (CRP) and soluble IL-6 receptor. Taken together, these data suggest that ABX may increase inflammation and microbial translocation but that ABX and FMT treatment did not have substantial or lasting effects on overall animal health and welfare and did not result in overtly increased systemic immune activation.
TABLE 1.
Clinical effects of FMT in SIV-infected rhesus macaques
| Characteristic | Valuea at: |
|||
|---|---|---|---|---|
| Wk −3 (pre-ABX) | Wk −1 (post-ABX) | Wk 2 (post-FMT) | Wk 6 (post-FMT) | |
| Animal wt (kg) | 10.12 ± 1.51 | 9.87 ± 1.53 | 9.54 ± 1.43b | 9.75 ± 1.41 |
| CBCc (1,000 cells/μl) | ||||
| Neutrophils | 2.67 ± 0.9 | 2.85 ± 0.98 | 4.16 ± 1.93 | 4.14 ± 0.89 |
| Monocytes | 0.66 ± 0.26 | 0.65 ± 0.31 | 0.62 ± 0.25 | 0.53 ± 0.17b |
| Lymphocytes | 2.16 ± 0.91 | 1.87 ± 0.73 | 2.03 ± 0.8 | 1.9 ± 1.01 |
| Basophils | 0.05 ± 0.02 | 0.04 ± 0.03 | 0.05 ± 0.03 | 0.07 ± 0.03 |
| Eosinophils | 0.11 ± 0.05 | 0.12 ± 0.06 | 0.09 ± 0.02 | 0.10 ± 0.04 |
| Viral load (no. of copies/ml) | 123.2 ± 64.12 | 319.2 ± 376.5 | 315.5 ± 286.8 | 109.8 ± 85.22 |
The data represent means ± standard deviations.
P < 0.05 by paired t test compared with the value pre-ABX as the baseline value.
CBC, complete blood count.
Increased peripheral Th17 and Th22 frequencies and decreased gut CD4+ T cell activation after fecal microbial transplantation.
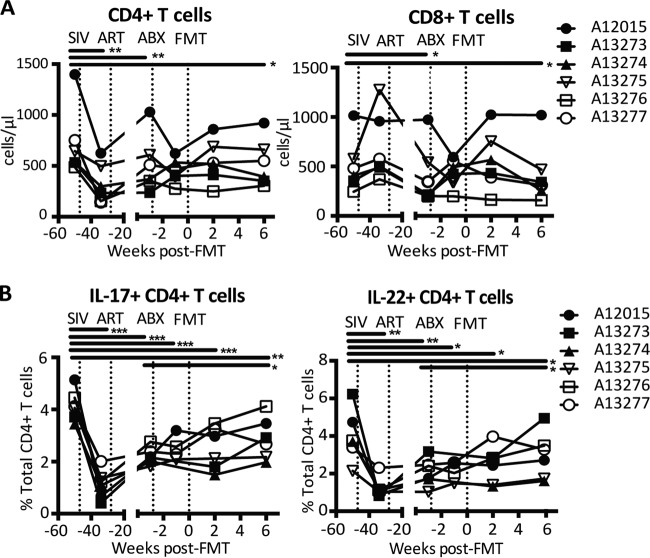
Th17 and Th22 cells, important components of mucosal immunity, are significantly depleted in HIV and SIV infection, and this depletion is associated with reduced barrier integrity, microbial translocation, and systemic immune activation (32–34). Previous studies have demonstrated a connection between commensal bacteria and the ability of probiotic therapy to enhance intestinal Th17 frequencies (15, 35, 36). Thus, we hypothesized that alteration of the microbiome through FMT might restore Th17 and Th22 frequencies. Indeed, while we observed no significant effects on total peripheral CD4+ and CD8+ T cell frequencies, peripheral Th17 and Th22 frequencies increased significantly at 6 weeks post-FMT compared to those pre-ABX (Fig. 3). Although the Th17 and Th22 frequencies remained significantly lower than those pre-SIV infection, the increases observed suggest that FMT has promising potential as a therapy for altered T cell subset homeostasis in HIV infection.
FIG 3.
Increased amounts of peripheral Th17 and Th22 cells after fecal microbial transplantation. (A) Absolute amounts of peripheral CD4+ and CD8+ T cells, calculated using the percentage of total CD45+ cells obtained by flow cytometry and the total leukocyte count obtained by complete blood count analysis. (B) Percentages of IL-22-producing (IL-22+) and IL-17-producing (IL-17+) CD4+ T cells in PBMCs measured via flow cytometry after stimulation with PMA-ionomycin. CD4+ T cells were identified as CD45+ CD3+ CD4+ CD8− cells. Asterisks indicate significant differences determined by a paired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
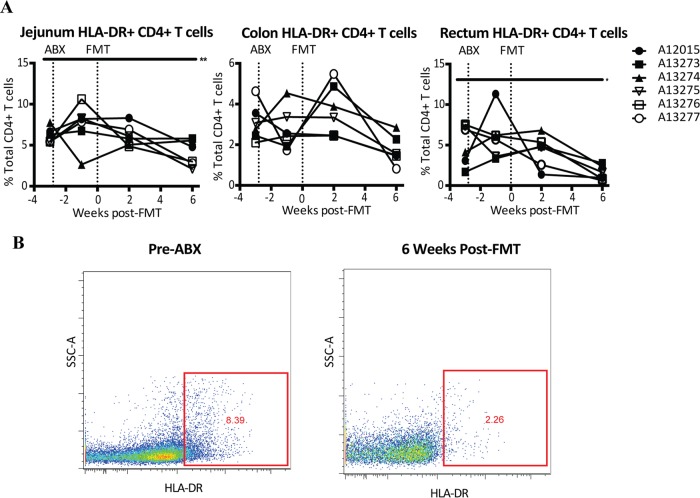
T cell activation is associated with CD4+ T cell loss and disease progression in HIV infection (37, 38). We measured the effect of FMT on intestinal T cell activation and found a decrease in the levels of activation, as measured by determination of the levels of HLA-DR expression, of jejunum, colon, and rectum CD4+ T cells but not CD8+ T cells at 6 weeks post-FMT, although the decrease in the colon was not statistically significant (Fig. 4). These data suggest that FMT might have a beneficial effect on intestinal T cell activation.
FIG 4.
Reduced CD4+ T cell activation after fecal microbial transplantation. (A) HLA-DR-positive (HLA-DR+) CD4+ T cells in colon, jejunum, and rectum biopsy specimens. CD4+ T cells were identified as CD45+ CD3+ CD4+ CD8− cells. Asterisks indicate significant differences determined by a paired t test (*, P < 0.05; **, P < 0.01). (B) Representative flow plots showing HLA-DR expression on CD4+ T cells from the jejunum of an SIV-infected rhesus macaque before ABX (left) and 6 weeks post-FMT (right). SSC, side scatter.
Altered immune parameters are associated with altered microbial frequencies.
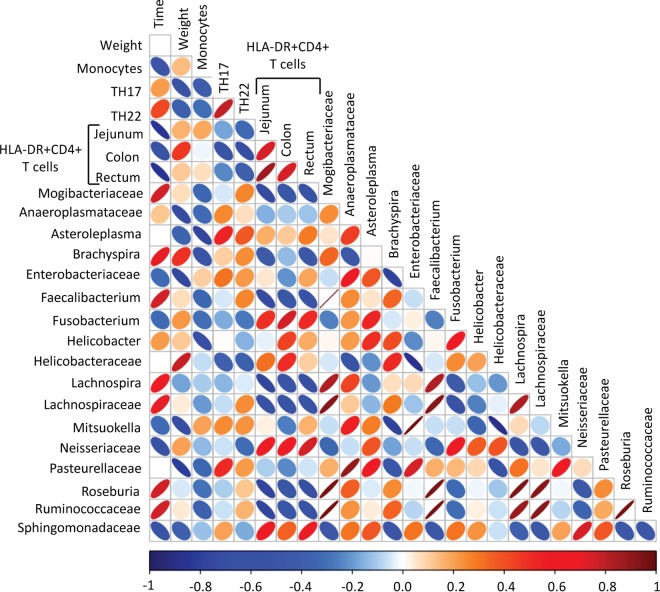
Finally, to investigate if specific alterations to the microbiome were associated with changes in immunological and physiological parameters, we analyzed the correlations between all identified taxonomic groups and the immunological parameters significantly altered by FMT. Using these data, we created a correlation matrix illustrating the taxonomic groups that correlated most strongly with the other parameters measured in our study (Fig. 5). Several strong correlations were observed, particularly among lower-abundance taxonomic groups. Specifically, animal weight was positively associated with the presence of members of the family Helicobacteraceae but negatively associated with the presence of members of the families Anaeroplasmataceae and Enterobacteriaceae, and Pasteurellaceae and the genus Mitsuokella. HLA-DR expression on CD4+ T cells was positively associated with the presence of the genus Fusobacterium and members of the families Neisseriaceae and Sphingomonadaceae and was negatively associated with the presence of members of the genus Lachnospira, the family Lachnospiraceae, the genus Roseburia, and the family Ruminococcaceae. We observed no strong correlations between the presence of any one specific bacterial taxon and peripheral Th17 or Th22 frequencies. Although none of these associations remained significant after correction for multiple comparisons, these data suggest that alterations to the microbiome, particularly in the low-abundance taxons, are potential contributors to the physiological and immunological changes observed in response to ABX and FMT treatment.
FIG 5.
Correlation matrix of taxonomic groups that most strongly correlated with immunological or physiological parameters. Colors indicate Spearman correlation coefficient values ranging from −1 (dark blue) to 1 (dark red). The ellipse shape indicates the relative spread of data points and the slope of the correlation, with a wider ellipse indicating a greater relative spread and a narrower ellipse indicating less of a spread.
DISCUSSION
In chronic HIV infection, an altered gut microbiome is associated with mucosal dysfunction, systemic inflammation, and disease progression (9, 10, 12, 39). FMT is emerging as an effective treatment option for antibiotic-refractory intestinal infections, such as C. difficile infections, as well as for chronic inflammatory conditions of the gut that are known to be associated with dysbiosis (24, 25, 40). ABX is often used prior to transplantation, although the overall impact of this strategy on the efficacy of therapy is largely unknown (41). Here we have reported on the effects of ABX and FMT on the microbiome, health, inflammation, and immunity of 6 SIV-infected rhesus macaques. Overall, we found that FMT is safe and well tolerated and may have beneficial effects on immunity.
Interestingly, after FMT, the microbiome community structure reverted to a state that was strikingly similar to that pre-FMT. This likely indicates that the transplant donors had a microbiota similar to that of the SIV-infected animals, even though the donors were healthy, SIV-uninfected animals. This may be because all animals were housed at WaNPRC and fed the same diet, which may outweigh the effects of SIV infection in these colonies. Another alternative is that the bacteria in the FMT did not colonize and the microbiota rapidly reverted to pre-FMT communities. However, given the extensive selection for a diverse community of anaerobic Gram-negative bacteria induced by antibiotics, this is unlikely, and furthermore, previous studies have demonstrated that the recipient microbiome resembles the donor microbiome post-FMT (42).
Despite the reversion to a microbiome similar to that pre-FMT, we observed enhanced Th17 and Th22 frequencies post-FMT treatment. Induction of Th17 and Th22 cells is important, given that these cells are essential in responding to bacterial and fungal pathogens and critical in maintaining the homeostasis of epithelial barriers (43–45). Additionally, during HIV and SIV infection, IL-17- and IL-22-producing cells are lost, and this is associated with damage to epithelial barrier integrity, inflammation, and pathogenic effects (32–34, 46). Although we observed no correlations between Th17 and Th22 frequencies and any specific bacterial population, the alterations observed may have been induced by minor populations or combinations of populations not represented in the overall microbiome composition detected. Additionally, we observed decreased intestinal CD4+ T cell activation post-FMT treatment. Importantly, HLA-DR expression on CD4+ T cells of the gut correlated strongly with several lower-abundance bacterial genera. The associations between bacterial genera and activated CD4+ T cells were similar between cells isolated from different GI tissues, suggesting either that FMT altered the microbiome at other intestinal sites in a manner similar to that in which it did in the colon or that changes in colonic microbial communities have widespread effects on gastrointestinal immunity. Further studies on microbiome alterations at different gastrointestinal sites after FMT should be performed to address this; however, the data suggest that FMT may provide a novel therapeutic intervention to enhance gastrointestinal integrity through enhanced T cell immunity.
Of interest, while FMT tended to improve health in the animals, antibiotic treatment appeared to have an inflammatory effect. The increase in that amounts of proinflammatory adaptive immune-related cytokines indicates a potential T or NK cell response to the altered microbiota induced by antibiotics (47, 48). In addition, the level of sCD14, a marker of microbial translocation, increased in 5 of the 6 animals post-ABX in our study and remained elevated through 6 weeks post-FMT. Although we cannot conclude that these changes were solely induced by ABX due to the lack of an ABX-only control group, a recently published study with chronically infected pigtail macaques similarly observed an increase in sCD14 levels in 2 of the 3 animals after treatment with the antibiotic rifaximin and the anti-inflammatory agent sulfasalazine (49). To our knowledge, no other studies have assessed inflammatory cytokines after ABX in infected macaques. The drastic shift in the microbiota from an aerobic to an anaerobic dominant community observed in the gut post-ABX could potentially influence the systemic inflammatory response. The overall change to an anaerobic environment in the intestine would directly impact oxygen levels, the gut pH, the microbial biofilm structure, and the microbial by-products produced, stimulating an immune response. Importantly, ABX differentially affected the animals in our study, with only minor microbiome changes being observed in 2 of the animals, while the other 4 animals had dramatic microbiome shifts. This demonstrates that different individuals can respond differently to microbiome-altering therapies. Further studies to assess how commonly used antibiotics may affect inflammation and immunity and the long-term consequences on the microbiome are warranted.
Finally, our study provides evidence that alterations to the gastrointestinal microbiome are associated with changes in immunological parameters. Microbial diversity has previously been demonstrated to be an important factor in protection from intestinal pathogens and inflammatory disorders. Additionally, previous studies of dysbiosis in IBD have shown associations between some low-abundance genera, such as Roseburia and Faecalibacterium, and intestinal inflammation (50, 51). Indeed, in our study, physiological and immune parameters altered by ABX and/or FMT treatment correlated most strongly across all sampling time points with lower-abundance taxonomic groups in the colon. Interestingly, we also observed a negative correlation between Roseburia and CD4+ T cell activation in the gut, further suggesting that this bacterial community may have beneficial anti-inflammatory properties. While this was a small study with few animals, these data warrant further investigation into how overall changes in the microbiome, particularly in terms of diversity and changes in minor populations, affect immunity.
Overall, this pilot study provides evidence that FMT would be a safe treatment for microbial dysbiosis in individuals with HIV infection and might potentially have beneficial effects on the immune dysfunction associated with chronic infection. One caveat of this small pilot study was our inability to include ABX-only, FMT-only, and no-treatment control groups. Without results for these control groups, we are unable to assess the relative contribution of ABX and FMT treatment to the reported observations. Although the changes observed after ABX but before FMT are logically attributable to ABX, a larger follow-up study that includes these control groups should be performed to confirm these findings. This will be particularly important to further understand how ABX and FMT treatment may have differentially contributed to the transient weight loss and increase in sCD14 levels observed in this study. Future FMT studies with macaques could also consider the use of prescreening of potential macaque donors to identify donor feces with a high microbial diversity or enriched for beneficial bacterial communities, the use of a combination of probiotics and fecal material, or the use of fecal material from healthy humans to better represent the human microbiota. Additionally, a recent report demonstrated that altering the microbiome with antibiotic treatment with a luminal antibiotic (rifaximin) together with an anti-inflammatory drug (sulfasalazine) during acute SIV infection resulted in beneficial reductions in immune activation and microbial translocation, while treatment during chronic infection had no discernible effect (49). This suggests that future studies assessing the benefits of FMT in lentiviral infection may result in increased benefits if it is administered during the acute phase of infection. Finally, while future preclinical studies are of interest to determine the mechanisms underlying the decreased inflammation and enhanced cellular effects of FMT, ultimately, clinical trials with HIV-infected individuals would be most informative as to the beneficial effects of FMT for the treatment of HIV-associated microbial dysbiosis. Importantly, the pilot studies described here provide evidence that FMT in immunodeficient primates infected with lentivirus did not have negative side effects but had overall positive effects, suggesting that clinical trials may be safe and efficacious.
Supplementary Material
ACKNOWLEDGMENTS
We thank all veterinary staff of the Washington National Primate Research Center for the animal studies.
This work was supported by grant 1K22AI098440-01 and start-up funds to N.R.K. from the University of Washington and the Washington National Primate Research Center. Follow-up studies were supported by the National Institutes of Health (1R01AI120712-01 to N.R.K.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00099-16.
REFERENCES
- 1.Klatt NR, Chomont N, Douek DC, Deeks SG. 2013. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol Rev 254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, Neaton JD, Brenchley JM, Deeks SG, Sereti I, Douek DC, INSIGHT SMART Study Group. 2011. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somsouk M, Estes JD, Deleage C, Dunham RM, Albright R, Inadomi JM, Martin JN, Deeks SG, McCune JM, Hunt PW. 2015. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS 29:43–51. doi: 10.1097/QAD.0000000000000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, Robinson J, Huang Y, Epling L, Martin JN, Deeks SG, Meinert CL, Van Natta ML, Jabs DA, Lederman MM. 2014. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 210:1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canary LA, Vinton CL, Morcock DR, Pierce JB, Estes JD, Brenchley JM, Klatt NR. 2013. Rate of AIDS progression is associated with gastrointestinal dysfunction in simian immunodeficiency virus-infected pigtail macaques. J Immunol 190:2959–2965. doi: 10.4049/jimmunol.1202319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. [DOI] [PubMed] [Google Scholar]
- 7.Burgener A, McGowan I, Klatt NR. 2015. HIV and mucosal barrier interactions: consequences for transmission and pathogenesis. Curr Opin Immunol 36:22–30. doi: 10.1016/j.coi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, Smith DM, Landay AL, McManus MC, Robertson CE, Frank DN, McCarter MD, Wilson CC. 2015. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol 9:24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, Robertson CE, Frank DN, Wilson CC. 2014. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, Deeks SG, Hunt PW, Lynch SV, McCune JM. 2013. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zevin AS, McKinnon L, Burgener A, Klatt NR. 2015. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 11:182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, Cox S, Engen P, Chakradeo P, Abbasi R, Gorenz A, Burns C, Landay A. 2014. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gori A, Tincati C, Rizzardini G, Torti C, Quirino T, Haarman M, Ben Amor K, van Schaik J, Vriesema A, Knol J, Marchetti G, Welling G, Clerici M. 2008. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol 46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. 2015. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol 8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortiz AM, Klase ZA, DiNapoli SR, Vujkovic-Cvijin I, Carmack K, Perkins MR, Calantone N, Vinton CL, Riddick NE, Gallagher J, Klatt NR, McCune JM, Estes JD, Paiardini M, Brenchley JM. 19 August 2015. IL-21 and probiotic therapy improve Th17 frequencies, microbial translocation, and microbiome in ARV-treated, SIV-infected macaques. Mucosal Immunol doi: 10.1038/mi.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klatt NR, Canary LA, Sun X, Vinton CL, Funderburg NT, Morcock DR, Quinones M, Deming CB, Perkins M, Hazuda DJ, Miller MD, Lederman MM, Segre JA, Lifson JD, Haddad EK, Estes JD, Brenchley JM. 2013. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. J Clin Invest 123:903–907. doi: 10.1172/JCI66227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.d'Ettorre G, Ceccarelli G, Giustini N, Serafino S, Calantone N, De Girolamo G, Bianchi L, Bellelli V, Ascoli-Bartoli T, Marcellini S, Turriziani O, Brenchley JM, Vullo V. 2015. Probiotics reduce inflammation in antiretroviral treated, HIV-infected individuals: results of the “Probio-HIV” clinical trial. PLoS One 10:e0137200. doi: 10.1371/journal.pone.0137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villar-Garcia J, Hernandez JJ, Guerri-Fernandez R, Gonzalez A, Lerma E, Guelar A, Saenz D, Sorli L, Montero M, Horcajada JP, Knobel Freud H. 2015. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr 68:256–263. doi: 10.1097/QAI.0000000000000468. [DOI] [PubMed] [Google Scholar]
- 19.Brandt LJ, Aroniadis OC. 2013. An overview of fecal microbiota transplantation: techniques, indications, and outcomes. Gastrointest Endosc 78:240–249. doi: 10.1016/j.gie.2013.03.1329. [DOI] [PubMed] [Google Scholar]
- 20.Mattila E, Uusitalo-Seppala R, Wuorela M, Lehtola L, Nurmi H, Ristikankare M, Moilanen V, Salminen K, Seppala M, Mattila PS, Anttila VJ, Arkkila P. 2012. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology 142:490–496. doi: 10.1053/j.gastro.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 21.McKenney PT, Pamer EG. 2015. From hype to hope: the gut microbiota in enteric infectious disease. Cell 163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stripling J, Kumar R, Baddley JW, Nellore A, Dixon P, Howard D, Ptacek T, Lefkowitz EJ, Tallaj JA, Benjamin WH Jr, Morrow CD, Rodriguez JM. 2015. Loss of vancomycin-resistant Enterococcus fecal dominance in an organ transplant patient with Clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect Dis 2:ofv078. doi: 10.1093/ofid/ofv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoruts A, Weingarden AR. 2014. Emergence of fecal microbiota transplantation as an approach to repair disrupted microbial gut ecology. Immunol Lett 162(2 Pt A):77–81. doi: 10.1016/j.imlet.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman-Moraco RJ, Mehta AK, Lyon GM, Kraft CS. 2014. Fecal microbiota transplantation for refractory Clostridium difficile colitis in solid organ transplant recipients. Am J Transplant 14:477–480. doi: 10.1111/ajt.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colman RJ, Rubin DT. 2014. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 8:1569–1581. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tauxe WM, Dhere T, Ward A, Racsa LD, Varkey JB, Kraft CS. 2015. Fecal microbiota transplant protocol for Clostridium difficile infection. Lab Med 46:e19–e23. doi: 10.1309/LMCI95M0TWPDZKOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol 82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Mirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sporer B, Koedel U, Paul R, Eberle J, Arendt G, Pfister HW. 2004. Vascular endothelial growth factor (VEGF) is increased in serum, but not in cerebrospinal fluid in HIV associated CNS diseases. J Neurol Neurosurg Psychiatry 75:298–300. doi: 10.1136/jnnp.2003.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, Tabb B, Canary LA, Dang Q, Hirsch VM, Alter G, Belkaid Y, Lifson JD, Silvestri G, Milner JD, Paiardini M, Haddad EK, Brenchley JM. 2012. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol 5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klatt NR, Brenchley JM. 2010. Th17 cell dynamics in HIV infection. Curr Opin HIV AIDS 5:135–140. doi: 10.1097/COH.0b013e3283364846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, Douek D, Martin JN, Deeks SG. 2008. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis 197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa AE, Carneiro J, Meier-Schellersheim M, Grossman Z, Victorino RM. 2002. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J Immunol 169:3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 39.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, Anton P, Braun J. 2013. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandt LJ, Aroniadis OC, Mellow M, Kanatzar A, Kelly C, Park T, Stollman N, Rohlke F, Surawicz C. 2012. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 107:1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- 41.Borody TJ, Khoruts A. 2012. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol 9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 42.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. 2010. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 43.Guglani L, Khader SA. 2010. Th17 cytokines in mucosal immunity and inflammation. Curr Opin HIV AIDS 5:120–127. doi: 10.1097/COH.0b013e328335c2f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sallusto F, Lanzavecchia A. 2009. Human Th17 cells in infection and autoimmunity. Microbes Infect 11:620–624. doi: 10.1016/j.micinf.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Mühl H, Scheiermann P, Bachmann M, Härdle L, Heinrichs A, Pfeilschifter J. 2013. IL-22 in tissue-protective therapy. Br J Pharmacol 169:761–771. doi: 10.1111/bph.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves RK, Rajakumar PA, Evans TI, Connole M, Gillis J, Wong FE, Kuzmichev YV, Carville A, Johnson RP. 2011. Gut inflammation and indoleamine deoxygenase inhibit IL-17 production and promote cytotoxic potential in NKp44+ mucosal NK cells during SIV infection. Blood 118:3321–3330. doi: 10.1182/blood-2011-04-347260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fehniger TA, Cooper MA, Nuovo GJ, Cella M, Facchetti F, Colonna M, Caligiuri MA. 2003. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood 101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 48.Fehniger TA, Shah MH, Turner MJ, VanDeusen JB, Whitman SP, Cooper MA, Suzuki K, Wechser M, Goodsaid F, Caligiuri MA. 1999. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: implications for the innate immune response. J Immunol 162:4511–4520. [PubMed] [Google Scholar]
- 49.Pandrea I, Xu C, Stock JL, Frank DN, Ma D, Policicchio BB, He T, Kristoff J, Cornell E, Haret-Richter GS, Trichel A, Ribeiro RM, Tracy R, Wilson C, Landay AL, Apetrei C. 2016. Antibiotic and antiinflammatory therapy transiently reduces inflammation and hypercoagulation in acutely SIV-infected pigtailed macaques. PLoS Pathog 12:e1005384. doi: 10.1371/journal.ppat.1005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. 2014. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 51.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.