ABSTRACT
Mouse polyomavirus (MPyV) is a ubiquitous persistent natural mouse pathogen. A glutamic acid (E)-to-glycine (G) difference at position 91 of the VP1 capsid protein shifts the profile of tumors induced by MPyV from an epithelial to a mesenchymal cell origin. Here we asked if this tropism difference affects the MPyV-specific CD8 T cell response, which controls MPyV infection and tumorigenesis. Infection by the laboratory MPyV strain RA (VP1-91G) or a strain A2 mutant with an E-to-G substitution at VP1 residue 91 [A2(91G)] generated a markedly smaller virus-specific CD8 T cell response than that induced by A2(VP1-91E) infection. Mutant A2(91G)-infected mice showed a higher frequency of memory precursor (CD127hi KLRG1lo) CD8 T cells and a higher recall response than those of A2-infected mice. Using T cell receptor (TCR)-transgenic CD8 T cells and immunization with peptide-pulsed dendritic cells, we found that early bystander inflammation associated with A2 infection contributed to recruitment of the larger MPyV-specific CD8 T cell response. Beta interferon (IFN-β) transcripts were induced early during A2 or A2(91G) infections. IFN-β inhibited replication of A2 and A2(91G) in vitro. Using mice lacking IFN-αβ receptors (IFNAR−/−), we showed that type I IFNs played a role in controlling MPyV replication in vivo but differentially affected the magnitude and functionality of virus-specific CD8 T cells recruited by A2 and A2(91G) viral infections. These data indicate that type I IFNs are involved in protection against MPyV infection and that their effect on the antiviral CD8 T cell response depends on capsid-mediated tropism properties of the MPyV strain.
IMPORTANCE Isolates of the human polyomavirus JC virus from patients with the frequently fatal demyelinating brain disease progressive multifocal leukoencephalopathy (PML) carry single amino acid substitutions in the domain of the VP1 capsid protein that binds the sialic acid moiety of glycoprotein/glycolipid receptors on host cells. These VP1 mutations may alter neural cell tropism or enable escape from neutralizing antibodies. Changes in host cell tropism can affect recruitment of virus-specific CD8 T cells. Using mouse polyomavirus, we demonstrate that a single amino acid difference in VP1 known to shift viral tropism profoundly affects the quantity and quality of the anti-polyomavirus CD8 T cell response and its differentiation into memory cells. These findings raise the possibility that CD8 T cell responses to infections by human polyomaviruses may be influenced by VP1 mutations involving domains that engage host cell receptors.
INTRODUCTION
Binding specificity among polyomaviruses is determined by interaction of the VP1 major capsid protein with host cell gangliosides having particular terminal sialic acid linkages (1). The gangliosides GD1a and GT1b are required for transport of mouse polyomavirus (MPyV) to the endoplasmic reticulum (2, 3). Discrete amino acid differences in the receptor binding site of VP1 impart important biological differences, including profound differences in pathogenicity (4). Replacement of the glycine (G) at position 91 of VP1 of the laboratory-derived small-plaque (SP) MPyV strain RA with glutamic acid (E), the amino acid at this position in the naturally occurring large-plaque (LP) strain PTA, was sufficient to convert it into a strain with an LP morphology and to alter the profile of induced tumors from a mesenchymal to an epithelial cell lineage (5). Alternatively, substitution of G for E at position 91 in VP1 in PTA had the opposite effect on plaque size and tumorigenicity (6, 7). In SP strains, VP1 capsids with G-91 interact with branched (α-2,6)-linked sialyloligosaccharides, which may act as pseudoreceptors by binding cell surface glycoproteins that divert virions into noninfectious pathways (8). An E at this position in VP1 leads to electrostatic repulsion of the (α-2,6)-linked sialic acids, thereby preventing binding of such branched structures by LP strains; however, binding to gangliosides with sialic acid (α-2,3)-linked to galactose is retained for virion uptake into an infectious pathway (9, 10). Interestingly, MPyVs isolated from feral mice have exclusively E-91 VP1s, an unexpected finding given that such LP viruses are potentially more oncogenic than G-91 SP viruses (11).
The human polyomavirus JC virus (JCV) is a frequent member of the human virome (12). Mutations of JCV capsid protein VP1 involving the sialic acid cell receptor binding domain are detected only in patients diagnosed with progressive multifocal leukoencephalopathy (PML), a frequently fatal demyelinating disease of the central nervous system whose incidence is increasing in individuals receiving immunomodulatory therapeutics for autoimmune diseases (13, 14). These VP1 mutations have been proposed to render JCV neurotropic but also appear to enable escape from humoral immunity (13). Accumulating evidence supports the likelihood that CD8 T cells are essential for controlling JCV infection, preventing PML, and promoting recovery from PML (15, 16). Whether changes in tropism caused by VP1 mutations may also affect anti-JCV CD8 T cell responses is not known.
The fate and function of pathogen-specific CD8 T cells are affected by the duration of antigen availability, antigen levels, and inflammatory factors (17–21). Memory CD8 T cells generated in response to infections that are cleared after acute infection self-renew in an antigen-independent manner. We previously reported that CD8 T cells recruited early in a high-dose MPyV infection undergo exhaustion during the persistent phase of infection (22); however, the less inflammatory environment associated with early infection after low-dose MPyV inoculation favors differentiation of memory CD8 T cells with improved effector function (23). Different pathogen-induced inflammatory environments play an important role in controlling proliferation and effector and memory differentiation of CD8 T cells (17, 24). In the case of lymphocytic choriomeningitis virus (LCMV), changes in virus tropism caused by point mutations in the polymerase and glycoprotein capsid proteins resulted in dramatic differences in viral loads and inflammatory environments and led to dysfunction and deletion of virus-specific CD8 T cells (i.e., T cell exhaustion) (25–27). Type I interferons (IFN-I), interleukin-10 (IL-10), and IL-12 are among the inflammatory factors that contribute to proliferation and differentiation of CD8 T cells during early infection (24, 28–30), and they also shape memory CD8 T cell differentiation (31, 32).
In this study, we demonstrate that a single amino acid change (E to G) at position 91 in VP1 alters the quantity and quality of the anti-MPyV CD8 T cell response. Moreover, we found that IFN-I is induced by MPyV infection and regulates the magnitude and function of virus-specific CD8 T cells but does so in a manner dependent on this E-to-G variation in VP1. These findings have important implications regarding the impact of VP1 mutations on recruitment of protective CD8 T cells against human polyomavirus infections.
MATERIALS AND METHODS
Viruses.
Stocks of MPyV (A2 and RA) were prepared in baby mouse kidney cells as described previously (33). Site-directed mutagenesis using previously published primers (7) was used to replace the codon for E with that for G at position 91 of VP1 in the A2 genome; this mutant virus is designated A2(91G). The MPyV.LT206 mutant virus was created by replacing the sequence encoding the Db-restricted LT359–368 CD8 T cell epitope from MPyV (SAVKNYCSKL) with that of the Db-restricted CD8 T cell epitope LT206–215 from simian virus 40 (SV40) (SAINNYAQKL). A2 and MPyV.LT206 have similar growth kinetics in vitro and in vivo (22). A recombinant vesicular stomatitis virus carrying the LT359–368 sequence (rVSV-LT359) was generated as described previously (23).
Quantitation of MPyV by plaque assay and qPCR.
Infectious virus was titrated by plaque assay on BALB/3T3 clone A31 cells as previously described (34), with the following modifications. Subconfluent A31 cell monolayers in 6-well tissue culture plates were inoculated with 200 μl of viral lysate at 4°C for 1 h, rinsed twice and cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen Life Technologies) containing 5% fetal bovine serum (FBS), penicillin-streptomycin (100 U/ml; Mediatech), and 0.5% low-melting-temperature agarose for 5 to 6 days, fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS), and stained with 0.03% methylene blue in PBS, and plaques were then counted. For one-step growth curve assay of viruses, normal mouse mammary gland (NMuMG) epithelial cells (American Type Culture Collection) were infected with MPyV [A2, RA, or A2(91G)] at a multiplicity of infection (MOI) of 1 and cultured in DMEM containing 10% FBS. Cells and supernatant were harvested at the indicated days postinfection (p.i.), and after freezing and thawing, the viral lysate was titrated by plaque assay. For quantitative PCR (qPCR), DNA was extracted from organs or cell cultures by use of a DNA extraction kit (Promega), and viral genome copies were quantified by qPCR as previously described (35).
Mice and infections.
Female C57BL/6Ncr (B6) mice were purchased from the National Cancer Institute (Frederick, MD). B6.129S2-Ifnar1tm1Agt/Mmjax (IFNAR−/−) mice from The Jackson Laboratory were backcrossed with C57BL/6 mice for 5 generations. Thy1.1 TCR-I-transgenic mice, bearing the T cell receptor (TCR) specific for LT206–215 of SV40, were described previously (36). Adult mice (8 to 12 weeks old) were inoculated in the hind footpads with 1.0 × 106 PFU of MPyV. For recall experiments, mice persistently infected (>30 days) with MPyV received 1 × 106 PFU of rVSV-LT359 through hind footpad injection. All protocols using mice were approved by the Institutional Animal Care and Use Committee of The Pennsylvania State University College of Medicine.
Flow cytometric analysis.
Red blood cell (RBC)-lysed blood and spleen samples were stained for 30 min at 4°C with monoclonal antibodies (MAbs) against the following molecules: CD44 (clone IM7; BD), CD127 (clone A7R34; Biolegend), killer cell lectin-like receptor G1 (KLRG1) (clone 2F1; BD), and CD8α (clone 53-6.7; eBioscience). H-2Db LT359 and H-2Db LT206 tetramers (35) were provided by the NIH Tetramer Core Facility (Atlanta, GA). Samples were acquired on an LSRII instrument (BD), and data were analyzed using FlowJo software (TreeStar, Inc., Ashland, OR). For intracellular cytokine staining (ICS), cells were stimulated for 5 h with the LT359–368 peptide (1 μM) in the presence of Golgiplug (BD). Cells were surface stained, permeabilized, and fixed by use of a Cytofix/Cytoperm kit (BD) and then stained with MAbs specific for IFN-γ (clone XMG1.2; eBioscience), IL-2 (clone JES6-5H4; BD), and tumor necrosis factor alpha (TNF-α) (clone TN3-19.12; BD). Fluorescence-minus-one (FMO) samples were used to set gates for each surface molecule examined (i.e., CD127, KLRG1, CD8, and CD44); for ICS, unstimulated samples served as negative controls, and phorbol 12-myristate 13-acetate (PMA) (50 ng/ml)- and ionomycin (ION) (2 μg/ml)-treated samples served as positive controls. Nucleated cells from blood and spleen samples were counted with a TC20 automated cell counter (Bio-Rad). The number of CD8 T cells in blood was determined by use of a flow cytometry absolute count standard (Bands Laboratories, Inc.).
Adoptive T cell transfer.
CD8 T cells were purified from Thy1.1+ TCR-I-transgenic mice by use of a negative-selection CD8 T cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. For all cell transfer experiments, >95% of donor TCR-I cells were CD44lo. One hundred TCR-I cells were transferred 1 day prior to infection. Recipient mice were inoculated with 5 × 105 PFU of MPyV.LT206 or MPyV.LT206 (91G). TCR-I cells in 100 μl of blood were quantified by flow cytometry at day 8 p.i. For analysis of cell division, 1 × 106 TCR-I cells were labeled with 5 μM carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen) for 20 min at room temperature and then injected intravenously (i.v.) into the tail vein at day 4 p.i. The extent of CFSE dilution was determined by flow cytometry at day 6 p.i.
Dendritic cell immunization.
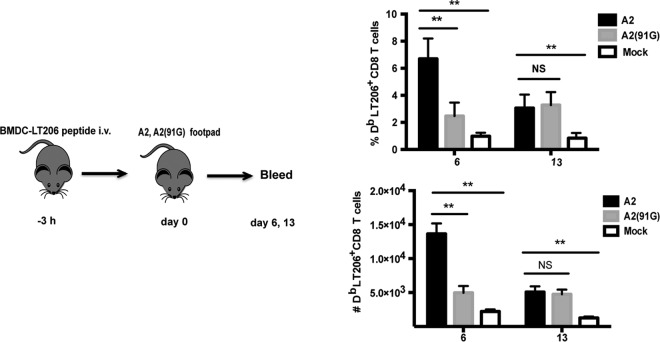
Bone marrow-derived dendritic cells (BMDCs) were cultured in vitro as previously described (37) and matured by exposure to 100 ng/ml lipopolysaccharide (LPS; Sigma-Aldrich) at day 7 of culture. Cells were harvested the next day and assessed by flow cytometry. More than 90% of cells were F4/80lo MHC-IIhi CD11chi. BMDCs were pulsed with the LT206 peptide (10 μM) at 37°C for 1.5 h and then washed, and 2.0 × 105 cells were injected i.v. into the tail vein in B6 mice. Three hours later, mice were injected with 1 × 106 PFU MPyV A2 or A2(91G) in the hind footpads. The frequency of DbLT206 tetramer+ CD8+ cells in blood was analyzed by flow cytometry.
Quantitation of cytokine mRNAs by qPCR.
Total RNA was isolated from spleens by use of a SimplyRNA kit (Promega) according to the manufacturer's instructions. One microgram of RNA was used to synthesize cDNA by use of a high-capacity cDNA reverse transcription kit (Thermo Scientific) according to the manufacturer's instructions. Transcripts for cytokine genes (IL-4, IL-10, IL-12, IFN-β, TNF-α, and IFN-γ) were quantified by qPCR with SYBR green master mix (Roche), using an ABI Prism 5700 sequence detection system (Applied Biosystems). The oligonucleotide primers used for qPCR are shown in Table 1. Primers for IL-4 (38), IL-12 p40 (39), IL-12 p35 (40), IFN-γ (41), TNF-α (42), and 18S rRNA (43) were previously published. Primers for IFN-β were validated by IDT. Primers for IL-10 were designed by use of the NCBI Primer Blast tool and then validated. Relative fold changes were determined using the threshold cycle (2−ΔΔCT) method, with the 18S rRNA gene used as a reference gene for normalization, as previously described (44). Briefly, amounts of cytokine mRNA were normalized to the amount of internal 18S rRNA. The fold change of cytokine mRNA for each infected mouse was calculated by dividing the amount of cytokine mRNA in infected mice by that in uninfected mice.
TABLE 1.
Primers used for quantitative RT-PCR
| Target | Forward primer | Reverse Primer |
|---|---|---|
| IL-4 | TCGGCATTTTGAACGAGGTC | GAAAAGCCCGAAAGAGTCTC |
| IL-10 | ACCTGGTAGAAGTGATGCCC | GACACCTTGGTCTTGGAGCTT |
| IL-12 p35 | AAATGAAGCTCTGCATCCTGC | TCACCCTGTTGATGGTCACG |
| IL-12 p40 | CCACTCACATCTGCTGCTCCACAA | CAGTTCAATGGGCAGGGTCTCCTC |
| IFN-β | ACTCATGAAGTACAACAGCTACG | GGCATCAACTGACAGGTCTT |
| TNF-α | TACTGAACTTCGGGGTGATTGGTCC | CAGCCTTGTCCCTTGAAGAGAACC |
| IFN-γ | TGAACGCTACACACTGCATCTTGG | CGACTCCTTTTCCGCTTCCTGAG |
| 18S rRNA | CACTTTTGGGGCCTTCGTGT | AGGCCCAGAGACTCATTTCTT |
Immunofluorescence (IF) assay.
Primary mouse embryonic fibroblast (MEF) cells prepared from 13-day-old B6 mice were pretreated with 500 U/ml recombinant mouse IFN-β purified from mammalian cell culture (PBL Interferon Source, Piscataway, NJ) for 24 h. Cells were infected with MPyV at an MOI of 0.01 and cultured with or without IFN-β (500 U/ml). At day 3 p.i., cells were fixed at room temperature for 10 min with 2% paraformaldehyde in PBS, stained with the pan-MPyV T antigen-specific F4 MAb (45) followed by goat anti-mouse IgG conjugated to Alexa 594 (Invitrogen), and mounted with Prolong reagent with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen).
Statistical analyses.
Statistical significance was determined by unpaired, two-tailed Student's t test, assuming equal variance. A P value of <0.05 was considered statistically significant. The 50% effective concentration of peptide (EC50) for the IFN-γ stimulation assay was calculated according to nonlinear trajectory analysis by using GraphPad Prism software (San Diego, CA).
RESULTS
E-to-G variation at position 91 of VP1 changes viral spread and the magnitude of the MPyV-specific CD8 T cell response.
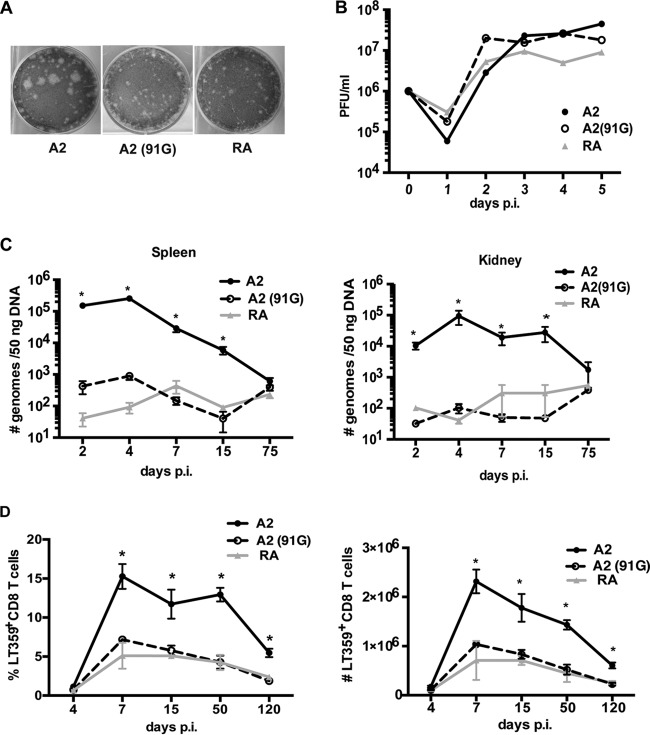
The magnitude of the CD8 T cell response correlates with the initial viral load and duration of infection in various viral infection models (46, 47). LP strains of MPyV disseminate more extensively than do SP strains (5, 6). Administration of a low dose of the LP strain A2 delays the kinetics of expansion, but not the magnitude, of the MPyV-specific CD8 T cell response (23). We therefore asked whether B6 mice infected with an LP strain (A2) or an SP strain (RA) showed differences in viral dissemination and in the magnitude of the strongly dominant Db-restricted CD8 T cell response to amino acids 359 to 368 of the large T (LT) antigen, designated DbLT359 (48). To control for differences in the noncoding control region and the few differences in coding regions between strains A2 and RA, we used site-directed mutagenesis to change the glutamic acid (E) at position 91 of VP1 in A2 to glycine (G), as previously described (7). This A2(91G) mutant virus converted the parental A2 virus from LP to SP, confirming previous reports (6, 7) that this single amino acid difference in VP1 is solely responsible for the small-plaque morphology of the RA strain (Fig. 1A). The mutant A2(91G) exhibited in vitro growth kinetics similar to those of A2 and SP strain RA (Fig. 1B). However, viral loads in the spleens and kidneys of mice infected by the RA or A2(91G) virus showed minimal differences over the course of infection (Fig. 1C) and were over 100-fold lower than those in mice infected by A2 virus during the acute phase of infection. The frequency and magnitude of the DbLT359-specific CD8 T cell response in mice infected by the RA or A2(91G) virus were significantly lower than those in mice infected by the A2 virus, but notably, the kinetics of T cell expansion and contraction in response to infection by each of these viruses were similar (Fig. 1D). To address the possibility that less replication by A2(91G) may itself be responsible for the smaller magnitude of the anti-MPyV CD8 T cell response, mice were inoculated with a 10-fold higher dose of A2(91G) than that of the A2 strain. We found that the kinetics of viral replication and numbers of DbLT359-specific CD8 T cells remained significantly lower than those for A2-infected mice (data not shown). Given that low-dose A2 virus inoculation yields a different DbLT359-specific CD8 T cell response profile (delayed kinetics of expansion but an equal peak magnitude during acute infection) (23), these results indicated that the tropism difference imparted by the E-to-G switch at position 91 of VP1 negatively affected the size of the MPyV-specific CD8 T cell response.
FIG 1.
E-to-G substitution at position 91 of VP1 limits viral spread and the magnitude of the MPyV-specific CD8 T cell response. (A) Plaque formation at day 6 p.i. by A2, A2(91G), and RA strain viruses on A31 cells. (B) One-step growth curve assay of A2, RA, and A2(91G) in NMuMG cells infected at an MOI of 1. Viral titers were measured by plaque assay at the indicated days p.i. (C) Viral DNAs in spleens and kidneys were measured by qPCR at the indicated days p.i. after A2, RA, or A2(91G) inoculation (1.0 × 106 PFU) into hind footpads. (D) Frequencies (left) and numbers (right) of splenic DbLT359 tetramer+ CD8+ cells at the indicated days p.i. Data are the means ± standard errors of the means (SEM) for at least three mice per group and are representative of three experiments. *, P < 0.05.
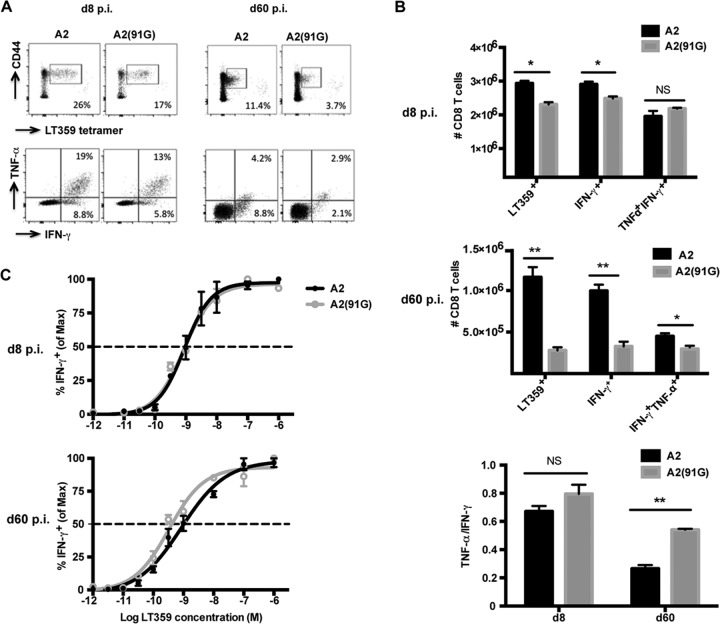
MPyV-specific CD8 T cells recruited by A2(91G) virus infection exhibit superior functionality.
During persistent infection with LCMV strain cl13, a high viral load correlates with a loss of CD8 T cell polycytokine functionality and proliferative potential (47). We next asked whether the 2-log difference in viral loads during acute infection between the SP viruses RA and A2(91G) and the LP virus A2 affected the functional competence of the MPyV-specific CD8 T cell response. During the acute (day 8 p.i.) and persistent (day 60 p.i.) phases of infection, A2(91G) virus-infected mice showed a lower frequency and number of splenic DbLT359-specific CD8 T cells than those for A2 virus-infected mice, as detected by DbLT359 tetramers and ex vivo LT359 peptide-stimulated intracellular IFN-γ and TNF-α production (Fig. 2A). Comparison of the capacity of LT359-specific CD8 T cells to coproduce IFN-γ and TNF-α (Fig. 2B, top and middle panels), however, revealed that MPyV-specific CD8 T cells recruited by A2(91G) infection had an elevated capacity to coproduce these cytokines during persistent infection (Fig. 2B, bottom panel). LT359 peptide titration further showed that MPyV-specific CD8 T cells recruited by A2(91G) infection during persistent infection (day 60 p.i.) exhibited a higher antigen sensitivity (approximately 3-fold, as shown by EC50 values in the legend to Fig. 2), a difference not seen for cells recruited during acute infection (Fig. 2C). It merits highlighting that this difference in functional avidity by DbLT359-specific CD8 T cells, albeit small, was not seen in mice given low- versus high-dose A2 virus inocula (23).
FIG 2.
A2(91G) infection recruits memory CD8 T cells having higher polycytokine functionality and functional avidity. (A) Representative dot plots of splenic DbLT359 tetramer+ CD8 T cells surface stained with anti-CD44 and of intracellular costaining with anti-IFN-γ and anti-TNF-α of anti-CD8 surface-stained spleen cells after LT359 peptide stimulation at days 8 and 60 p.i. (no IFN-γ+ or TNF-α+ cells were detected in the absence of LT359 peptide [data not shown]). (B) Numbers of DbLT359 tetramer+, IFN-γ+, and IFN-γ+ TNF-α+ CD8 T cells at days 8 and 60 p.i. (top and middle) and ratios of TNF-α+ to IFN-γ+ CD8 T cells (bottom). Values represent the means and SEM for three experiments using 3 or 4 mice per group, *, P < 0.05; **, P < 0.005; NS, not significant. (C) LT359 peptide dose-response curves for intracellular IFN-γ staining of splenic CD8 T cells from A2- and A2(91G)-infected mice at day 8 p.i. (top) and day 60 p.i. (bottom). EC50 values were as follows: at day 8 p.i., 9.0 × 10−10 M for A2 and 8.5 × 10−10 M for A2(91G) (P = 0.96); and at day 60 p.i., 8.71 × 10−10 M for A2 and 3.3 × 10−10 M for A2(91G) (P = 0.017). Experiments were repeated twice, with 3 mice per group.
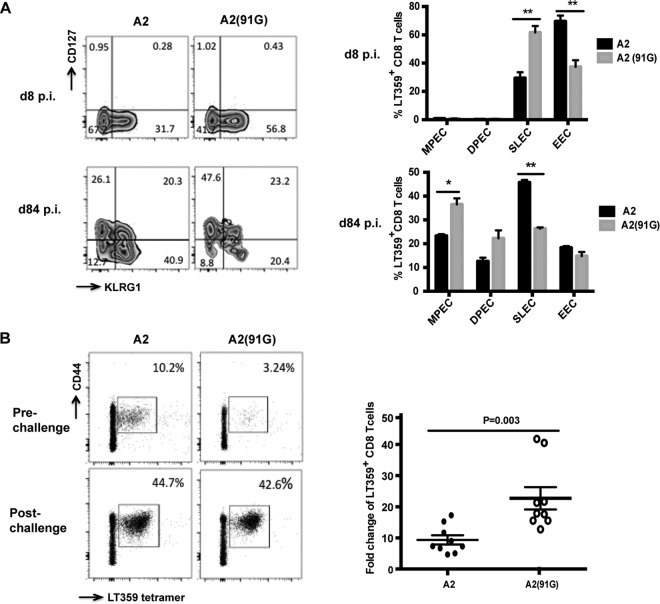
MPyV-specific CD8 T cells generated in response to A2 and A2(91G) infections also exhibited phenotypic differences indicative of skewed memory T cell differentiation. Memory precursors and terminally differentiated CD8 T cells can be distinguished based on their expression of CD127 (IL-7Rα) (49, 50) and killer cell lectin-like receptor G1 (KLRG1) (51). Phenotypic dissection using these markers demarcates CD127lo KLRG1lo cells as early effector cells (EECs), CD127lo KLRG1hi cells as short-lived effector cells (SLECs), CD127hi KLRG1hi cells as double-positive effector cells (DPECs), and CD127hi KLRG1lo cells as memory precursor effector cells (MPECs). Using these markers, we observed that acute infection with A2(91G) generated a significantly higher frequency of SLECs and a lower frequency of EECs than did A2 infection. In persistently infected mice, however, A2(91G) recruited a higher frequency of MPECs and a lower frequency of SLECs than those with the A2 strain (Fig. 3A). The higher virus levels during acute A2 infection appear to favor differentiation of EECs over SLECs, whereas in persistently infected mice, where levels of A2 and A2(91G) are similar (Fig. 1C), A2 infection biases cells toward SLEC differentiation and A2(91G) infection tilts them toward MPEC differentiation.
FIG 3.
A2(91G) infection-recruited CD8 T cells exhibit a skewed CD127hi KLRG1lo phenotype and have a larger recall response. (A) At days 8 and 84 p.i., spleen cells were surface stained with DbLT359 tetramers and MAbs to KLRG1 and CD127. (Left) Contour plots representative of 3 to 5 mice per time point and three independent experiments; (right) frequencies of LT359-specific CD8 T cells with the MPEC (KLRG1lo CD127hi), DPEC (KLRG1hi CD127hi), SLEC (KLRG1hi CD127lo), and EEC (KLRG1lo CD127lo) phenotypes. Values indicate means and SEM. *, P < 0.05; **, P < 0.005. (B) At 80 days p.i. with A2 or A2(91G), B6 mice were challenged with rVSV-LT359. (Left) Representative mean frequencies of DbLT359 tetramer+ CD44+ CD8 T cells in blood before and 5 days after challenge; (right) pre- versus postchallenge fold expansion, with each dot corresponding to an individual mouse and horizontal lines indicating means ± SEM. Data were collected from two independent experiments (n = 4 or 5 mice for each group).
We next tested the prediction that this MPEC bias by MPyV-specific CD8 T cells in mice persistently infected with A2(91G) was associated with an augmented recall capability upon antigen challenge. A2- and A2(91G)-infected mice at day 84 p.i. were thus infected with a recombinant VSV strain encoding the LT359 epitope (22). As shown in Fig. 3B, at 5 days postchallenge, DbLT359-specific CD8 T cells were expanded to a significantly greater extent in A2(91G)-infected mice than in A2-infected mice. Taken together, the data show that E-to-G variation at position 91 of VP1 is associated with superior functionality of the MPyV-specific CD8 T cell response during persistent infection.
Inflammatory environments created by A2 versus A2(91G) infection differentially affect the MPyV-specific CD8 T cell response.
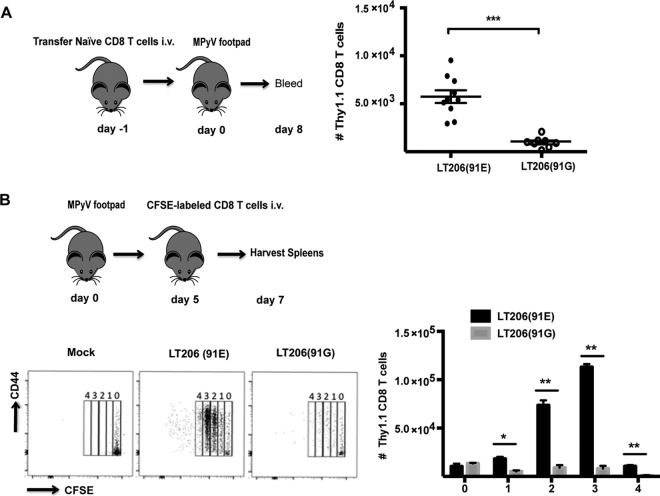
Early inflammation during viral infection plays a role in regulating CD8 T cell proliferation (17). Thus, we asked whether the inflammatory environment associated with A2 infection and A2(91G) infection affected the proliferation of naive virus-specific CD8 T cells. We introduced the G-91 VP1 mutation into the previously characterized MPyV.LT206 virus, an A2 mutant in which the LT359 peptide sequence was replaced with that of the SV40 LT206 epitope recognized by TCR-I-transgenic CD8 T cells (22, 36). We designated this virus LT206(91G). MPyV.LT206 and LT206(91G) have replication kinetics similar to those of A2 and A2(91G), respectively (data not shown). Adoptive transfer of Thy1.1+ TCR-I cells into B6 (Thy1.2) mice prior to infection recapitulated the observation made in Fig. 1D that peak accumulation of virus-specific CD8 T cells during acute infection was higher for MPyV.LT206(E-91) than for LT206(91G) (Fig. 4A). Using CFSE-labeled donor TCR-I cells, we determined that this higher-magnitude expansion could be accounted for by more efficient expansion of anti-MPyV CD8 T cells in mice infected by the E-91 VP1 than the G-91 VP1 virus (Fig. 4B).
FIG 4.
Naive MPyV-specific CD8 T cells are recruited more efficiently by infection with A2(91E) virus than by that with A2(91G) virus. (A) Experimental setup. One hundred naive Thy1.1 TCR-I cells were injected i.v. into the tail vein in B6 (Thy1.2) mice 1 day before inoculation with 5 × 105 PFU of MPyV.LT206(91E) or MPyV.LT206(91G). The number of TCR-I cells in blood was determined by flow cytometry at day 8 p.i. Data were collected from three experiments with three mice per group. Each dot represents an individual mouse, with horizontal lines indicating means ± SEM. ***, P < 0.0005. (B) (Top) Experimental setup. A total of 1 × 106 naive TCR-I cells labeled with CFSE were transferred i.v. into B6 mice inoculated 5 days earlier with MPyV.LT206(91E) or MPyV.LT206(91G). (Bottom left) After 40 h, the number of cell divisions of donor TCR-I cells was determined based on CFSE dilution 2 days after cell transfer. (Bottom right) Mean numbers of cell divisions of TCR-I cells (and SEM) for three mice per group from two independent experiments. **, P < 0.005.
To determine whether the A2 infection-associated inflammatory environment also contributed to more efficient recruitment of naive virus-specific CD8 T cells, we immunized mice with LT206 peptide-pulsed BMDCs in the setting of infection by A2 or A2(91G) virus, both of which lack the DbLT206 epitope. No cross-reactivity is seen by CD8 T cells specific for the DbLT206 and DbLT359 epitopes (22; data not shown). This approach allowed us to eliminate differences in cognate viral antigen levels resulting from A2 versus A2(91G) infection. As shown in Fig. 5, the inflammatory environment associated with A2 infection drove a significantly larger DbLT206 CD8 T cell expansion than that with A2(91G) infection by day 6 p.i. These results raised the possibility that A2 and A2(91G) elicited different types or amounts of cytokines early in infection that affected the expansion of MPyV-specific CD8 T cells.
FIG 5.
Inflammation associated with A2 and A2(91G) infections differentially promotes CD8 T cell expansion. (Left) Experimental setup. A total of 2 × 105 BMDCs pulsed with LT206 peptide were injected i.v. into the tail vein in B6 mice. Three hours later, mice were inoculated with 1.0 × 106 PFU of A2 or A2(91G) in the hind footpads. (Right) The frequency of DbLT206 tetramer+ CD8 T cells in blood was analyzed at the indicated days p.i. Data were combined from three independent experiments with three mice per group. Values indicate means and SEM. **, P < 0.005; NS, not significant.
Type I IFN controls MPyV replication in vitro and in vivo.
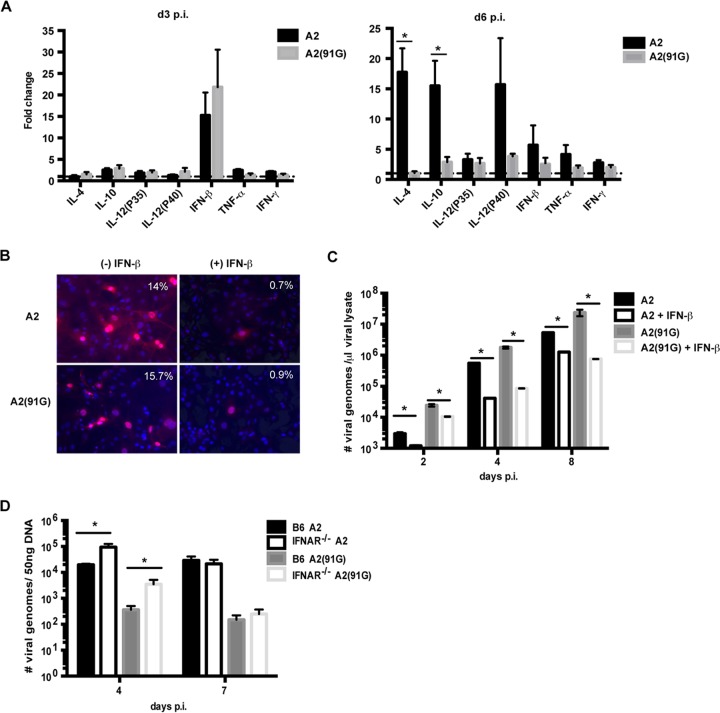
Using quantitative PCR, we assessed the mRNA levels for candidate cytokines (IL-4, IL-10, IL-12, IFN-β, TNF-α, and IFN-γ) potentially elicited early during MPyV infection and compared the levels of these transcripts in MPyV-infected and uninfected mice. Of the cytokine transcripts examined at days 3 and 6 p.i., IFN-β mRNA levels were elevated in infected mice at both time points, although they were markedly higher at day 3 p.i.; no significant difference was observed in induction of IFN-β transcripts between A2 and A2(91G) virus-infected mice (Fig. 6A). Interestingly, by day 6 p.i., mice infected with A2 virus showed a dramatic upregulation of transcripts for IL-12 p40, the subunit shared by IL-12 and IL-23, as well as transcripts for IL-4 and IL-10; strong induction of transcripts for these cytokines was not seen in A2(91G)-infected mice (Fig. 6A). This difference in profile for these cytokines between A2 and A2(91G) during acute infection may influence the effector versus memory differentiation of MPyV-specific CD8 T cells. To determine whether type I interferons inhibited MPyV replication, primary mouse embryonic fibroblasts (MEFs) were infected with either A2 or mutant A2(91G) virus in the presence or absence of mouse IFN-β. IFN-β inhibited infection by both the A2 and A2(91G) viruses. Approximately 15% of MEFs among untreated cells were T antigen-positive, while fewer than 1% of cells among IFN-β-treated cells were positive (Fig. 6B). The antiviral effect of IFN-β was confirmed by enumeration of MPyV genome copies by qPCR (Fig. 6C). IFN-β-mediated antiviral effects were also observed in vivo. In spleens of B6 versus IFNAR−/− mice infected with A2 or A2(91G), viral levels in IFNAR−/− mice were approximately 1 log higher than those in B6 mice at day 4 p.i. (Fig. 6D), a time point preceding detection of MPyV-specific CD8 T cells (52). These results indicate that IFN-I controls viral replication early in the course of MPyV infection and that it does so equivalently for infections with A2 and A2(91G).
FIG 6.
Type I interferons inhibit viral replication in vitro and in vivo. (A) Fold changes of the indicated cytokine mRNAs from spleens of infected mice at days 3 and 6 p.i. normalized to transcripts from spleens of uninfected mice. Data were collected from three independent experiments with three mice per group. Values indicate means and SEM. *, P < 0.05. (B) MEFs were infected with A2 or A2(91G) virus at an MOI of 0.01, with and without addition of IFN-β. MEFs were stained with a pan-T-antigen MAb and counterstained with DAPI at 24 h p.i. Representative percentages of T antigen+ cells among DAPI+ cells (>250 cells counted) from two experiments are shown. (C) Viral genome copies in the cell lysates used for panel B were quantified by qPCR at the indicated time points. Values indicate means and SEM for triplicates in one experiment. This experiment was repeated twice. (D) Viral loads in spleens of infected B6 and IFNAR−/− mice were measured by qPCR at days 4 and 7 p.i. Values indicate means and SEM for 4 or 5 mice from each group. *, P < 0.05.
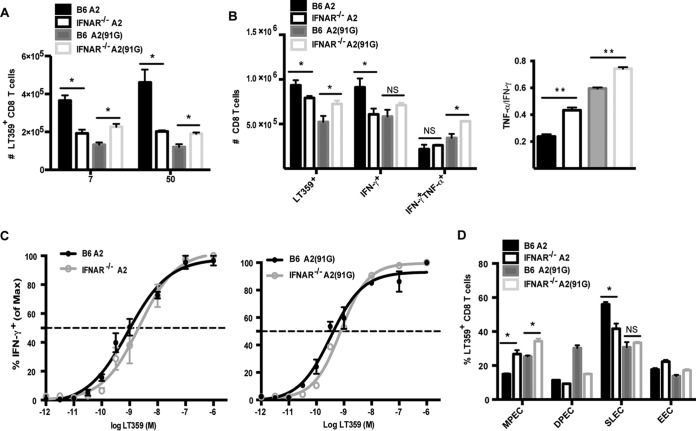
Next, we compared MPyV-specific CD8 T cell responses in B6 versus IFNAR−/− mice infected by the A2 or A2(91G) virus. As shown in Fig. 7A and B, the absence of IFN-I receptors had opposite effects on the anti-MPyV CD8 T cell responses recruited in response to A2 and A2(91G) infections. Although the size of the DbLT359-specific CD8 T cell response was lower in A2-infected IFNAR−/− mice than B6 mice, the reverse was seen for A2(91G) infection; this difference was seen in both acutely and persistently infected mice (Fig. 7A and B). IFN-I receptor deficiency was generally associated with an augmented cytokine-producing capability, which was particularly evident for polycytokine production by CD8 T cells during persistent infection (Fig. 7B). Interestingly, memory CD8 T cells in mice persistently infected with either A2 or A2(91G) virus exhibited a shift toward decreased viral antigen sensitivity in the absence of IFN-I receptors (Fig. 7C and as shown by EC50 values in the legend to Fig. 7), as well as skewed differentiation toward the MPEC phenotype (Fig. 7D). Thus, MPyV infections of wild-type and IFNAR−/− mice, irrespective of the 91E-to-G VP1 tropism difference, inversely affected the effector function and functional avidity of the virus-specific CD8 T cells.
FIG 7.
Type I interferons regulate CD8 T cell proliferation, functionality, and memory differentiation in a virus strain-dependent manner. B6 mice and IFNAR−/− mice were infected with 1.0 × 106 PFU of A2 or A2(91G) virus in the hind footpads. (A) The frequency of DbLT359 tetramer+ CD8 T cells in blood was measured by flow cytometry at days 7 and 50 p.i. Data were collected from two independent experiments (3 to 5 mice/group). (B) Splenocytes from mice inoculated 55 days earlier with A2 or A2(91G) were surface stained with anti-CD44, anti-CD8α, and DbLT359 tetramers or stained for surface CD8α and intracellular IFN-γ and TNF-α after 5 h of stimulation with LT359 peptide or without peptide (data not shown; used to set gates). (Left) Numbers of splenic DbLT359 tetramer+ CD8 T cells and LT359 peptide-stimulated IFN-γ+ and TNF-α+ IFN-γ+ CD8 T cells. (Right) Ratios of TNF-α+ to IFN-γ+ LT359 CD8 T cells. (C) LT359 peptide dose-response curves for intracellular IFN-γ staining of splenic CD8 T cells at day 55 p.i. The EC50 for A2-infected B6 mice was 8.87 × 10−10 M, and that for A2-infected IFNAR−/− mice was 2.25 × 10−9 M (P = 0.04). The EC50 for A2(91G)-infected B6 mice was 2.3 × 10−10 M, and that for A2(91G)-infected IFNAR−/− mice was 7.63 × 10−10 M (P = 0.0022). (D) At day 50 p.i., splenic DbLT359 tetramer+ CD8 T cells were stained for KLRG1 and CD127; designations based on these molecules are described in the legend to Fig. 3A. Data are representative of three experiments using 3 or 4 mice/group. Values indicate means and SEM. *, P < 0.05; **, P < 0.005; NS, not significant.
DISCUSSION
In this study, we compared the CD8 T cell responses to MPyV strains with different receptor binding specificities and affinities. The single-amino-acid E-to-G exchange at position 91 in VP1 causes MPyV virions to bind not only to terminally unbranched sialic acid true cellular receptors but also to branched-chain sialic acid “pseudoreceptors,” resulting in attenuation of viral spread and tumorigenicity (6, 9). We validated that the E-to-G substitution at VP1 position 91 dampened viral replication in vivo and that it did so in adult B6 mice, which are highly resistant to MPyV tumorigenesis (53). Here we found that this VP1 difference limited expansion of MPyV-specific CD8 T cells and endowed them with superior function (more polycytokine production, higher functional avidity, and a larger recall response) during persistent infection. We further found that IFN-I was induced early during MPyV infection and differentially regulated the magnitude and functionality of virus-specific CD8 T cells in a manner dependent on the tropism of viruses with an E versus G at VP1 position 91.
CD8 T cell differentiation is programmed by three major factors: the strength and duration of peptide major histocompatibility complex (MHC)-TCR engagement, the availability and constellation of costimulatory molecules from antigen-presenting cells, and the profile of inflammatory cytokines. Accumulating literature indicates that the pathogen-induced inflammatory environment plays a central role in guiding the pathway for memory CD8 T cell differentiation (17, 24). Our previous study showed that early virus-associated bystander events induced by a low-dose inoculum of MPyV A2 generated virus-specific CD8 T cells with a superior capacity to produce IFN-γ. CD8 T cells primed by low-dose MPyV also expressed higher levels of CD127 and the antiapoptotic protein Bcl-2, which are required for memory CD8 T cell survival during persistent infection (23). The low-dose A2 strain inoculation delayed the kinetics but did not change the magnitude of the CD8 T cell response compared to high-dose inoculation of the A2 strain virus (23). In sharp contrast, infection with the VP1 mutant A2(91G) virus, which spread poorly compared to the parental A2(91E) virus, generated an MPyV-specific CD8 T cell response with the same kinetics as that of the A2(91E) virus, albeit of lower magnitude. Thus, the lower virus infection levels and lesser spread by A2(91G) were of themselves insufficient factors to account for the less efficient recruitment of anti-MPyV CD8 T cells. Using an adoptive T cell transfer approach and dendritic cell immunization, we demonstrated that the early inflammatory environment caused by a change in viral receptor binding contributed to a different magnitude and functionality of virus-specific CD8 T cells. IFN-I and IL-12 have been described as a “third signal” to promote full activation and development of CD8 T cells (54–56). Through cytokine profiling, we found that the transcripts of IFN-β were markedly upregulated during early acute infection by both A2(91E) and A2(91G) viruses, whereas mRNAs for IL-12 (and/or IL-23) were increased later in acute infection only by A2(91E) virus.
Type I IFNs are comprised of a family of highly related molecules that program a state of resistance to intracellular pathogens and serve to alarm cells of both innate and adaptive immunity of the presence of infections (57). In our study, we showed that MPyV replication was directly inhibited by IFN-β in vitro and that viral levels were higher in IFNAR−/− mice than in wild-type mice. IFN-I has been shown to promote IFN-γ production by CD4 T cells through STAT4 activation (58), to enhance the terminal differentiation of dendritic cells (59), and to augment the proliferation of CD8 T cells (55). We showed that the tropism change due to the E-to-G difference at position 91 of VP1, while associated with a lower-magnitude CD8 T cell response, was equally capable of eliciting IFN-β. This discrepancy might be explained by the timing of exposure of naive CD8 T cells to IFN-I. IFN-I may mitigate CD8 T cell proliferation when T cells encounter IFN-I before engaging their cognate antigen (60). Because A2(91G) disseminates less efficiently than A2, naive anti-MPyV CD8 T cells recruited in response to infection by A2(91G) may bind IFN-I before TCR activation. Thus, it is conceivable that polyomavirus VP1 variants that differ in type and/or affinity for host cell receptor glycans may alter the fate of the antiviral CD8 T cell response by changing when naive T cells engage IFN-I relative to antigen. This possibility is supported by evidence that innate inflammatory signals induced by different pathogens differentially dictate the IFN-I dependence of CD8 T cell expansion and memory differentiation (24). An increase in IL-12 (and/or IL-23) during acute infection by A2(91E) virus may also have promoted MPyV-specific CD8 T cell expansion and effector differentiation.
IFN-I plays an important role in the effector and memory fate decision process via regulating the secretion of particular cytokines (e.g., IL-15) (61–63) and the expression of the transcription factors T-bet (51) and eomesodermin (64). IFN-I receptor signaling regulates the expression of CD127 and KLRG1, which are molecules used to demarcate subsets of effector and memory T cells (49, 51). Our data showed that the E-to-G change at position 91 of VP1 shifted CD8 T cell differentiation toward SLECs during acute infection but toward MPECs during persistent infection. In contrast, IFN-I signaling skewed virus-specific CD8 T cell differentiation toward SLECs during persistent infection by A2 virus. The increased polycytokine functionality by MPyV-specific memory CD8 T cells in mice lacking IFN-I receptors may be related to their lower peak expansion and effector differentiation during acute infection. IFN-I receptor signaling during persistent infection may also impair memory CD8 T cell functionality, as recently reported for chronic LCMV infection, where it has been proposed to limit immunopathology (65–67). IFN-I receptor signaling by CD8 T cells during acute LCMV infection upregulated T-bet and directed differentiation toward SLECs, while a subset of CD8 T cells exhibited the MPEC phenotype during chronic LCMV infection in both IFN-I receptor-sufficient and -deficient mice (68). While a mechanistic understanding of how IFN-I modulates CD8 T cell differentiation remains unknown, our results reinforce the concept that the impact of IFN-I on memory differentiation is pathogen dependent.
All MPyV isolates from feral mice have a glutamic acid at VP1 position 91 (11). From the perspective of virus-host coevolution, it is seemingly counterintuitive that mice in the wild harbor MPyV strains with a VP1 amino acid shared by highly tumorigenic laboratory strains. Tumor induction by MPyV, however, is relegated to neonatal infection of virus-free colonies of mice of tumor-susceptible strains or infection of immunodeficient naive adult mice (69). In natural mouse populations, MPyV establishes a silent persistent infection. It has been proposed that differences in regulatory sequences among MPyV isolates from feral mice might offset the viral oncogenicity by changing host range properties of the virus. The results presented here give the additional explanation that the relative impairment in virus-specific CD8 T cell immunity associated with infection by E91-VP1 strains in feral mice favors higher viral loads and intermouse transmission.
CD8 T cells play an essential role in controlling persistent infection by polyomavirus (34). The high frequency of amino acid mutations in VP1 close to or at the receptor binding site in isolates of JCV from PML patients suggests that subtle changes in binding affinity for sialylated receptors can significantly affect viral pathogenicity (70). Recent evidence indicates that mutations in VP1 may serve to enable escape from neutralizing JCV antibodies (71, 72). PML nonsurvivors had selectively impaired JCV-specific CD8 T cell responses (16, 73). We recently reported that brain-resident MPyV-specific memory CD8 T cells expressed higher-affinity TCRs than those of splenic CD8 T cells (74). Whether IFN-I is induced by JCV infection and affects the functional integrity of CD8 T cells in PML patients in response to VP1 variant viruses is an open question. IFN-I receptor blockade has been reported to reverse CD8 T cell exhaustion and to control chronic LCMV infection (67, 75). Our study suggests that the IFN-I pathway may similarly warrant consideration as a therapeutic target for persistent polyomavirus infection.
ACKNOWLEDGMENTS
We thank Ge Jin for excellent technical support and animal handling and Taryn Mockus for a critical review of the manuscript. We thank Ziaur Rahman (Department of Microbiology and Immunology, Penn State College of Medicine) for generously providing IFNAR−/− mice. We acknowledge the technical support of Nate Sheaffer, Jade Vogel, and Joe Bednarcyk of the Flow Cytometry Core Facility of The Penn State College of Medicine (Hershey, PA).
REFERENCES
- 1.Neu U, Stehle T, Atwood WJ. 2009. The Polyomaviridae: contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 384:389–399. doi: 10.1016/j.virol.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA. 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J 22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You J, O'Hara SD, Velupillai P, Castle S, Levery S, Garcea RL, Benjamin T. 2015. Ganglioside and non-ganglioside mediated host responses to the mouse polyomavirus. PLoS Pathog 11:e1005175. doi: 10.1371/journal.ppat.1005175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer PH, Bronson RT, Fung SC, Freund R, Stehle T, Harrison SC, Benjamin TL. 1995. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J Virol 69:7925–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubensky TW, Freund R, Dawe CJ, Benjamin TL. 1991. Polyomavirus replication in mice: influences of VP1 type and route of inoculation. J Virol 65:342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer PH, Cui C, Liu WR, Stehle T, Harrison SC, DeCaprio JA, Benjamin TL. 1999. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J Virol 73:5826–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freund R, Garcea RL, Sahli R, Benjamin TL. 1991. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J Virol 65:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian M, Tsai B. 2010. Lipids and proteins act in opposing manners to regulate polyomavirus infection. J Virol 84:9840–9852. doi: 10.1128/JVI.01093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buch MH, Liaci AM, O'Hara SD, Garcea RL, Neu U, Stehle T. 2015. Structural and functional analysis of murine polyomavirus capsid proteins establish the determinants of ligand recognition and pathogenicity. PLoS Pathog 11:e1005104. doi: 10.1371/journal.ppat.1005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stehle T, Harrison SC. 1996. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure 4:183–194. doi: 10.1016/S0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 11.Carroll J, Dey D, Kreisman L, Velupillai P, Dahl J, Telford S, Bronson R, Benjamin T. 2007. Receptor-binding and oncogenic properties of polyoma viruses isolated from feral mice. PLoS Pathog 3:e179. doi: 10.1371/journal.ppat.0030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelik L, Reid C, Testa M, Brickelmaier M, Bossolasco S, Pazzi A, Bestetti A, Carmillo P, Wilson E, McAuliffe M, Tonkin C, Carulli JP, Lugovskoy A, Lazzarin A, Sunyaev S, Simon K, Cinque P. 2011. Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. J Infect Dis 204:103–114. doi: 10.1093/infdis/jir198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayliss J, Harrison E, McLean CA. 2011. Progressive multifocal leukoencephalopathy development is associated with mutations in JC virus capsid protein VP1 that change the receptor specificity of the virus. J Infect Dis 204:1643–1644. doi: 10.1093/infdis/jir611. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Blondel G, Bauer J, Cuvinciuc V, Uro-Coste E, Debard A, Massip P, Delisle MB, Lassmann H, Marchou B, Mars LT, Liblau RS. 2013. In situ evidence of JC virus control by CD8+ T cells in PML-IRIS during HIV infection. Neurology 81:964–970. doi: 10.1212/WNL.0b013e3182a43e6d. [DOI] [PubMed] [Google Scholar]
- 16.Gheuens S, Bord E, Kesari S, Simpson DM, Gandhi RT, Clifford DB, Berger JR, Ngo L, Koralnik IJ. 2011. Role of CD4+ and CD8+ T-cell responses against JC virus in the outcome of patients with progressive multifocal leukoencephalopathy (PML) and PML with immune reconstitution inflammatory syndrome. J Virol 85:7256–7263. doi: 10.1128/JVI.02506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obar JJ, Jellison ER, Sheridan BS, Blair DA, Pham QM, Zickovich JM, Lefrancois L. 2011. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J Immunol 187:4967–4978. doi: 10.4049/jimmunol.1102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair DA, Turner DL, Bose TO, Pham QM, Bouchard KR, Williams KJ, McAleer JP, Cauley LS, Vella AT, Lefrancois L. 2011. Duration of antigen availability influences the expansion and memory differentiation of T cells. J Immunol 187:2310–2321. doi: 10.4049/jimmunol.1100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban SL, Welsh RM. 2014. Out-of-sequence signal 3 as a mechanism for virus-induced immune suppression of CD8 T cell responses. PLoS Pathog 10:e1004357. doi: 10.1371/journal.ppat.1004357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller SN, Ahmed R. 2009. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc Natl Acad Sci U S A 106:8623–8628. doi: 10.1073/pnas.0809818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wherry EJ, Ahmed R. 2004. Memory CD8 T-cell differentiation during viral infection. J Virol 78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JJ, Pack CD, Lin E, Frost EL, Albrecht JA, Hadley A, Hofstetter AR, Tevethia SS, Schell TD, Lukacher AE. 2012. CD8 T cells recruited early in mouse polyomavirus infection undergo exhaustion. J Immunol 188:4340–4348. doi: 10.4049/jimmunol.1103727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews NP, Pack CD, Vezys V, Barber GN, Lukacher AE. 2007. Early virus-associated bystander events affect the fitness of the CD8 T cell response to persistent virus infection. J Immunol 178:7267–7275. doi: 10.4049/jimmunol.178.11.7267. [DOI] [PubMed] [Google Scholar]
- 24.Thompson LJ, Kolumam GA, Thomas S, Murali-Krishna K. 2006. Innate inflammatory signals induced by various pathogens differentially dictate the IFN-I dependence of CD8 T cells for clonal expansion and memory formation. J Immunol 177:1746–1754. doi: 10.4049/jimmunol.177.3.1746. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed R, Hahn CS, Somasundaram T, Villarete L, Matloubian M, Strauss JH. 1991. Molecular basis of organ-specific selection of viral variants during chronic infection. J Virol 65:4242–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matloubian M, Kolhekar SR, Somasundaram T, Ahmed R. 1993. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol 67:7340–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baca Jones C, Filippi C, Sachithanantham S, Rodriguez-Calvo T, Ehrhardt K, von Herrath M. 2014. Direct infection of dendritic cells during chronic viral infection suppresses antiviral T cell proliferation and induces IL-10 expression in CD4 T cells. PLoS One 9:e90855. doi: 10.1371/journal.pone.0090855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. 2006. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med 12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Z, Casey KA, Jameson SC, Curtsinger JM, Mescher MF. 2009. Programming for CD8 T cell memory development requires IL-12 or type I IFN. J Immunol 182:2786–2794. doi: 10.4049/jimmunol.0803484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keppler SJ, Rosenits K, Koegl T, Vucikuja S, Aichele P. 2012. Signal 3 cytokines as modulators of primary immune responses during infections: the interplay of type I IFN and IL-12 in CD8 T cell responses. PLoS One 7:e40865. doi: 10.1371/journal.pone.0040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Way SS, Havenar-Daughton C, Kolumam GA, Orgun NN, Murali-Krishna K. 2007. IL-12 and type-I IFN synergize for IFN-γ production by CD4 T cells, whereas neither are required for IFN-γ production by CD8 T cells after Listeria monocytogenes infection. J Immunol 178:4498–4505. doi: 10.4049/jimmunol.178.7.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukacher AE, Wilson CS. 1998. Resistance to polyoma virus-induced tumors correlates with CTL recognition of an immunodominant H-2Dk-restricted epitope in the middle T protein. J Immunol 160:1724–1734. [PubMed] [Google Scholar]
- 34.Moser JM, Altman JD, Lukacher AE. 2001. Antiviral CD8+ T cell responses in neonatal mice: susceptibility to polyoma virus-induced tumors is associated with lack of cytotoxic function by viral antigen-specific T cells. J Exp Med 193:595–606. doi: 10.1084/jem.193.5.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemball CC, Lee ED, Vezys V, Pearson TC, Larsen CP, Lukacher AE. 2005. Late priming and variability of epitope-specific CD8+ T cell responses during a persistent virus infection. J Immunol 174:7950–7960. doi: 10.4049/jimmunol.174.12.7950. [DOI] [PubMed] [Google Scholar]
- 36.Staveley-O'Carroll K, Schell TD, Jimenez M, Mylin LM, Tevethia MJ, Schoenberger SP, Tevethia SS. 2003. In vivo ligation of CD40 enhances priming against the endogenous tumor antigen and promotes CD8+ T cell effector function in SV40 T antigen transgenic mice. J Immunol 171:697–707. doi: 10.4049/jimmunol.171.2.697. [DOI] [PubMed] [Google Scholar]
- 37.Bender A, Sapp M, Schuler G, Steinman RM, Bhardwaj N. 1996. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J Immunol Methods 196:121–135. doi: 10.1016/0022-1759(96)00079-8. [DOI] [PubMed] [Google Scholar]
- 38.Le Moine A, Flamand V, Demoor FX, Noel JC, Surquin M, Kiss R, Nahori MA, Pretolani M, Goldman M, Abramowicz D. 1999. Critical roles for IL-4, IL-5, and eosinophils in chronic skin allograft rejection. J Clin Invest 103:1659–1667. doi: 10.1172/JCI5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malmgaard L, Paludan SR, Mogensen SC, Ellermann-Eriksen S. 2000. Herpes simplex virus type 2 induces secretion of IL-12 by macrophages through a mechanism involving NF-κB. J Gen Virol 81:3011–3020. doi: 10.1099/0022-1317-81-12-3011. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Guan X, Tamura T, Ozato K, Ma X. 2004. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J Biol Chem 279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 41.Hein J, Schellenberg U, Bein G, Hackstein H. 2001. Quantification of murine IFN-γ mRNA and protein expression: impact of real-time kinetic RT-PCR using SYBR green I dye. Scand J Immunol 54:285–291. doi: 10.1046/j.1365-3083.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- 42.Park SW, Kim M, Kim M, D'Agati VD, Lee HT. 2011. Sphingosine kinase 1 protects against renal ischemia-reperfusion injury in mice by sphingosine-1-phosphate1 receptor activation. Kidney Int 80:1315–1327. doi: 10.1038/ki.2011.281. [DOI] [PubMed] [Google Scholar]
- 43.Soni C, Domeier PP, Wong EB, Shwetank, Khan TN, Elias MJ, Schell SL, Lukacher AE, Cooper TK, Rahman ZS. 2015. Distinct and synergistic roles of FcγRIIB deficiency and 129 strain-derived SLAM family proteins in the development of spontaneous germinal centers and autoimmunity. J Autoimmun 63:31–46. doi: 10.1016/j.jaut.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Pallas DC, Schley C, Mahoney M, Harlow E, Schaffhausen BS, Roberts TM. 1986. Polyomavirus small t antigen: overproduction in bacteria, purification, and utilization for monoclonal and polyclonal antibody production. J Virol 60:1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akondy RS, Johnson PL, Nakaya HI, Edupuganti S, Mulligan MJ, Lawson B, Miller JD, Pulendran B, Antia R, Ahmed R. 2015. Initial viral load determines the magnitude of the human CD8 T cell response to yellow fever vaccination. Proc Natl Acad Sci U S A 112:3050–3055. doi: 10.1073/pnas.1500475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wherry EJ, Blattman JN, Ahmed R. 2005. Low CD8 T-cell proliferative potential and high viral load limit the effectiveness of therapeutic vaccination. J Virol 79:8960–8968. doi: 10.1128/JVI.79.14.8960-8968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kemball CC, Pack CD, Guay HM, Li ZN, Steinhauer DA, Szomolanyi-Tsuda E, Lukacher AE. 2007. The antiviral CD8+ T cell response is differentially dependent on CD4+ T cell help over the course of persistent infection. J Immunol 179:1113–1121. doi: 10.4049/jimmunol.179.2.1113. [DOI] [PubMed] [Google Scholar]
- 49.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. 2000. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol 1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 50.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 51.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. 2007. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukacher AE, Moser JM, Hadley A, Altman JD. 1999. Visualization of polyoma virus-specific CD8+ T cells in vivo during infection and tumor rejection. J Immunol 163:3369–3378. [PubMed] [Google Scholar]
- 53.Freund R, Dubensky T, Bronson R, Sotnikov A, Carroll J, Benjamin T. 1992. Polyoma tumorigenesis in mice: evidence for dominant resistance and dominant susceptibility genes of the host. Virology 191:724–731. doi: 10.1016/0042-6822(92)90248-N. [DOI] [PubMed] [Google Scholar]
- 54.Curtsinger JM, Lins DC, Mescher MF. 2003. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J Exp Med 197:1141–1151. doi: 10.1084/jem.20021910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D, Mescher MF. 2005. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol 174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt CS, Mescher MF. 1999. Adjuvant effect of IL-12: conversion of peptide antigen administration from tolerizing to immunizing for CD8+ T cells in vivo. J Immunol 163:2561–2567. [PubMed] [Google Scholar]
- 57.Huber JP, Farrar JD. 2011. Regulation of effector and memory T-cell functions by type I interferon. Immunology 132:466–474. doi: 10.1111/j.1365-2567.2011.03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O'Shea JJ, Biron CA. 2002. Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science 297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 59.Luft T, Pang KC, Thomas E, Hertzog P, Hart DN, Trapani J, Cebon J. 1998. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol 161:1947–1953. [PubMed] [Google Scholar]
- 60.Marshall HD, Urban SL, Welsh RM. 2011. Virus-induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J Virol 85:5929–5939. doi: 10.1128/JVI.02516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berard M, Brandt K, Bulfone-Paus S, Tough DF. 2003. IL-15 promotes the survival of naive and memory phenotype CD8+ T cells. J Immunol 170:5018–5026. doi: 10.4049/jimmunol.170.10.5018. [DOI] [PubMed] [Google Scholar]
- 62.Mattei F, Schiavoni G, Belardelli F, Tough DF. 2001. IL-15 is expressed by dendritic cells in response to type I IFN, double-stranded RNA, or lipopolysaccharide and promotes dendritic cell activation. J Immunol 167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X, Sun S, Hwang I, Tough DF, Sprent J. 1998. Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity 8:591–599. doi: 10.1016/S1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 64.Martinet V, Tonon S, Torres D, Azouz A, Nguyen M, Kohler A, Flamand V, Mao CA, Klein WH, Leo O, Goriely S. 2015. Type I interferons regulate eomesodermin expression and the development of unconventional memory CD8+ T cells. Nat Commun 6:7089. doi: 10.1038/ncomms8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Honke N, Shaabani N, Merches K, Gassa A, Kraft A, Ehrhardt K, Haussinger D, Lohning M, Dittmer U, Hengel H, Recher M, Lang PA, Lang KS. 2016. Immunoactivation induced by chronic viral infection inhibits viral replication and drives immunosuppression through sustained IFN-I responses. Eur J Immunol 46:372–380. doi: 10.1002/eji.201545765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Odorizzi PM, Wherry EJ. 2013. Immunology. An interferon paradox. Science 340:155–156. doi: 10.1126/science.1237568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiesel M, Crouse J, Bedenikovic G, Sutherland A, Joller N, Oxenius A. 2012. Type-I IFN drives the differentiation of short-lived effector CD8+ T cells in vivo. Eur J Immunol 42:320–329. doi: 10.1002/eji.201142091. [DOI] [PubMed] [Google Scholar]
- 69.Lukacher AE, Ma Y, Carroll JP, Abromson-Leeman SR, Laning JC, Dorf ME, Benjamin TL. 1995. Susceptibility to tumors induced by polyoma virus is conferred by an endogenous mouse mammary tumor virus superantigen. J Exp Med 181:1683–1692. doi: 10.1084/jem.181.5.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stroh LJ, Maginnis MS, Blaum BS, Nelson CD, Neu U, Gee GV, O'Hara BA, Motamedi N, DiMaio D, Atwood WJ, Stehle T. 2015. The greater affinity of JC polyomavirus capsid for α2,6-linked lactoseries tetrasaccharide c than for other sialylated glycans is a major determinant of infectivity. J Virol 89:6364–6375. doi: 10.1128/JVI.00489-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray U, Cinque P, Gerevini S, Longo V, Lazzarin A, Schippling S, Martin R, Buck CB, Pastrana DV. 2015. JC polyomavirus mutants escape antibody-mediated neutralization. Sci Transl Med 7:306ra151. doi: 10.1126/scitranslmed.aab1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jelcic I, Combaluzier B, Jelcic I, Faigle W, Senn L, Reinhart BJ, Stroh L, Nitsch RM, Stehle T, Sospedra M, Grimm J, Martin R. 2015. Broadly neutralizing human monoclonal JC polyomavirus VP1-specific antibodies as candidate therapeutics for progressive multifocal leukoencephalopathy. Sci Transl Med 7:306ra150. doi: 10.1126/scitranslmed.aac8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khanna N, Wolbers M, Mueller NJ, Garzoni C, Du Pasquier RA, Fux CA, Vernazza P, Bernasconi E, Viscidi R, Battegay M, Hirsch HH, Swiss HIV Cohort Study. 2009. JC virus-specific immune responses in human immunodeficiency virus type 1 patients with progressive multifocal leukoencephalopathy. J Virol 83:4404–4411. doi: 10.1128/JVI.02657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frost EL, Kersh AE, Evavold BD, Lukacher AE. 2015. Cutting edge: resident memory CD8 T cells express high-affinity TCRs. J Immunol 195:3520–3524. doi: 10.4049/jimmunol.1501521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, Karp CL, Brooks DG. 2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]