Abstract
Thyroid cancer has the fastest rising incidence rates and is the fifth most common cancer in women. There are four main types of which the papillary and follicular types together account for >90%, followed by medullary cancers (3%−5%) and anaplastic carcinomas (<3%). For individuals who present with early stage disease of papillary and follicular cancers, there are no accurate markers to predict whether they will develop metastatic or recurrent disease. Our immediate goal is to molecularly differentiate follicular cancer subtypes for enhanced classification. Promoter methylation status of genes with reported associations in thyroid cancer (CASP8, CDKN2A, DAPK1, ESR1, NIS, RASSF1 and TIMP3) were examined in a cohort of follicular thyroid cancers comprising of 26 Hurthle and 27 Classic subtypes utilizing quantitative methylation-specific PCR. RASSF1 was differentially methylated in Classic tumor tissue compared to Hurthle (p<0.001). Methylation of RASSF1 pointed to racial group differences between African Americans and Caucasian Americans (p=0.05). Extra thyroidal extension was found to be associated with DAPK1 (p=0.014) and ESR1 (p=0.036) methylation. Late stage disease was associated with older age (p<0.001) and methylation of DAPK1 (p=0.034) and ESR1 (p=0.035). The methylation status of RASSF1, DAPK1 and ESR1 suggests the utility of methylation markers to molecularly differentiate thyroid cancer subtypes for enhanced classification and early detection of thyroid cancer.
Keywords: follicular thyroid cancer, DNA methylation, QMSP, Hurthle cell cancer
1. Introduction
Thyroid cancer (TC) is the most common endocrine malignancy, accounting for 95% of all endocrine malignancies (Boufraqech, Patel, Xiong, & Kebebew, 2013), and which has an increasing incidence for 1999-2008: going up by an estimated 6.2% per year for men and 7.3% per year for women (Simard, Ward, Siegel, & Jemal, 2012). Unfortunately, neoplasias of the thyroid have been largely ignored because of the overall favorable prognosis of papillary thyroid cancers (PTC) and follicular thyroid cancers (FTC), with 5-year survival rates of approximately 98% when submitted to timely and appropriate treatment (SEER Cancer Statistics Factsheets: Thyroid Cancer. 2014). Despite this favorable prognosis, the increasing incidence is of concern. Moreover, no significant progress has been made to improve survival. Those diagnosed at late stages have a devastating 5-year survival rate of under 60% (Weber & Eng, 2005). Also, the recurrence rate of thyroid cancer is high (30%) (Mazzaferri & Kloos, 2001), and only one third of patients with distant metastases respond to radio-iodine (131I) therapy with complete remission (Schlumberger, 1998). In addition, there is a lack of accurate preoperative markers or molecular-based predictive models to differentiate benign nodules (follicular adenomas [FA]) from follicular cancer. Thus, despite advances on scientific and clinical fronts, many advanced thyroid cancers remain incurable.
The majority of thyroid nodules are benign so their clinical importance is related to excluding thyroid cancer. Fine-needle aspiration (FNA) biopsy is the standard diagnostic tool for preoperative diagnosis (Segev, Clark, Zeiger, & Umbricht, 2003). However, it has some limitations, especially in the presence of follicular lesions. A single false-negative FNA can delay surgical treatment by 28 months even with clinical evidence suggesting malignancy. This may lead to higher rates of vascular and capsular invasion, persistent disease at follow up, and advanced stage disease making it incurable (Yeh, Demircan, Ituarte, & Clark, 2004). For individuals who present with early stage disease of either PTC or FTC, there is no accurate marker(s) to predict whether they will develop metastatic or recurrent disease.
New approaches are necessary to identify potential novel diagnostic and prognostic markers, which would allow for more accurate early diagnoses, with personalized clinical management and surveillance. The present challenge is to obtain reliable preoperative markers that differentiate benign and malignant thyroid nodules, to detect early thyroid cancer, to improve discrimination of adenoma, PTC, and FTC so as to delineate thyroid subtypes in order to improve patient management. In attempting to tackle these challenges, this study’s goal was to identify DNA methylation markers that molecularly differentiate FTC subtypes, specifically FTC-Classic and FTC-Hurthle, for enhanced classification which can eventually lead to early detection of FTC.
2. Method
2.1 Cohort
This retrospective study cohort of 53 follicular thyroid cancers (26 FTC-Hurthle and 27 FTC-Classic subtypes) comprised of 14 males and 39 females ranging in age from 18 to 86 years with a race distribution of 34 Caucasian American (CA), 14 African American (AA), and 5 unknown race (Table 1).
Table 1.
Cohort Characteristics
| Characteristics | FTC-Hurthle (N=26) | FTC-Classic (N=27) | Total (N=53) |
|---|---|---|---|
| Gender | |||
| Male | 7 | 7 | 14 |
| Female | 19 | 20 | 39 |
| Race | |||
| CA | 17 | 17 | 34 |
| AA | 6 | 8 | 14 |
| Unknown | 3 | 2 | 5 |
| Stage | |||
| 1 | 9 | 13 | 22 |
| 2 | 9 | 3 | 12 |
| 3 | 6 | 8 | 14 |
| 4 | 1 | 2 | 3 |
| Unknown | 1 | 1 | 2 |
FTC - Follicular thyroid cancer; CA - Caucasian American; AA - African American.
The cohort was drawn from a large, clinically well-characterized multiethnic (33% AA) primary care patient population in the Detroit, Michigan area. Patients 18 years or older diagnosed with a primary thyroid cancer in the Henry Ford Health System between 1983 and 2012 with available tumor tissue blocks were identified through tumor registry records. All tumors were staged using the American Joint Committee on Cancer (AJCC) TNM system (American Joint Committee on Cancer, 2010). Cases staged as 1, 2 and unknown stage (2 cases) were grouped as Early stage while stages 3 and 4 were grouped together as Late stage disease. Risk factor information of race (as self reported), age, gender, and marital status was obtained through medical record abstraction. Promoter methylation status of CASP8, CDKN2A, DAPK1, ESR1, NIS, RASSF1 and TIMP3, genes with reported associations in thyroid cancer, were examined utilizing quantitative methylation-specific PCR (QMSP). This study was approved by the Henry Ford Health System Institutional Review Board committee.
2.2 DNA Extraction
Whole 5 micron tissue sections or microdissected thyroid lesions and adjacent normal when present were processed for DNA extraction as previously described (Chen, Sawhney, Khan, Benninger, & Hou, 2007).
2.3 Bisulfite Modification and Quantitative Methylation-Specific Polymerase Chain Reaction (QMSP) Assay
Genomic DNA (~100 ng) from formalin-fixed paraffin embedded thyroid tissue and control universal methylated DNA (Chamicon International, Inc) were modified using the EZ-96 DNA Methylation Lightning Kit (Zymo Research, Orange, CA) during which methylated DNA is protected and unmethylated cytosine is converted to uracil (Chen et al., 2007).
QMSP detects the presence of tumor-specific DNA with a sensitivity of 1 cell in 1000 normal cells (Harden, Tokumaru, Westra, Goodman, & Ahrendt, 2003). This PCR approach is more sensitive than conventional PCR and more specific due to the use of an internally binding, fluorogenic hybridization probe (Eads, Danenberg, Kawakami, Saltz, & Blake, 2000; Lo, Wong, Zhang, Tein, & Ng, 1999). An advantage of QMSP is that it can measure the amount of methylation in a sample. Primers and probes (Table 2) were specifically designed to amplify CASP8, CDKN2A, DAPK1, ESR1, NIS, RASSF1 and TIMP3 genes. Primers and probes to an internal reference gene (ACTB) were run in parallel to standardize the input DNA.
Table 2.
QMSP Primers and Probes
| ACTB | |
| Forward Primer | TGGTGATGGAGGAGGTTTAGTAAGT |
| Reverse Primer | AACCAATAAAACCTACTCCTCCCTTAA |
| Probe | ACCACCACCCAACACACAATAACAAACACA |
| CDKN2A | |
| Forward Primer | TTATTAGAGGGTGGGGCGGATCGC |
| Reverse Primer | GACCCCGAACCGCGACCGTAA |
| Probe | AGTAGTATGGAGTCGGCGGCGGG |
| CASP8 | |
| Forward Primer | TAGGGGATTCGGAGATTGCGA |
| Reverse Primer | AAACCGTATATCTACATTCGAAACGA |
| Probe | CCCGCTCCACCCTTTCCTAACACCA |
| DAPK1 | |
| Forward Primer | GGATAGTCGGATCGAGTTAACGTC |
| Reverse Primer | CCCTCCCAAACGCCGA |
| Probe | TTCGGTAATTCGTAGCGGTAGGGTTTGG |
| ESR1 | |
| Forward Primer | GGCGTTCGTTTTGGGATTG |
| Reverse Primer | GCCGACACGCGAACTCTAA |
| Probe | CGATAAAACCGAACGACCCGACGA |
| NIS | |
| Forward Primer | ATAGGGAGGTCGATACGGATATC |
| Reverse Primer | GAAAAAACAAAACGAAAAAAACG |
| Probe | TAGACGGAGCGGGGATAGGTTGTCGAGT |
| RASSF1 | |
| Forward Primer | GCGTTGAAGTCGGGGTTC |
| Reverse Primer | CCCGTACTTCGCTAACTTTAAACG |
| Probe | ACAAACGCGAACCGAACGAAACCA |
| TIMP3 | |
| Forward Primer | GCGTCGGAGGTTAAGGTTGTT |
| Reverse Primer | CTCTCCAAAATTACCGTACGCG |
| Probe | AACTCGCTCGCCCGCCGAA |
PCR was carried out using the EpiTect MethyLight PCR Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol in 384-well plate using the 7900HT Sequence detector (Applied Biosystems). Each plate included multiple water blanks and serial dilutions of a universal methylated positive control DNA (100ng/μl, Chemicon, Temecula, CA) which was bisulfite converted in the lab for constructing the standard curve. These were run with the samples on each plate in duplicate. To determine the relative levels of methylated promoter DNA in each sample, the values of the gene of interest were compared with the values of the internal reference gene (ACTB) to obtain a ratio that is then multiplied by 100 to give a percentage value (gene of interest/reference gene X 100). The methylation results obtained by QMSP in this cohort are considered as a binary event in which any quantity of methylation in a sample is considered as positive (> 0.0) (Carvalho, Henrique, Jeronimo, Nayak, & Reddy, 2011).
2.4 Statistical Analysis
Comparisons were made between AA and CA, early and late stage disease and between patients with and without extra thyroidal extension findings, by t-test (for age), chi square tests (for categorical variables), and by Wilcoxon rank sum tests (for QMSP values). The distributions of QMSP values for the two groups were displayed using dot plots. In a few instances of patients who did not have values available for race, stage or extra thyroidal extension, comparisons were performed using all usable data for each comparison and the sample sizes available for each analysis were noted and reported.
3. Results
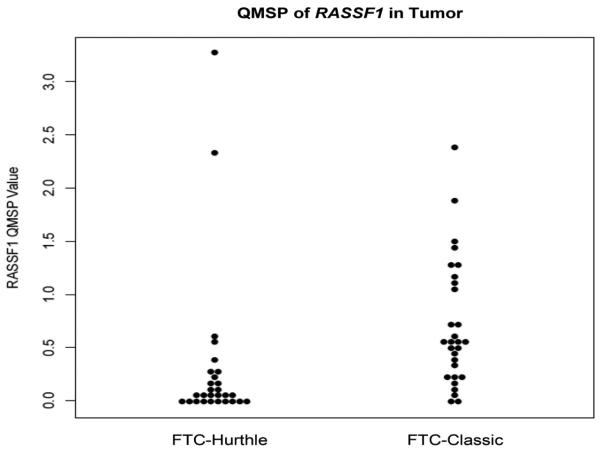
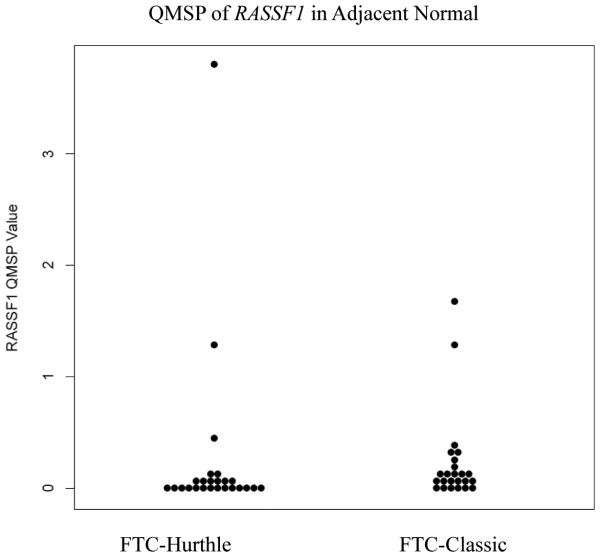
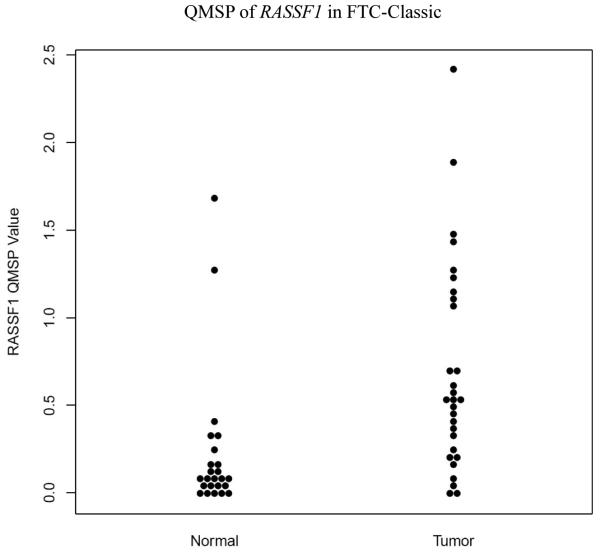
The retrospective cohort comprised of 26 FTC-Hurthle and 27 FTC-Classic thyroid cancer subtypes. Comparisons between FTC-Classic and FTC-Hurthle tumor tissue demonstrated significant differences in RASSF1 methylation levels (p<0.001, Table 3, Figure 1) with higher levels in the FTC-Classic tumor tissue. Comparisons between their adjacent normal tissues were also significant (p=0.01). Within each FTC subtype, the tumor tissue demonstrated higher methylation levels compared to the adjacent normal, but was significant for the FTC-Classic subtype (p<0.001, Table 4, Figures 2 and 3). DAPK1, ESR1, NIS and TIMP3 demonstrated methylation (QMSP values >0.0) but their levels were not statistically significant between subtypes. CASP8 and CDKN2A were not methylated in any samples (all QMSP values were 0). Comparisons between AA and CA, early and late stage disease and between patients with and without extra thyroidal extension findings for age, gender, other categorical variables (angiolymphatic invasion and capsular invasion), and QMSP values were performed (Table 3). RASSF1 methylation levels pointed to race group differences with higher mean methylation values in AA than CA (0.637 vs 0.422, p=0.05). Extra thyroidal extension is associated with DAPK1 (p=0.014) and ESR1 (p=0.036) methylation. Late stage disease is associated with older age (p<0.001, average age 66.7 years) and methylation of DAPK1 (p=0.034) and ESR1 (p=0.035). Angiolymphatic invasion, capsular invasion and gender (data not shown) did not show any significant association with gene methylation, age, race, stage or extra thyroidal extension.
Table 3.
Sample Sizes, Methylation Means and Standard Deviations by Race, Stage, Extra Thyroidal Extension and FTC type
| Race: AA versus CA | Variable | AA (N= 14) | CA (N= 34) | p-value |
|---|---|---|---|---|
| RASSF1 | 14 (0.637 ± 0.531) | 34 (0.422 ± 0.702) | 0.054 | |
| DAPK1 | 14 (0.000 ± 0.000) | 34 (0.001 ± 0.003) | 0.61 | |
| ESR1 | 14 (0.000 ± 0.001) | 34 (0.005 ± 0.015) | 0.575 | |
| Stage: Early vs Late | Early (N= 36) | Late (N= 15) | ||
| AGE | 36 (51.4 ± 13.0) | 15 (66.7 ± 13.6) | <.001 | |
| DAPK1 | 36 (0.000 ± 0.000) | 15 (0.001 ± 0.005) | 0.034 | |
| ESR1 | 36 (0.000 ± 0.001) | 15 (0.017 ± 0.038) | 0.035 | |
| RASSF1 | 36 (0.419 ± 0.511) | 15 (0.634 ± 0.777) | 0.877 | |
| Extra Thyroidal Extension | No Extension (N= 41) | Extension Present (N= 6) | ||
| DAPK1 | 41 (0.000 ± 0.000) | 6 (0.003 ± 0.007) | 0.014 | |
| ESR1 | 41 (0.002 ± 0.008) | 6 (0.036 ± 0.057) | 0.036 | |
| RASSF1 | 41 (0.470 ± 0.602) | 6 (0.703 ± 0.693) | 0.306 | |
| FTC Hurthle vs FTC Classic | FTC Hurthle (N= 26) | FTC Classic (N= 27) | ||
| RASSF1 | 26 (0.340 ± 0.751) | 27 (0.730 ± 0.605) | <.001 | |
| DAPK1 | 26 (0.001 ± 0.004) | 27 (0.000 ± 0.000) | 0.159 | |
| ESR1 | 26 (0.001 ± 0.002) | 27 (0.011 ± 0.030) | 0.154 |
AA - African American; CA - Caucasian American; Early Stage - Stages 1, 2, and Unknown; Late Stage - Stages 3 and 4; FTC - Follicular Thyroid Cancer
Figure 1.
QMSP of RASSF1 in Tumor (FTC-Hurthle vs FTC-Classic): Dot plot chart of QMSP values for FTC-Hurthle and FTC-Classic cases demonstrating differences in methylation levels of RASSF1 (p<0.001) between the two tumor groups. (FTC - follicular thyroid cancer)
Table 4.
Sample Sizes, Methylation Means, Standard Deviations and Wilcoxon rank sum p-values for RASSF1 in FTC-Hurthle and FTC-Classic lesions
| FTC-Hurthle | FTC-Classic | p-value | |
|---|---|---|---|
| Tumor Tissue | 28 (0.067 ± 0.728) | 29 (0.539 ± 0.596) | <0.001 |
| Adjacent Normal Tissue | 25 (0.022 ± 0.780) | 24 (0.084 ± 0.404) | 0.01 |
| Tumor vs Adjacent Normal p-value | 0.258 | <0.001 |
Figure 2.
QMSP of RASSF1 in Adjacent Normal (FTC-Hurthle vs FTC-Classic): Dot plot chart of QMSP values for FTC-Hurthle and FTC-Classic cases demonstrating differences in methylation levels of RASSF1 (p=0.01) between the adjacent normal lesions for the two groups. (FTC - follicular thyroid cancer)
Figure 3.
QMSP of RASSF1 in FTC-Classic (Tumor vs Adjacent Normal): Dot plot chart of QMSP values for FTC-Classic cases demonstrating differences in methylation levels of RASSF1 (p <0.001) between the adjacent normal and tumor lesions. (FTC - follicular thyroid cancer)
4. Discussion
Early detection of cancer before metastasis is important for patients and clinicians as it can improve prognosis, patient quality of life and provide additional treatment options. One of the best ways for early detection of cancer is through the use of biomarkers. Several immunohistochemical and molecular markers for thyroid tumorigenesis have been proposed and some have been validated on a large scale for use in routine practice.
Various molecular alterations (mutations and/or gene rearrangements) have been described in the development of thyroid cancers. Genetic rearrangements which result in activation of the RET proto oncogene was the first recognized molecular event found in PTC especially in those exposed to ionizing radiation (Eberhardt, 2003). Chromosomal rearrangement resulting in a fusion gene, PAX8/PPARγ, may be involved in follicular adenoma (FA) to FTC progression (Eberhardt, 2003). Currently, several types of gene mutation and expression panels are being tested for introduction into clinical practice. Mutational markers for FNA biopsy include BRAF and RAS point mutations and RET/PTC and PAX8/PPARg rearrangements, and possible TRK rearrangement (Nikiforov, Ohori, Hodak, Carty, & LeBeau, 2011; Nikiforov, Steward, Robinson-Smith, Haugen, & Klopper, 2009; Cantara, Capezzone, Marchisotta, Capuano, & Busonero, 2010). A limitation of genetic testing for molecular alterations is the low incidence of mutations, resulting in low sensitivity for diagnosing thyroid cancer in indeterminate nodules (Patel, Goyal, & Goldenberg, 2014). The Afirma Gene Expression Classifier, an mRNA based test utilizing FNA, analyzes the expression of 142 genes (167 RNA transcripts) using a proprietary algorithm to assign indeterminate thyroid nodules into either benign or suspicious groups (Chudova, Wilde, Wang, Wang, & Rabbee, 2010) and is currently commercially available. However, it is costly and does not discriminate amongst various thyroid lesions. A major limitation of the Afirma test is its low specificity (52%) in indeterminate nodules (Patel et al., 2014). The main concern for many endocrinologists is that it misclassifies lesions as suspicious about half the time.
Epigenetic alterations in the form of aberrant DNA methylation of tumor suppressor genes have been reported in many cancers. Methylation of DAPK1 and RASSF1 genes is known to strongly contribute to carcinogenesis, metastasis and treatment failure in various types of cancer (Wong, Chang, Tang, Wei, & Kwong, 2002; Supic, Kozomara, Brankovic-Magic, Jovic, & Magic, 2009). Hypermethylation of RASSF1, a known tumor suppressor gene, has been described in 75% of FTC, a smaller percentage of benign adenomas (44%), and PTC (20%) (Xing, Cohen, Mambo, Tallini, & Udelsman, 2004) indicating that this may be an early step in follicular cell derived thyroid tumorigenesis. In a pilot study of 21 thyroid cases by our group (Stephen, Chitale, Narra, Chen, & Sawhney, 2011), RASSF1, CASP8 and NIS were frequently methylated in normal thyroid samples, hyperthyroid nodules and in thyroid cancer cases (matched for presence of normal and tumor tissue from the same biopsy specimen), suggesting these as early changes in thyroid tumorigenesis regardless of cell type. In our current study, RASSF1 demonstrates significant differential methylation between FTC-Classic and FTC-Hurthle, with higher methylation levels in FTC-Classic, suggesting its utilization as a molecular marker to differentiate these two subtypes.
RASSF1 encodes a signaling protein that functions through a pathway involving RAS, a component of PI3K/Akt pathway. The main mechanism of RASSF1 inactivation appears to be through promoter methylation and loss of heterozygosity (LOH); mutational inactivation is very rare. It is thought that RASSF1 may belong to the class of haplo-insufficient tumor suppressor genes that promotes tumor formation through the inactivation of only one allele (Schagdarsurengin, Gimm, Hoang-Vu, Dralle, & Pfeifer, 2002). Genetic alterations in genes along pathways involving RASSF1’s tumor suppressor role are also a consideration. Schagdarsurengin et al. (2002) examined the methylation status of RASSF1 by methylation-specific PCR (MSP) in nine thyroid cancer cell lines and 38 primary thyroid tumors and found methylation in 70% of the FTC samples. The expression status of RASSF1 by RT-PCR demonstrated a correlation with RASSF1 hypermethylation and loss of transcription. Nakamura et al. (Nakamura, Carney, Jin, Kajita, & Pallares, 2005) also identified RASSF1 methylation in thyroid cancers (PTC, FTC, medullary and anaplastic thyroid cancers, and hyalinizing trabecular tumors) and in benign follicular adenomas (FA) utilizing MSP. They found concordance between methylation of RASSF1 and decreased protein expression in the thyroid tumors. Although 100% of their FTC samples demonstrated methylation of RASSF1, its methylation status was not used to differentiate the FTC subtypes as there were only 4 cases.
There have been several reports on RASSF1 promoter hypermethylation in thyroid tumors (Xing et al., 2004; Schagdarsurengin et al., 2002) including comparisons between FTCs and their corresponding normal tissues utilizing non-quantitative methods such as MSP. Lee et al. (Lee, Geli, Larsson, Wallin, & Karimi, 2008), investigating genome-wide methylation changes utilizing Long Interspersed Nucleotide Elements-1 (LINE-1) and LUminometric methylation assay (LUMA) in 21 FTCs, observed no changes between FTCs and their corresponding normal thyroid tissues. However, RASSF1 promoter methylation, determined by pyrosequencing and validated by MSP, was observed in 86% of their FTCs with 33% of its matched normals demonstrating methylation ≥10%. Our study is novel in that it compares the FTC subtypes, FTC-Classic and FTC-Hurthle, and their corresponding adjacent normal thyroid tissue utilizing quantitative MSP. We identified a significant difference in RASSF1 methylation levels between FTC-Classic and FTC-Hurthle tumor tissues (p<0.001) as well as between the adjacent normal tissues (p=0.01) of these two subtypes. This suggests that RASSF1 methylation may be used to distinguish these two FTC subtypes. When comparing RASSF1 methylation levels between the tumor tissue and adjacent normal within an FTC subtype in the current study, as expected, the tumor tissue demonstrated higher methylation levels. This was significantly different for the FTC-Classic subtype and its adjacent normal (p<0.001), but the FTC-Hurthle subtype and its adjacent normal showed no differences in methylation levels. A pilot study (Stephen et al., 2011) by our group which looked at various thyroid lesions (normal thyroid, hyperthyroid, FTC, FTC-Hurthle and PTC) found CASP8, RASSF1and NIS to be frequently methylated in normal thyroid, hyperthyroid lesions, thyroid cancer and their adjacent normal. Using larger cohorts, we plan further studies to investigate the differences in methylation status between normal thyroid tissue and FTC so as to identify methylation markers that distinguish normal thyroid tissue from FTC. In our study, significant differential methylation of RASSF1 between FTC-Classic and FTC-Hurthle tumor suggests that DNA methylation markers may be useful in discriminating among thyroid cancer subtypes.
DAPK1, ESR1, NIS and TIMP3 also demonstrated methylation (>0.0) in our FTC samples but were not statistically significant between the Hurthle and Classic subtypes. Death-associated protein kinase 1, DAPK1, which is located at 9q34.1 is a positive mediator of interferon γ-induced programmed cell death in HeLa cells (Cohen, Feinstein, & Kimchi, 1997). DAPK1 methylation frequently occurs in head and neck cancers (Worsham, Chen, Meduri, Nygren, & Errami, 2006), non-small cell lung carcinoma (Esteller, Sanchez-Cespedes, Rosell, Sidransky, & Baylin, 1999), gastric and colorectal carcinomas (Lee, Leung, Chan, Ng, & Tong, 2002; Satoh, Toyota, Itoh, Kikuchi, & Obata, 2002). In HNSCC, DAPK1 methylation is associated with metastasis to lymph nodes and advanced stage disease (Sanchez-Cespedes, Esteller, Wu, Nawroz-Danish, & Yoo, 2000). In this study, we found an association of late stage disease with DAPK1 methylation (p=0.034) in addition to an association with extra thyroidal extension (p=0.014) suggesting that DAPK1 methylation maybe a marker of advanced disease and metastasis in follicular thyroid cancers.
ESR1 is located at 6q25.1 and is involved in hormone binding, DNA binding, and activation of transcription (Ponglikitmongkol, Green, & Chambon, 1988). ESR1 has metastasis-suppressor properties, especially in breast cancer, suggesting a tumor-suppressor role (Issa, Ottaviano, Celano, Hamilton, & Davidson, 1994). Thus, methylation mediated silencing of ESR1 can lead to spread of a tumor. A previous study by our group on laryngeal squamous cell carcinomas identified aberrant methylation of ESR1 as an independent predictor of late stage diagnosis (Stephen, Chen, Shah, Havard, & Kapke, 2010). In the current follicular thyroid cancer study, extra thyroidal extension (p=0.036) and late stage disease (p=0.035) are associated with ESR1 methylation suggesting loss of its tumor-suppressor role. ESR1 may also be involved in age-dependent increase in cancer incidence as age-dependent methylation has been detected in colon mucosa (Issa et al., 1994), cardiovascular system (Post, Goldschmidt-Clermont, Wilhide, Heldman, & Sussman, 1999), ulcerative colitis (Issa, Ahuja, Toyota, Bronner, & Brentnall, 2001), and prostate cancer. Comparisons of angiolymphatic invasion, capsular invasion and gender were also performed with respect to QMSP values, age, race, stage and extra thyroidal extension. However, they were not significantly associated with methylation of any genes or other variables.
Our long term goal is to identify methylation markers with utility in FNAs to differentiate benign thyroid, adenomas and cancer subtypes so as to determine course of management (surgery vs no surgery). Although FNA is the standard diagnostic tool for preoperative diagnosis (Segev et al., 2003), it has limitations in the presence of follicular lesions. The FTC-Hurthle variant is more easily diagnosed on FNA histology due to the presence of Hurthle cells compared to other FTC subtypes, but a definitive diagnosis of FTC, in general, requires the presence of capsular and vascular invasion which cannot be appreciated on an FNA (Ruggeri, Campenni, Baldari, Trimarchi, & Trovato, 2008). In addition, the FNA is less sensitive in detecting classic FTC and Hurthle cell carcinomas when compared to PTC (Yeh et al., 2004). Recently, a 21 gene candidate panel (inclusive of the 7 genes in this study) was evaluated for presence of methylation in matched post-surgical FNA and corresponding fresh thyroid tissue from 2 cases using QMSP. We found that 6 genes, including RASSF1, showed concordant presence of methylation results (Table 5). The latter suggests support for methylation markers as an adjunct to histopathology diagnoses of FNAs.
Table 5.
Methylation in Matched Post-Surgical FNA and Thyroid Tissue
| Genes | Thy-FNA1 | Thy-T1 | Thy-FNA2 | Thy-T2 |
|---|---|---|---|---|
| NIS | 0.028122 | 0.02179 | 0 | 0 |
| RASSF1 | 0.253875 | 0.24554 | 0.02155 | 0.03766 |
| TSHR | 0.095524 | 0.20984 | 0.0844 | 0.1315 |
| SERPINB5 | 0.500365 | 0.58745 | 1.36001 | 0.59044 |
| SLC26A4 | 0.119488 | 0.03552 | 0.00523 | 18.7248 |
| TPO | 0.595375 | 0.65587 | 1.09508 | 0.89846 |
FNA – fine needle aspiration; T – matched fresh thyroid tumor.
The incidence rates of thyroid cancer are almost twice as high in Caucasian Americans (CA) as in African Americans (AA) (SEER, 2014). This study was able to draw from a sizable pool of primary care patients serviced by Health Alliance Plan (HAP) and benefits from HFHS’ unique strength of a large pool of African American patients who make up about 33% of the total patient population. Of the 48 cases with self-reported race as AA and CA, over half were CA (34 vs 14), reflective of the racial incidence disparity reported for this cancer. The observed significant differential methylation of RASSF1 (p=0.05) in AA vs CA suggests epigenetic influences, offering opportunities for further study to examine the role of DNA methylation in thyroid cancer racial disparities.
5. Conclusion
Together, in this study, RASSF1, DAPK1 and ESR1 suggest utility of methylation markers to molecularly differentiate follicular thyroid cancer subtypes for enhanced classification. Classification based on promoter methylation profiling may have preferred utility over expression profiling since such DNA-based markers are less subject to problems of tissue preservation, potential pitfalls of tissue heterogeneity, and are easily detected by PCR-based methods. Aberrant DNA methylation profiles that differentiate benign and malignant thyroid nodules may help identify potential novel diagnostic and prognostic markers, which would allow accurate early diagnosis, with personalized clinical management and surveillance. The potential reversibility of DNA methylation holds promise for these markers as potential targets for novel alternative demethylating treatments.
Acknowledgments
Dr. Stephen had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. This study was supported by NIH R03 CA159426.
Footnotes
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- American Joint Committee on Cancer . AJCC Cancer Staging Manual. 7th. XV. Springer; 2010. p. 1. 1158/1078-0432.CCR-11-0324. [Google Scholar]
- Boufraqech M, Patel D, Xiong Y, Kebebew E. Diagnosis of thyroid cancer: state of art. Expert Opin Med Diagn. 2013;7(4):331–342. doi: 10.1517/17530059.2013.800481. doi:10.1517/17530059.2013.800481 http://dx.doi.org/10.1517/17530059.2013.800481. [DOI] [PubMed] [Google Scholar]
- Cantara S, Capezzone M, Marchisotta S, Capuano S, Busonero G, Toti P, Pacini F. Impact of proto-oncogene mutation detection in cytological specimens from thyroid nodules improves the diagnostic accuracy of cytology. J Clin Endocrinol Metab. 2010;95(3):1365–1369. doi: 10.1210/jc.2009-2103. doi:jc.2009-2103 [pii] 10.1210/jc.2009-2103 http://dx.doi.org/10.1210/jc.2009-2103. [DOI] [PubMed] [Google Scholar]
- Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, Califano JA. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res. 2011;17(14):4782–4789. doi: 10.1158/1078-0432.CCR-11-0324. doi:1078-0432.CCR-11-0324 [pii] http://dx.doi.org/10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, Sethi S, Worsham MJ. Methylation of multiple genes as diagnostic and therapeutic markers in primary Head and Neck Squamous Cell Carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133(11):1131–1138. doi: 10.1001/archotol.133.11.1131. http://dx.doi.org/10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- Chudova D, Wilde JI, Wang ET, Wang H, Rabbee N, Egidio CM, Kennedy GC. Molecular classification of thyroid nodules using high-dimensionality genomic data. J Clin Endocrinol Metab. 2010;95(12):5296–5304. doi: 10.1210/jc.2010-1087. doi:jc.2010-1087 [pii] 10.1210/jc.2010-1087 http://dx.doi.org/10.1210/jc.2010-1087. [DOI] [PubMed] [Google Scholar]
- Cohen O, Feinstein E, Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. Embo J. 1997;16(5):998–1008. doi: 10.1093/emboj/16.5.998. http://dx.doi.org/10.1093/emboj/16.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. doi:gnd033 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt N. Biomarkers in Thyroid Neoplasia; American Thyroid Association 75th Annual Meeting; Palm Beach, FL: Sep, 2003. pp. 16–21. 2003. [Google Scholar]
- Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59(1):67–70. [PubMed] [Google Scholar]
- Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9(4):1370–1375. [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. http://dx.doi.org/10.1038/ng0894-536 PMid:7951326. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- Lee JJ, Geli J, Larsson C, Wallin G, Karimi M, Zedenius J, Foukakis T. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. International journal of oncology. 2008;33(4):861–869. [PubMed] [Google Scholar]
- Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, To KF. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002;8(6):1761–1766. [PubMed] [Google Scholar]
- Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, Huang DP. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61(10):3877–3881. [PubMed] [Google Scholar]
- Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res. 1999;59(16):3899–3903. [PubMed] [Google Scholar]
- Mazzaferri EL, Kloos RT. Clinical review 128: Current approaches to primary therapy for papillary and follicular thyroid cancer. J Clin Endocrinol Metab. 2001;86(4):1447–1463. doi: 10.1210/jcem.86.4.7407. http://dx.doi.org/10.1210/jcem.86.4.7407. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Carney JA, Jin L, Kajita S, Pallares J, Zhang H, Lloyd RV. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Laboratory investigation; a journal of technical methods and pathology. 2005;85(9):1065–1075. doi: 10.1038/labinvest.3700306. doi:10.1038/labinvest.3700306 http://dx.doi.org/10.1038/labinvest.3700306. [DOI] [PubMed] [Google Scholar]
- Nikiforov YE, Steward DL, Robinson-Smith TM, Haugen BR, Klopper JP, Zhu Z, Nikiforova MN. Molecular testing for mutations in improving the fine-needle aspiration diagnosis of thyroid nodules. J Clin Endocrinol Metab. 2009;94(6):2092–2098. doi: 10.1210/jc.2009-0247. doi:jc.2009-0247 [pii] 10.1210/jc.2009-0247 http://dx.doi.org/10.1210/jc.2009-0247. [DOI] [PubMed] [Google Scholar]
- Nikiforov YE, Ohori NP, Hodak SP, Carty SE, LeBeau SO, Ferris RL, Nikiforova MN. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 2011;96(11):3390–3397. doi: 10.1210/jc.2011-1469. doi:jc.2011-1469 [pii] 10.1210/jc.2011-1469 http://dx.doi.org/10.1210/jc.2011-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HH, Goyal N, Goldenberg D. Imaging, genetic testing, and biomarker assessment of follicular cell-derived thyroid cancer. Annals of medicine. 2014;46(6):409–416. doi: 10.3109/07853890.2014.923739. http://dx.doi.org/10.3109/07853890.2014.923739. [DOI] [PubMed] [Google Scholar]
- Ponglikitmongkol M, Green S, Chambon P. Genomic organization of the human oestrogen receptor gene. Embo J. 1988;7(11):3385–3388. doi: 10.1002/j.1460-2075.1988.tb03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–991. doi: 10.1016/s0008-6363(99)00153-4. http://dx.doi.org/10.1016/S0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- Ruggeri RM, Campenni A, Baldari S, Trimarchi F, Trovato M. What is New on Thyroid Cancer Biomarkers. Biomark Insights. 2008;3:237–252. doi: 10.4137/bmi.s669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60(4):892–895. [PubMed] [Google Scholar]
- Satoh A, Toyota M, Itoh F, Kikuchi T, Obata T, Sasaki Y, Imai K. DNA methylation and histone deacetylation associated with silencing DAP kinase gene expression in colorectal and gastric cancers. Br J Cancer. 2002;86(11):1817–1823. doi: 10.1038/sj.bjc.6600319. http://dx.doi.org/10.1038/sj.bjc.6600319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagdarsurengin U, Gimm O, Hoang-Vu C, Dralle H, Pfeifer GP, Dammann R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002;62(13):3698–3701. [PubMed] [Google Scholar]
- Schlumberger MJ. Papillary and follicular thyroid carcinoma. N Engl J Med. 1998;338(5):297–306. doi: 10.1056/NEJM199801293380506. http://dx.doi.org/10.1056/NEJM199801293380506. [DOI] [PubMed] [Google Scholar]
- SEER Cancer Statistics Factsheets: Thyroid Cancer National Cancer Institute. 2014 http://seer.cancer.gov/statfacts/html/thyro.html. Accessed April 30 2014.
- Segev DL, Clark DP, Zeiger MA, Umbricht C. Beyond the suspicious thyroid fine needle aspirate. Acta Cytol. 2003;47(5):709–722. doi: 10.1159/000326594. A review. http://dx.doi.org/10.1159/000326594. [DOI] [PubMed] [Google Scholar]
- Simard EP, Ward EM, Siegel R, Jemal A. Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J Clin. 2012 doi: 10.3322/caac.20141. doi:10.3322/caac.20141 http://dx.doi.org/10.3322/caac.20141. [DOI] [PubMed]
- Stephen JK, Chen KM, Shah V, Havard S, Kapke A, Lu M, Worsham MJ. DNA hypermethylation markers of poor outcome in laryngeal cancer. Clin Epigenetics. 2010;1(1-2):61–69. doi: 10.1007/s13148-010-0005-3. doi:10.1007/s13148-010-0005-3 http://dx.doi.org/10.1007/s13148-010-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen JK, Chitale D, Narra V, Chen KM, Sawhney R, Worsham MJ. DNA methylation in thyroid tumorigenesis. Cancers (Basel) 2011;3(2):1732–1743. doi: 10.3390/cancers3021732. doi:10.3390/cancers3021732 http://dx.doi.org/10.3390/cancers3021732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supic G, Kozomara R, Brankovic-Magic M, Jovic N, Magic Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol. 2009;45(12):1051–1057. doi: 10.1016/j.oraloncology.2009.07.007. doi:10.1016/j.oraloncology.2009.07.007 http://dx.doi.org/10.1016/j.oraloncology.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Weber F, Eng C. Gene-expression profiling in differentiated thyroid cancer--a viable strategy for the practice of genomic medicine? Future Oncol. 2005;1(4):497–510. doi: 10.2217/14796694.1.4.497. http://dx.doi.org/10.2217/14796694.1.4.497 PMid:16556026 [DOI] [PubMed] [Google Scholar]
- Wong TS, Chang HW, Tang KC, Wei WI, Kwong DL, Sham JS, Kwong YL. High frequency of promoter hypermethylation of the death-associated protein-kinase gene in nasopharyngeal carcinoma and its detection in the peripheral blood of patients. Clin Cancer Res. 2002;8(2):433–437. [PubMed] [Google Scholar]
- Worsham MJ, Chen KM, Meduri V, Nygren AO, Errami A, Schouten JP, Benninger MS. Epigenetic events of disease progression in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2006;132(6):668–677. doi: 10.1001/archotol.132.6.668. http://dx.doi.org/10.1001/archotol.132.6.668 PMid:16785414. [DOI] [PubMed] [Google Scholar]
- Xing M, Cohen Y, Mambo E, Tallini G, Udelsman R, Ladenson PW, Sidransky D. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64(5):1664–1668. doi: 10.1158/0008-5472.can-03-3242. http://dx.doi.org/10.1158/0008-5472.CAN-03-3242 PMid:14996725. [DOI] [PubMed] [Google Scholar]
- Yeh MW, Demircan O, Ituarte P, Clark OH. False-negative fine-needle aspiration cytology results delay treatment and adversely affect outcome in patients with thyroid carcinoma. Thyroid. 2004;14(3):207–215. doi: 10.1089/105072504773297885. http://dx.doi.org/10.1089/105072504773297885. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94(2):212–217. doi: 10.1002/ijc.1466. http://dx.doi.org/10.1002/ijc.1466. [DOI] [PubMed] [Google Scholar]