Abstract
Manganese superoxide dismutase (MnSOD) is a mitochondrially localized primary antioxidant enzyme, known to be essential for the survival of aerobic life and to play important roles in tumorigenesis. Here, we show that MnSOD deficiency in skin tissues of MnSOD-heterozygous knockout (Sod2+/−) mice leads to increased expresson of uncoupling proteins (UCPs). When MnSOD is deficient, superoxide radical and its resulting reactive oxygen species (ROS) activate ligand binding to peroxisome proliferator-activated receptor alpha (PPARα), suggesting that the activation of PPARα signaling is a major mechanism underlying MnSOD-dependent UCPs expression that consequently triggers the PI3K/Akt/mTOR pathway, leading to increased aerobic glycolysis. Knockdown of UCPs and mTOR suppresses lactate production and increases ATP levels, suggesting that UCPs contribute to increased glycolysis. These results highlight the existence of a free radical-mediated mechanism that activates mitochondria uncoupling to reduce ROS production, which precedes the glycolytic adaptation described as the Warburg Effect.
Keywords: MnSOD, UCPs, Redox homeostasis, Energetic metabolism, glycolysis
INTRODUCTION
MnSOD, an important primary antioxidant enzyme located in mitochondria, is essential for the removal of superoxide radicals constantly being generated by the electron transport chain (ETC). Abnormalities in MnSOD function are involved in the pathogenesis of many diseases, including cancer.1 Homozygous knockout Sod2−/− mice are small at birth and eventually develop severe pathology in heart and brain, leading to early death.2 Heterozygous knockout Sod2+/− mice display a variety of clinically silent mitochondrial abnormalities underlying mitochondrial stress, including reduced complex I and II activities, increased permeability transition,3 ultrastructural abnormalities, and enhanced lipid peroxidation.4 In contrast, Sod2 transgenic mice decrease lipid peroxidation,5 reduce ROS generated in mitochondria,6 and protect against apoptotic cell death.7
Careful regulation of MnSOD is critical for maintenance of cellular ROS homeostasis and bioenergetic balance.8 A large body of evidence has demonstrated that Sod2 gene expression is deregulated in cancers.9 Unlike what occurs in normal cells, the level of MnSOD oscillates during cancer development, which is downregulated in early-stage tumor cells but is upregulated in late-stage tumor cells.10,11,12 However, the mechanisms by which MnSOD deficiency leads to cancer development are still not fully elucidated.
Normal differentiated cells rely on mitochondrial oxidative phosphorylation (mtOXPHOS) to produce ATP using several types of bioenergetic molecules as fuel: glucose, fatty acids and glutamine. Cancer cells, however, exhibit increased uptake of these molecules through aerobic glycolysis.13 The altered metabolism in cancers occurs through oncogenic transformation of multiple signaling pathways, including PI3K-Akt and Myc, which allows cancer cells to constitutively take up and metabolize cellular bioenergetic fuels in excess of their biosynthetic capacity.14,15,16
During mitochondrial respiration, ETC activity creates a proton gradient across the inner mitochondrial membrane, which is essential for coupling oxygen consumption to ATP production. Uncoupling proteins (UCPs) are inner mitochondrial membrane proteins sustaining an inducible proton conductance, which dissipates proton gradient potential and accelerates metabolism for production of heat instead of ATP.17 UCP1, present in brown adipocytes in mammals, sustains a free fatty acid-induced and purine nucleotide-inhibited proton conductance, which is required for adaptive thermogenesis.18 UCP2 and UCP3, two UCP1 homologs, share the same function of causing proton leakage and uncoupling mitochondrial respiration, though they are more widely distributed in bodily tissues.19 As a consequence of the energy dissipation they facilitate, an agreed role of UCPs could be described as being protection against ROS generation by decreasing mitochondrial respiration.20
In the present study, we investigated the mechanistic basis underlying alteration of bioenergetic metabolism under MnSOD-deficient conditions. We found that adaptive induction of UCP1 and UCP2 in the skin of heterozygous Sod2+/− knockout mice was closely correlated to PPARα-mediated transcriptional activation. Our results suggest that UCPs are important intermediaries in the cellular bioenergetic switch from mtOXPHOS to the glycolytic pathway.
RESULTS
Adaptive increases of p53 and UCPs in skin tissues of MnSOD-deficient mice
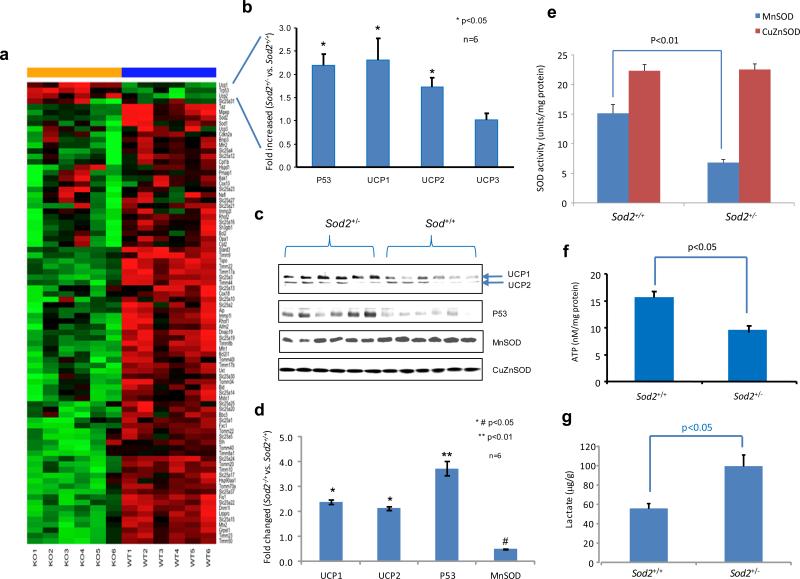
Our previous studies have demonstrated that MnSOD deficiency plays a causal role in mouse skin carcinogenesis when p53 is activated and translocated into mitochondria.10,21 To examine how mitochondrial function changes when MnSOD is deficient, we determined the expression profiles of genes coding for mitochondrial proteins in the skin of Sod2+/− and Sod2+/+ mice using a mouse mitochondrial qRT-PCR array (Figure 1a-b). The results show reduced expression levels of these genes in Sod2+/− mice as compared to Sod2+/+ mice, suggesting that MnSOD is critical for maintenance of mitochondrial function. However, in contrast to most mitochondrial proteins, the levels of p53 and UCPs (UCP1 and UCP2) increased in the skin cells of Sod2+/− mice, which was also confirmed by Western blots (Figure 1c-d). The reduced MnSOD activity was confirmed by a SOD enzyme assay (Figure 1e). Quantifications of ATP and lactate productions indicated that ATP level was significantly lower, but lactate level was increased more in Sod2+/− skin tissues than in Sod2+/+ (Figure 1f-g). The presence of elevated UCPs in MnSOD-deficient mice predicts that decreased proton gradient through uncoupling leads to reduced ETC-driven ATP production and increased glycolysis.
Figure 1.
Adaptive increased expression of p53 and UCPs in the skin of MnSOD heterozygous knockout (Sod2+/−) and wild-type (Sod2+/+) mice. (a) The skin tissues from 6 mice for each genotype were analyzed using a mouse mitochondrial qRT-PCR array. mRNA signatures are presented by a heatmap in which green indicates low expression levels and red indicates high expression levels. (b) Expression levels of p53 and UCP1/UCP2/UCP3 in the Sod2+/− mice relative to their levels in the Sod2+/+ mice. (c) The levels of these proteins in mouse skin tissues were assayed by Western blots. (d) Relative levels of proteins in the Western blots were plotted. (e) MnSOD and CuZnSOD activities were quantified. (f, g) ATP and lactate productions in the skin tissues were measured.
Decline in mitochondrial function in MnSOD-deficient cells
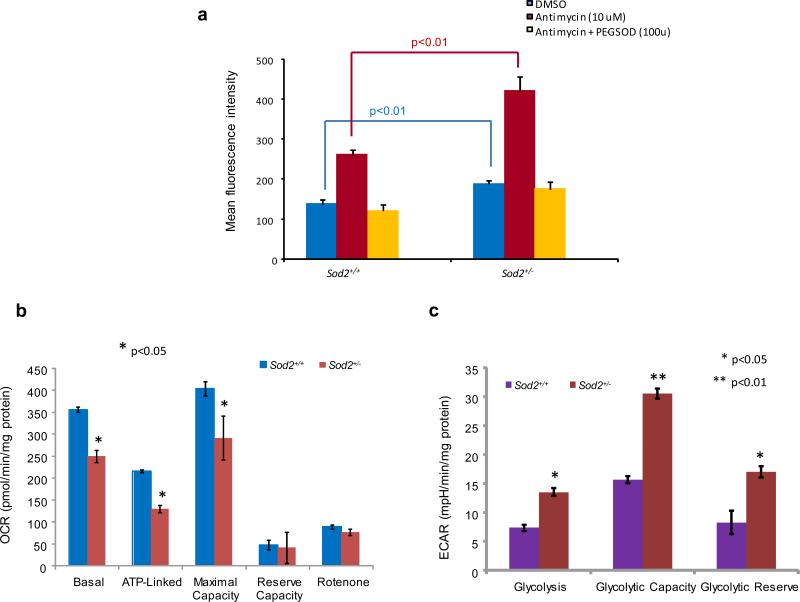
A mouse embryonic fibroblast primary cell culture system was established to further elucidate the effect of MnSOD deficiency on mitochondrial respiration. The levels of MnSOD, UCP1 and UCP2 were characterized in the primary cell cultures of Sod2+/− and Sod2+/+ mice (Supplementary Figure S1a). The levels of ATP and lactate in the primary cells from Sod2+/− mice were significantly lower than in the cells from Sod2+/+ mice (Supplementary Figure S1b-c). To determine if MnSOD deficiency led to accumulating superoxide radicals in primary cell cultures, superoxide anion was measured using dihydroethidium (DHE), a fluorescent cholesterol analog (Figure 2a). The levels of superoxide radicals in Sod2+/− cells were consistently and significantly higher than the levels in Sod2+/+ cells. Antimycin A, an inhibitor of ETC complex III, is able to inhibit mtOXPHOS, leading to generation of high levels of superoxide radicals. In turn, the radicals can be removed by adding PEG-SOD. To examine cellular bioenergetic metabolism by mtOXPHOS or aerobic glycolysis, the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) were analyzed using a Seahorse Bioscience FX OxygenFlux Analyzer. As shown in Figure 2b-c, basal, ATP-linked OCR and maximal capacity of oxygen utilization in Sod2+/− cells were significantly lower compared to Sod2+/+ cells, but no differences were observed in reserve capacity. When rotenone, an inhibitor of the NADH-coenzyme Q reductase (complex I) of ETC, was used to fully inhibit mitochondrial respiration, the OCR level fell below the basal OCR. In contrast, the ECAR showed that basal glycolysis, glycolytic capacity and glycolytic reserve in Sod2+/− cells were significantly higher than in Sod2+/+ cells. These results suggest that cellular bioenergetic metabolism apparently shifts from mtOXPHOS to a glycolytic pathway in MnSOD-deficient cells.
Figure 2.
Diminished mitochondrial function in the primary embryonic fibroblast cells from the MnSOD-deficient mice. (a) Superoxide radicals produced in the cells were analyzed using a DHE fluorescence probe. Antimycin was added to induce cellular superoxide radicals and PEG-SOD was used to catalyze the produced superoxide radicals. (b) Mitochondrial respiration was analyzed by quantification of the oxygen consumption rate (OCR). Rotenone was used to fully inhibit oxygen consumption. (c) Cellular glycolysis was analyzed by estimating the extracellular acidification rate. 2-Deoxy-D-glucose was added to inhibit the cellular glycolytic pathway.
UCP1 and UCP2 regulate bioenergetics adaptation
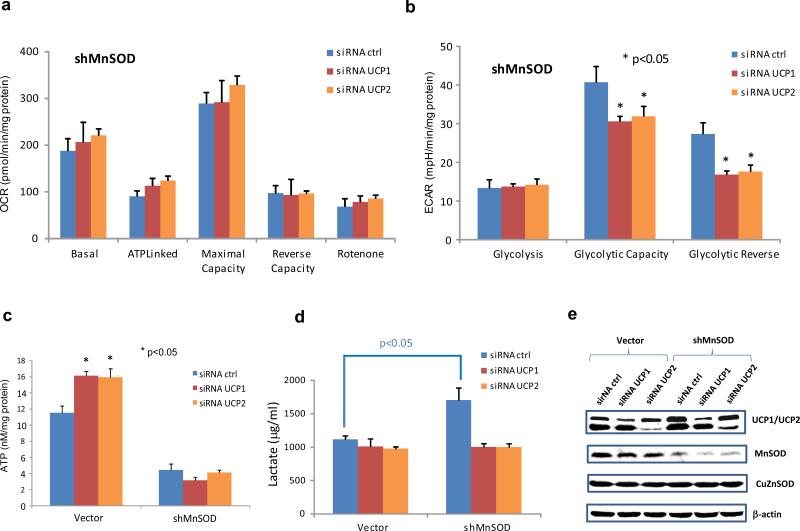
MnSOD deficiency led to increased levels of UCP1 and UCP2, which suggests that MnSOD regulates bioenergetics adaptation through modulation of UCPs. Previously, we generated a MnSOD-deficient HaCaT human keratinocyte cell line and established that Nox4-mediated EGFR activation is responsive to UV-inducible carcinogenesis.22 To directly test whether UCP1 and UCP2 affect cellular bioenergetic metabolism, UCP1 and UCP2 in the MnSOD-knockdown HaCaT cells were silenced by lentivirally expressed shRNAs. Silencing single UCP1 or UCP2 in HaCaT cells with a normal level of MnSOD did not alter OCR or ECAR of the cells (data not shown). ECAR in the MnSOD-knockdown HaCaT cells was reduced by either UCP1 silence or UCP2 silence, but no change in OCR was observed in the cells (Figure 3a-b). In addition, ATP production in the HaCaT cells increased when UCP1 and UCP2 were silenced (Figure 3c). However, the ATP levels did not change significantly in the MnSOD-knockdown HaCaT cells regardless of the UCP level, indicating that downregulation of UCP1 or UCP2 led to improvement of mitochondrial ATP production under the normal level of MnSOD. To probe the effects of UCP1 and UCP2 on the glycolytic pathway, cellular lactate concentrations were measured. As shown in Figure 3d, silencing UCP1 or UCP2 resulted in significant decreases in lactate production in the MnSOD-knockdown cells. Silencing UCP1 or UCP2 in the HaCaT cells was confirmed by Western blots (Figure 3e). These results suggest that UCP1 and UCP2 may serve as important bioenergetics regulators coordinating mtOXPHOS and glycolysis to maintain cellular ATP levels.
Figure 3.
The effects of UCP1 and UCP2 on glycolysis. (a, b) MnSOD-knockdown human keratinocyte HaCaT cells (shMnSOD) were transfected with siRNA to silence either UCP1 or UCP2 and alterations in OCR and ECAR were quantified. (c) ATP production was analyzed. (d) Cellular lactate concentration was measured. (e) Corresponding proteins levels were confirmed.
MnSOD deficiency leads to increased PPARα-mediated transcriptional activation of UCPs
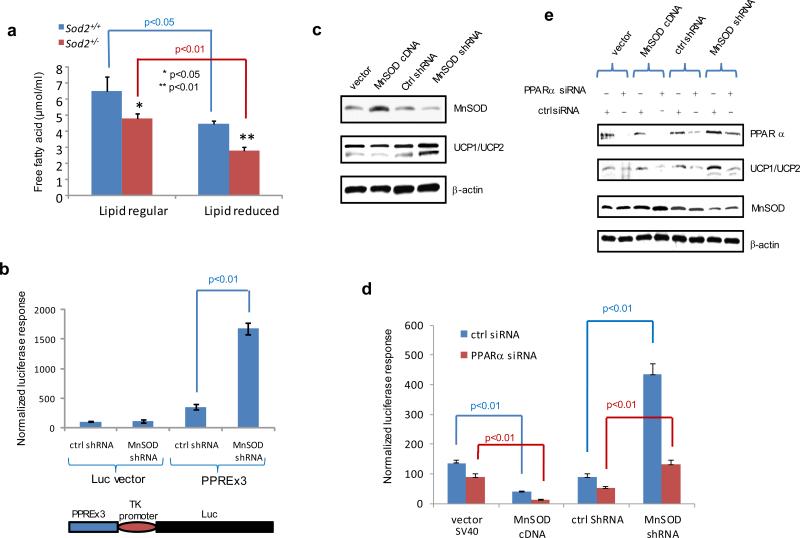
To examine the effects of MnSOD on metabolism, a metabolomics analysis of skin tissues from Sod2+/− and Sod2+/+ mice was conducted by Metabolomics Fiehn Laboratory. As shown in Supplementary Table S2, compared to Sod2+/+ control, Sod2+/− appears to predominantly use lipids as a main bioenergetic resource, and long-chain fatty acids were significantly reduced in Sod2+/− mice. Lipid concentrations in primary cell cultures from Sod2+/− and Sod2+/+ mice were measured to confirm the results from the metabolomics analysis (Figure 4a). Consistently, the level of total cellular lipids in the Sod2+/− cells was decreased compared to the Sod2+/+ cells, indicating that MnSOD deficiency leads to lipid utilization.
Figure 4.
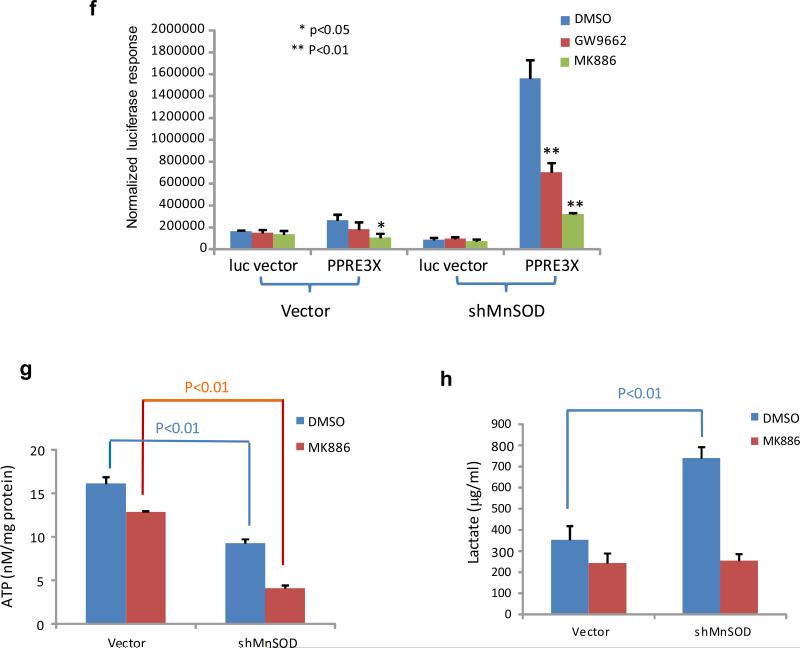
Free fatty acid-activated PPARα-mediated transcription of the UCP1 gene under MnSOD-deficient conditions. (a) The Sod2+/− and Sod2+/+ primary cells were cultured in DMEM and lipid-reduced DMEM media, and the concentrations of free fatty acids in the cells were measured. (b) PPAR elements located in the UCP1 promoter region were linked to the TK basal promoter to drive the luciferase reporter gene (bottom). The reporter construct was transiently transfected into mouse skin JB6 cells, and induction of the reporter gene was carried out in the presence or absence of MnSOD silencing. Reporter activity was normalized to cotransfected β-galactosidase activity. The empty pGL3 vector was also transfected as a control. (c) The JB6 cells were stably transfected with MnSOD cDNA or injected with MnSOD lentiviral shRNA. The levels of MnSOD and related UCP1/UCP2 proteins were measured. (d) The luciferase reporter construct was cotransfected with PPARα siRNA into the JB6 cells with different levels of MnSOD. A vector containing a SV40 promoter alone was used as a control. The luciferase reporter responses were analyzed. (e) The corresponding proteins in transfected JB6 cells were measured to confirm the reporter responses. (f) The reporter construct was transfected into JB6 cells, and then treated with PPARα inhibitor MK886 or PPARγ inhibitor GW9662; the normalized reporter responses were estimated. (g,h) After MnSOD-silenced JB6 cells (shMnSOD) were treated with MK886, the productions of ATP and lactate were measured.
Transcription of UCPs is regulated by the PPAR family23,24 and endogenous ligands for transcription factor PPARs include free fatty acids.25 Within the PPAR family, PPARα is most responsive to massive oxidative stress conditions.26 Upon ligand binding, PPARs:retinoid X receptor heterodimers stimulate UCP1 transcriptional activation.27 To verify the effect of MnSOD deficiency on PPARα-mediated transcriptional activation for upregulation of UCP1, a luciferase reporter, driven by the UCP1 enhancer region containing 3 copies of PPAR elements (PPREx3) linked to the TK basal promoter, was transfected into murine skin epidermal JB6 cells, in which MnSOD had been previously silenced using MnSOD siRNA. The reporter activity in the MnSOD-silenced cells was significantly higher than in the control cells without MnSOD silencing (Figure 4b). To further confirm that the level of MnSOD affects UCP expression, MnSOD expression in JB6 cells was manipulated by either transfecting the cells with MnSOD cDNA or infecting the cells with a lentivirus carrying MnSOD shRNA. The levels of MnSOD, UCP1 and UCP2 were quantified by Western blots in stably transfected cells. Reverse correlation between MnSOD and UCP1/UCP2 was verified as shown in Figure 4c. Subsequently, the PPREx3-driven luciferase reporter construct was transfected into cells with different levels of MnSOD with and without PPARα silencing. As shown in Figure 4d, the reporter activities were decreased in the MnSOD-overexpressed cells but increased in the MnSOD-silenced cells. Importantly, the reporter activities were decreased in all cell lines in which PPARα was silenced. Moreover, the results from Western blots confirm that the expression of UCP1 and UCP2 was inversely related to the cellular MnSOD levels and was further decreased by silencing PPARα in the cells (Figure 4e).
To further verify that PPARα is a major molecular mechanism by which MnSOD deficiency upregulates UCPs, the specific PPARα inhibitor MK886 was used to suppress PPARα activation. As shown in Figure 4f, the PPREx3-driven luciferase reporter activity was significantly reduced by MK886 as compared to PPARγ inhibitor GW9662 and no inhibitor treatment control. Furthermore, MK886 decreased lactate and ATP productions in the MnSOD-deficient JB6 cells (Figure 4g-h).
In addition to activation of mitochondrial uncoupling, another main energy resource for cell metabolism is the breaking down of fatty acids by β-oxidation in mitochondria. Thus, a decrease in fatty acids should promote cells to uptake glucose. Total RNA from primary cells isolated from Sod2+/+ and Sod2+/− mice cultured in both regular and lipid-reduced media was subjected to qRT-PCR using gene-specific primer sets for GAPDH and LDH-A (Supplementary Figure S3 a-b). GAPDH and LDH-A mRNA were increased slightly more in Sod2+/− cells cultured in the regular medium than in Sod2+/+ cells, but significantly increased in the cells cultured in the lipid-reduced media. The result is further confirmed by Western blots (Supplementary Figure S3c), suggesting that decreased lipids in the media leads cells, particularly Sod2+/− cells, to use glucose through the glycolytic pathway.
MnSOD deficiency activates the PI3K/Akt/mTOR pathway involved in adaptive glycolysis
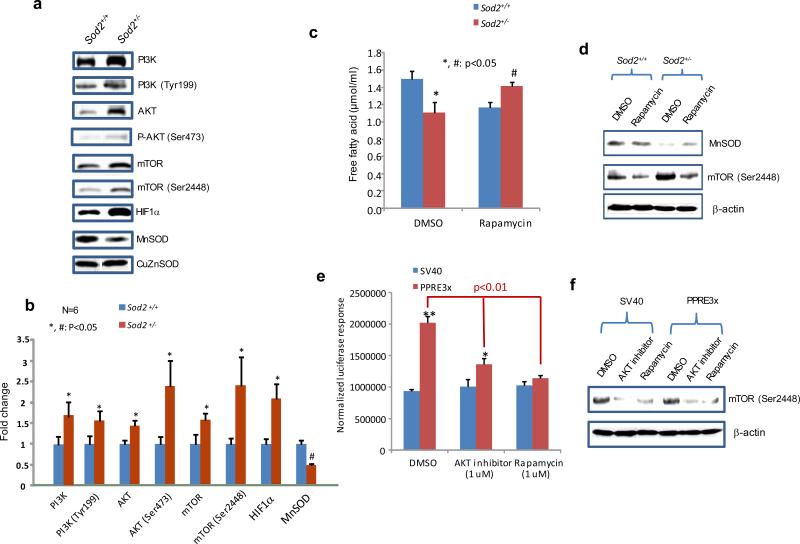
Impaired MnSOD expression and subsequent high levels of ROS have been reported to play a role in the development of neoplastic phenotypes.28 To determine how MnSOD deficiency affects the signaling pathways involved in energy production and carcinogenesis, skins from Sod2+/+ and Sod2+/− mice were analyzed for PI3K/Akt/mTOR signaling. As shown in Figure 5a, the levels of total protein and phosphorylated protein were remarkably higher in the Sod2+/− tissues than in the Sod2+/+ tissues. In addition, HIF1α, a glycolysis-associated protein, also increased in the Sod2+/− tissues. These results are reproducible in skin tissues obtained from multiple mice; the data were quantified and are plotted in Figure 5b. To verify whether the activation of mTOR links to MnSOD-mediated PPAR activation, primary fibroblast cells were treated with rapamycin, an mTOR inhibitor. The levels of free fatty acids in the Sod2+/+ cells were decreased when mTOR was inhibited but were significantly increased in the Sod2+/− cells (Figure 5c). Phosphorylated mTOR levels were increased in the Sod2+/− cells compared to the Sod2+/+ cells, but were eliminated after the cells were treated with rapamycin (Figure 5d). Furthermore, when murine JB6 skin cells were transfected with the PPREx3-driven luciferase reporter constructs and then treated with an Akt inhibitor and rapamycin, the reporter gene expression was suppressed by both the Akt inhibitor and rapamycin, but no change was observed in a control vector which lacked the PPAR enhancer (Figure 5e). Phosphorylation of mTOR was eliminated by treatment with either Akt inhibitor or rapamycin (Figure 5f).
Figure 5.
Adaptive activation of PI3K/Akt/mTOR signaling pathway in MnSOD-deficient conditions. (a) PI3K/Akt/mTOR proteins and their phosphorylated forms in the skin of Sod2+/− and Sod2+/+ mice were detected using specific antibodies. (b) After normalization to CuZnSOD, the average of images from 6 mice was plotted. (c) The mouse Sod2+/− and Sod2+/+ primary cell cultures were treated with rapamycin, an inhibitor of mTOR, and free fatty acids in the cells were measured. (d) The levels of MnSOD and phosphorylated mTOR in the cells were measured. (e) The reporter constructs containing PPAR elements or SV40 promoter alone were transiently transfected into mouse skin JB6 cells followed by treatment with an Akt inhibitor or rapamycin. The reporter responses were analyzed as described in Fig.4. (f) Inhibition of mTOR phosphorylation by the Akt inhibitor or rapamycin was confirmed.
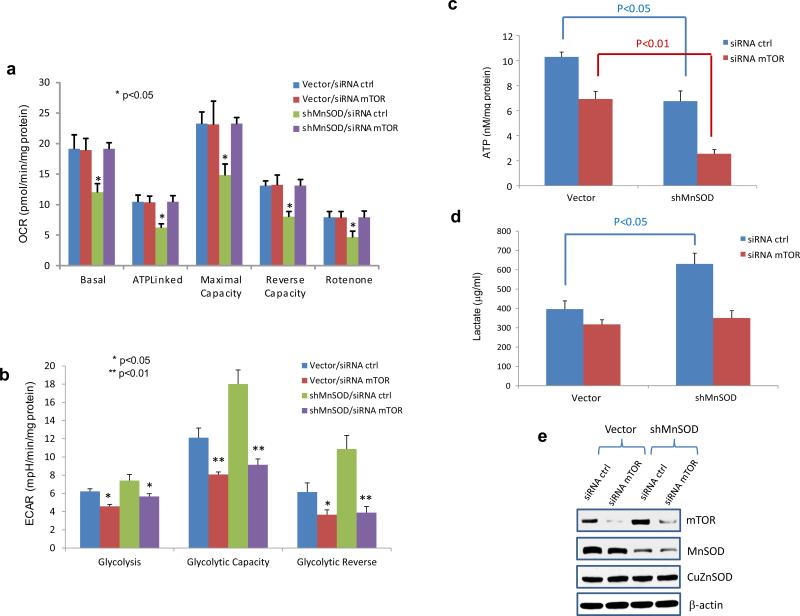
To further confirm that MnSOD deficiency mediated upregulation of UCPs alters cellule metabolism by triggering the mTOR signaling pathway, mTOR was subsequently silenced in MnSOD-knockdown JB6 cells. As shown in Figure 6a-b, silencing mTOR resulted in improvement of oxygen consumption and reduction of cellular glycolysis in the MnSOD-deficient cells. Furthermore, silencing mTOR subsequently reduced lactate and ATP productions, in particular in the MnSOD-deficient cells (Figure 6c-d). The levels of mTOR and related proteins in the cells were confirmed by Western blotting (Figure 6e). These results suggest that mTOR is linked to the glycolytic adaptation activated by MnSOD deficiency.
Figure 6.
Silencing mTOR attenuates altered metabolism in MnSOD-deficient cells (shMnSOD). MnSOD-knockdown JB6 cells and control cells were transfected with mTOR siRNA to further silence mTOR. (a) OCR, (b) ECAR, (c) ATP and (d) lactate were quantified. (e) The reduced protein levels of MnSOD and mTOR were confirmed.
DISCUSSION
The correlation between MnSOD and UCPs
Redox homeostasis is thought to regulate many cellular processes that are essential for maintenance of normal physiological conditions but are aberrantly modulated in cancers.29,30 Our data indicate that MnSOD deficiency leads to UCP expression, resulting in uncoupling mitochondrial respiration from ATP production and reduction of superoxide levels. These results are consistent with previous studies that demonstrated that mitochondrial uncoupling permits reentry of protons into the mitochondrial matrix, thereby preventing superoxide generation,31 and that superoxide-induced activation of UCPs serves as a negative feedback mechanism that limits further superoxide production.20 Thus, MnSOD activity modulates UCP levels.
However, Silva et al. previously reported that the levels of UCPs and mitochondrial ATP production rate were not altered in isolated mitochondria from Sod2 transgenic mice despite lowered superoxide levels, which indicates that a high level of MnSOD beyond the physiological level is not needed to further suppress UCPs.32 In the present study, we analyzed heterozygous knockout Sod2+/− mice using a mouse mitochondrial reverse transcriptional qPCR array. Our results clearly show that UCP1 and UCP2 are remarkably increased compared to the samples from Sod2+/+ control mice, despite the fact that most of the mitochondrial components related to mtOXPHOS were decreased in Sod2+/− mice (Figure 1). Data obtained from embryonic fibroblasts isolated from MnSOD-deficient mice and mouse skin keratinocytes further confirm an increase in superoxide radical/anion levels and subsequent increase in UCP transcription. Together, these results suggest that superoxide radical/anion concentrations lower than normal physiological conditions are not needed for the suppression of UCPs but that increased superoxide radical/anion under conditions of MnSOD deficiency can activate UCP induction. This study is the first to demonstrate a role for superoxide radical/anion in the expression of UCPs and it provides proof-of-concept data demonstrating that activation of UCPs results in reduction of excessive superoxide radicals produced under MnSOD-deficient conditions.
The molecular mechanism for UCPs induction
Eukaryotic cells have developed sophisticated strategies to combat myriad cellular metabolic stresses. Under stress, the main priority of cells is to conserve energy and divert the rest for survival. For instance, to generate heat to survive under conditions of severe cold, brown adipocytes accelerate the metabolism of cellular fatty acids through β-oxidation for mitochondrial respiration and then activate UCPs to uncouple ATP synthesis from mtOXPHOS. Free fatty acids not only fuel cells for thermogenesis in response to environmental stresses such as cold temperatures but also function as ligands for PPAR to transcriptionally activate UCP1.17,33 Our study demonstrates that MnSOD deficiency leads to increased utilization of free fatty acids to promote the UCP1/UCP2-activated bioenergetic shift from mtOXPHOS to glycolysis. PPARα binding to the UCP1 promoter region is essential for transcriptional activation under MnSOD-deficient conditions, indicating that PPAR-mediated UCP activation is an important mechanism for cellular responses to both oxidative and environmental stresses.
In contrast to UCP1, UCP2 and UCP3 are expressed in various tissues.19 Due to their homology with UCP1, UCP2 and UCP3 are also thermogenic and are involved in regulation of energy expenditure and body weight. In addition, the UCP1 homologues also appear to attenuate mitochondrial production of free radicals and protect against oxidative damage.34 It has been reported that PPARα expression is activated under glucose deprivation and massive oxidative stress conditions26,35 and that PPARα regulates UCP2 gene expression in the presence of free fatty acids.36 In the present study, we detected both UCP1 and UCP2 proteins in mouse skin cells, but not UCP3. UCP1 and UCP2 are regulated similarly by PPARα, suggesting that they share the same function.
The roles of MnSOD and UCPs in tumorigenesis
Due to the pivotal roles of UCPs in the regulation of energy efficiency and oxidative stress, UCPs are postulated to be critical in cancer development.37,38 UCP2, a unique homologue of the UCP family, is highly expressed in several types of cancer,37,39,40 which lowers mitochondrial potential by inducing proton conductance, resulting in limited superoxide generation but also inhibition of mtOXPHOS-mediated ATP synthesis.41 Under this condition, the effects of glycolysis on bioenergetics and biosynthesis are increased without cytotoxicity from high levels of mitochondrial ROS.42,43 Paradoxically, the cancer cells have increased ROS levels despite their high levels of UCP2. Accordingly, it has been shown that UCP2 functions as a metabolic sensor rather than as an uncoupler. Increased UCP2 expression not only leads to increased fatty acid oxidation, but it also exports pyruvate from mitochondria to the cytosol and limits glycolytic-derived pyruvate utilization.44,45,46 Recently, Vozza et al. reported that UCP2 functions as a metabolite transporter that limits oxidation of acetyl-CoA-producing substrates by removing C4 metabolites (TCA intermediates) from mitochondria.47 The present study demonstrates that increased UCPs in MnSOD-deficient cells lead to increased production of cellular energy via aerobic glycolysis, suggesting that adaptive activation of UCPs mediates the cellular energetic metabolism shift from mtOXPHOS to the glycolytic pathway.
Cancer cells have increased ROS levels, which are considered instrumental to tumor initiation and progression. ROS not only cause DNA damage and mutagenesis but also act as the secondary messenger to stimulate proliferation or inhibit apoptosis, conferring a growth advantage to established cancer cells. Since MnSOD is an important antioxidant enzyme that scavenges mitochondrial ROS, Sod2 has been characterized as a tumor suppressor gene.9,28 Previously, we demonstrated that removing ROS by overexpression of the Sod2 gene efficiently inhibits neoplastic transformation and DMBA-TPA-induced mouse skin carcinoma.10,12 The present study elucidates that silencing MnSOD leads to an increase in ROS generation and activation of glycolysis-derived energy production, which is mediated by the activation of several glycolytic/oncogenic biomarkers including HIF1α, PI3K, Akt and mTOR.
Altered metabolism is a hallmark of cancer cells. Otto Warburg discovered that cancer cells predominantly produce energy by aerobic glycolysis rather than by mtOXPHOS. Originally, it was thought that the Warburg Effect was caused by a mitochondrial defect, but accumulating data demonstrate that damaged mitochondria do not explain aerobic glycolysis in tumor cells. Indeed, most tumor cells have intact mitochondria that are able to carry out OXPHOS. Currently, reprogramming of cellular energetic metabolism directed by oncogenic signaling networks is considered to be the cause of the Warburg Effect.48 Our study reveals that MnSOD deficiency leads to mitochondrial uncoupling and subsequently activates the PI3K/Akt/mTOR that is essential for oncogenic signaling via altering bioenergetic metabolism, suggesting that redox homeostasis regulators such as MnSOD can modulate oncogenesis by promoting the transcription of uncoupling proteins that is a precursor to glycolytic metabolism. Our results predict that antioxidant-deficient mitochondria may serve as an early cellular signaling mechanism that contributes to the Warburg Effect.
MATERIALS AND METHODS
Mouse skin tissues and embryonic fibroblast primary cell cultures
Heterozygous MnSOD knockdown mice (Sod2+/−) were generated and genotyped as described previously.2,49 The animal experimental procedures used in this study were approved by the Institutional Animal Care and Use Committee of the University of Kentucky. Mouse skin tissues were prepared as described previously10. For mouse embryonic fibroblast primary cell cultures, fetuses were harvested from uteruses 13 days after pregnancy. The embryo head, liver and blood clots were removed and tissues were washed several times with 1 ml of PBS. The tissues were minced and incubated in 2 ml of 0.25% trypsin-EDTA for 10 min and vigorously pipetted for preparation of signal cell suspension. The cells were cultured in Dulbecco's modified Eagle's medium with standard fetal bovine serum (FBS) or lipid reduced (0.1 μm sterile-filtered) FBS (Gemini Bio-products, Sacramento, CA, USA). The cultures were split every 3-4 days during the early passages (p1-p6).
Quantification analyses of superoxide generation, oxygen consumption rate (OCR), extracellular acidification rate (ECAR) and ATP production
Dihydroethidium (Life Technologies, Grand Island, NY, USA) was used to quantify the levels of superoxide as described previously.50 XF extracellular flux assays (Seahorse Bioscience, North Billerica, MA, USA) were performed to simultaneously measure mitochondrial respiration (i.e., OCR) and glycolysis (i.e., ECAR) according to the manufacturer's protocols. An ATP determination kit (Life Technologies) was used to measure cellular levels of ATP by following procedures provided by the manufacturer.
Measurements of cellular free acids and lactate concentrations
Sod2+/+ and Sod2+/− mice skin tissues and embryonic fibroblast primary culture cells were harvested and 106 cells were homogenized in 200 μl of 1% (w/v) Triton X-100 in chloroform solution. The samples were centrifuged at 13,000 g for 10 min to remove insoluble phases. Organic phases were collected and chloroform was removed by vacuum dry for 30 min. The dried lipids were dissolved in 200 μl of fatty acid assay buffer and quantified using a Free Fatty Acid Quantitation Kit (Sigma, St. Louis, MO, USA) according to the manufacturer's protocols. A L-Lactate Assay Kit (Biomedical Research Service Center, SUNY at Buffalo, NY, USA) was used to measure lactate concentrations in the culture media and mouse skin tissues according to a protocol provided.
Mouse skin cell transfection
Murine epidermal JB6 and human keratinocyte HaCaT cells were cultured in Eagle's minimal essential medium containing 10% FBS. The cells were transfected with a PPRE3X-luciferase reporter construct (a kind gift from Dr. Ronald M. Evans, Howard Hughes Medical Institute, San Diego, CA, USA) using a Lipofectamine 2000 (Life Technologies). The luciferase activity was quantified using a luciferase assay kit (Promega, Madison, WI, USA) and normalized by β-galactosidase activity that was measured using chlorophenol red-β-D-galactopyranoside monosodium (Roche Molecular Biochemicals, Indianapolis, IN, USA). A lentiviral vector carrying MnSOD shRNA (Open Biosystems, Pittsburgh, PA, USA) was used to silence MnSOD in JB6 cells, and stable clones were generated by puromycin selection. UCP1, UCP2, PPARα, mTOR siRNAs (Dharmcon, Pittsburgh, PA, USA) were transiently transfected into HaCaT and JB6 cells to silence the expression of relating proteins using the manufacturer's cell transfection reagent. To inhibit PPAR-mediated UCP activation, the JB6 cells were treated with 10 μM PPARα inhibitor MK886 or 10 μM PPARγ inhibitor GW9662 (Santa Cruz Biotechnology, Inc., CA, USA). To inhibit the Akt-mTOR pathway, the JB6 cells were treated with either 1 μM Akt inhibitor (Selleckchem, Burlington, NC, USA) or 1 μM rapamycin (Selleckchem).
qRT-PCR
Total RNA was isolated from homogenized cells and mouse skin tissues using a Total RNA Isolation Kit (Life Technologies). A reverse transcription reaction was performed using a TaqMan Reverse Transcription Reagents kit (Applied Biosystems, Branchburg, NJ, USA). RT-PCR was performed with gene-specific primer-probe sets using a LightCycler 480 Real-Time PCR System (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer's protocols. A mouse mitochondrial qRT-PCR array (SABiosciences, Frederick, MD, USA) was used to detect gene expression profiles associated with mitochondrial function.
Western blot analysis
Homogenized cells and mouse skin tissues were electrophoresed on an 8% (w/v) SDS-PAGE gel, transferred onto a nitrocellulose membrane, and subsequently incubated with primary antibodies against UCP1, UCP2, p53, PPARα (Santa Cruz Biotechnology), MnSOD (Upstate Biotechnology, Inc., Lake Placid, NY, USA), CuZnSOD (eBioscience, Inc., San Diego, CA, USA), HIF1α (BD Biosciences, San Jose, CA, USA), and β-actin (Sigma-Aldrich Co., St. Louis, MO, USA); antibodies for signal pathways including PI3K/PI3K (Tyr199), Akt/Akt (Ser473) and mTOR/mTOR (Ser2448) (Cell Signaling Technology, Inc., Danvers, MA, USA). All secondary antibodies were obtained from Santa Cruz Biotechnology, Inc. Immunoblots were visualized using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech., Inc., Piscataway, NJ, USA).
Statistical data analyses
Multiple independent experiments were performed for each set of data presented. Images in Western blots were quantified using Carestream Molecular Imaging software (Carestream Health, Inc., Rochester, NY, USA). Statistical significance of data in the bar graphs was analyzed using one-way ANOVA and Tukey's Multiple Comparison Test, followed by data analysis with Graphpad Prism.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants CA 049797 and CA 143428, and an Edward P. Evans Foundation grant, training grant number T32 DK 007778 to Daret K. St. Clair; CA 143428 to William H. St. Clair; and National Nature Science Foundation of China grant 81372199 to Yong Xu. We would like to thank Dr. Ronald M. Evans, Howard Hughes Medical Institute, San Diego, California 92186, USA, for providing the PPREx3-TK-luciferase construct for this study.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- 2.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 3.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, et al. Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol. 2001;281:H1422–1432. doi: 10.1152/ajpheart.2001.281.3.H1422. [DOI] [PubMed] [Google Scholar]
- 4.Keller JN, Kindy MS, Holtsberg FW, St Clair DK, Yen HC, Germeyer A, et al. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagano Y, Matsui H, Shimokawa O, Hirayama A, Tamura M, Nakamura Y, et al. Rebamipide attenuates nonsteroidal anti-inflammatory drugs (NSAID) induced lipid peroxidation by the manganese superoxide dismutase (MnSOD) overexpression in gastrointestinal epithelial cells. J Physiol Pharmacol. 2012;63:137–142. [PubMed] [Google Scholar]
- 6.Kiningham KK, Oberley TD, Lin S, Mattingly CA, St Clair DK. Overexpression of manganese superoxide dismutase protects against mitochondrial-initiated poly(ADP-ribose) polymerase-mediated cell death. FASEB J. 1999;13:1601–1610. doi: 10.1096/fasebj.13.12.1601. [DOI] [PubMed] [Google Scholar]
- 7.Ilizarov AM, Koo HC, Kazzaz JA, Mantell LL, Li Y, Bhapat R, et al. Overexpression of manganese superoxide dismutase protects lung epithelial cells against oxidant injury. Am J Respir Cell Mol Biol. 2001;24:436–441. doi: 10.1165/ajrcmb.24.4.4240. [DOI] [PubMed] [Google Scholar]
- 8.Miriyala S, Holley AK, St Clair DK. Mitochondrial superoxide dismutase--signals of distinction. Anticancer Agents Med Chem. 2011;11:181–190. doi: 10.2174/187152011795255920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holley AK, Dhar SK, St Clair DK. Curbing cancer's sweet tooth: is there a role for MnSOD in regulation of the Warburg effect? Mitochondrion. 2013;13:170–188. doi: 10.1016/j.mito.2012.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Xue Y, Oberley TD, Kiningham KK, Lin SM, Yen HC, et al. Overexpression of manganese superoxide dismutase suppresses tumor formation by modulation of activator protein-1 signaling in a multistage skin carcinogenesis model. Cancer Res. 2001;61:6082–6088. [PubMed] [Google Scholar]
- 11.Oberley TD, Xue Y, Zhao Y, Kiningham K, Szweda LI, St Clair DK. In situ reduction of oxidative damage, increased cell turnover, and delay of mitochondrial injury by overexpression of manganese superoxide dismutase in a multistage skin carcinogenesis model. Antioxid Redox Signal. 2004;6:537–548. doi: 10.1089/152308604773934297. [DOI] [PubMed] [Google Scholar]
- 12.Dhar SK, Tangpong J, Chaiswing L, Oberley TD, St Clair DK. Manganese superoxide dismutase is a p53-regulated gene that switches cancers between early and advanced stages. Cancer Res. 2011;71:6684–6695. doi: 10.1158/0008-5472.CAN-11-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira LM. Cancer metabolism: the Warburg effect today. Exp Mol Pathol. 2010;89:372–380. doi: 10.1016/j.yexmp.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Wang J, Greisinger AJ, Grossman HB, Forman MR, Dinney CP, et al. Energy balance, the PI3K-AKT-mTOR pathway genes, and the risk of bladder cancer. Cancer Prev Res. 2010;3:505–517. doi: 10.1158/1940-6207.CAPR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 16.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarmuszkiewicz W, Woyda-Ploszczyca A, Antos-Krzeminska N, Sluse FE. Mitochondrial uncoupling proteins in unicellular eukaryotes. Biochim Biophys Acta. 2010;1797:792–799. doi: 10.1016/j.bbabio.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Klingenberg M. Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends Biochem Sci. 1990;15:108–112. doi: 10.1016/0968-0004(90)90194-g. [DOI] [PubMed] [Google Scholar]
- 19.Porter RK. Mitochondrial proton leak: a role for uncoupling proteins 2 and 3? Biochim Biophys Acta. 2001;1504:120–127. doi: 10.1016/s0005-2728(00)00246-2. [DOI] [PubMed] [Google Scholar]
- 20.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, et al. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 22.Holley AK, Xu Y, Noel T, Bakthavatchalu V, Batinic-Haberle I, St Clair DK. Manganese superoxide dismutase-mediated inside-out signaling in HaCaT human keratinocytes and SKH-1 mouse skin. Antioxid Redox Signal. 2014;20:2347–2360. doi: 10.1089/ars.2013.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 1996;16:3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubert J, Champigny O, Saint-Marc P, Negrel R, Collins S, Ricquier D, et al. Up-regulation of UCP-2 gene expression by PPAR agonists in preadipose and adipose cells. Biochem Biophys Res Commun. 1997;238:606–611. doi: 10.1006/bbrc.1997.7348. [DOI] [PubMed] [Google Scholar]
- 25.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen S, Cashman K, Thompson JG, Pantaleon M, Kaye PL. Glucose deprivation, oxidative stress and peroxisome proliferator-activated receptor-alpha (PPARA) cause peroxisome proliferation in preimplantation mouse embryos. Reproduction. 2009;138:493–505. doi: 10.1530/REP-09-0038. [DOI] [PubMed] [Google Scholar]
- 27.Teruel T, Hernandez R, Benito M, Lorenzo M. Rosiglitazone and retinoic acid induce uncoupling protein-1 (UCP-1) in a p38 mitogen-activated protein kinase-dependent manner in fetal primary brown adipocytes. J Biol Chem. 2003;278:263–269. doi: 10.1074/jbc.M207200200. [DOI] [PubMed] [Google Scholar]
- 28.Oberley LW, Oberley TD. Role of antioxidant enzymes in cell immortalization and transformation. Mol Cell Biochem. 1988;84:147–153. doi: 10.1007/BF00421049. [DOI] [PubMed] [Google Scholar]
- 29.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boss O, Hagen T, Lowell BB. Uncoupling proteins 2 and 3: potential regulators of mitochondrial energy metabolism. Diabetes. 2000;49:143–156. doi: 10.2337/diabetes.49.2.143. [DOI] [PubMed] [Google Scholar]
- 32.Silva JP, Shabalina IG, Dufour E, Petrovic N, Backlund EC, Hultenby K, et al. SOD2 overexpression: enhanced mitochondrial tolerance but absence of effect on UCP activity. EMBO J. 2005;24:4061–4070. doi: 10.1038/sj.emboj.7600866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Echtay KS. Mitochondrial uncoupling proteins--what is their physiological role? Free Radical Biol Med. 2007;43:1351–1371. doi: 10.1016/j.freeradbiomed.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Sluse FE, Jarmuszkiewicz W, Navet R, Douette P, Mathy G, Sluse-Goffart CM. Mitochondrial UCPs: new insights into regulation and impact. Biochim Biophys Acta. 2006;1757:480–485. doi: 10.1016/j.bbabio.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Okiyama W, Tanaka N, Nakajima T, Tanaka E, Kiyosawa K, Gonzalez FJ, et al. Polyenephosphatidylcholine prevents alcoholic liver disease in PPAR alpha-null mice through attenuation of increases in oxidative stress. J Hepatol. 2009;50:1236–1246. doi: 10.1016/j.jhep.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson AD, Shah YM, Matsubara T, Krausz KW, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha induction of uncoupling protein 2 protects against acetaminophen-induced liver toxicity. Hepatology. 2012;56:281–290. doi: 10.1002/hep.25645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baffy G. Uncoupling protein-2 and cancer. Mitochondrion. 2010;10:243–252. doi: 10.1016/j.mito.2009.12.143. [DOI] [PubMed] [Google Scholar]
- 38.Valle A, Oliver J, Roca P. Role of uncoupling proteins in cancer. Cancers. 2010;2:567–591. doi: 10.3390/cancers2020567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong Y, Fink BD, Dillon JS, Sivitz WI. Effects of adenoviral overexpression of uncoupling protein-2 and -3 on mitochondrial respiration in insulinoma cells. Endocrinology. 2001;142:249–256. doi: 10.1210/endo.142.1.7889. [DOI] [PubMed] [Google Scholar]
- 40.Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res. 2004;10:6203–6207. doi: 10.1158/1078-0432.CCR-04-0419. [DOI] [PubMed] [Google Scholar]
- 41.Joseph JW, Koshkin V, Saleh MC, Sivitz WI, Zhang CY, Lowell BB, et al. Free fatty acid-induced beta-cell defects are dependent on uncoupling protein 2 expression. J Biol Chem. 2004;279:51049–51056. doi: 10.1074/jbc.M409189200. [DOI] [PubMed] [Google Scholar]
- 42.Diano S, Horvath TL. Mitochondrial uncoupling protein 2 (UCP2) in glucose and lipid metabolism. Trends Mol Med. 2012;18:52–58. doi: 10.1016/j.molmed.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Samudio I, Fiegl M, McQueen T, Clise-Dwyer K, Andreeff M. The warburg effect in leukemia-stroma cocultures is mediated by mitochondrial uncoupling associated with uncoupling protein 2 activation. Cancer Res. 2008;68:5198–5205. doi: 10.1158/0008-5472.CAN-08-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouillaud F. UCP2, not a physiologically relevant uncoupler but a glucose sparing switch impacting ROS production and glucose sensing. Biochim Biophys Acta. 2009;1787:377–383. doi: 10.1016/j.bbabio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Criscuolo F, Mozo J, Hurtaud C, Nubel T, Bouillaud F. UCP2, UCP3, avUCP, what do they do when proton transport is not stimulated? Possible relevance to pyruvate and glutamine metabolism. Biochim Biophys Acta. 2006;1757:1284–1291. doi: 10.1016/j.bbabio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Pecqueur C, Bui T, Gelly C, Hauchard J, Barbot C, Bouillaud F, et al. Uncoupling protein-2 controls proliferation by promoting fatty acid oxidation and limiting glycolysis-derived pyruvate utilization. FASEB J. 2008;22:9–18. doi: 10.1096/fj.07-8945com. [DOI] [PubMed] [Google Scholar]
- 47.Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, et al. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci USA. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathupala SP, Ko YH, Pedersen PL. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta. 2010;1797:1225–1230. doi: 10.1016/j.bbabio.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Remmen H, Salvador C, Yang H, Huang TT, Epstein CJ, Richardson A. Characterization of the antioxidant status of the heterozygous manganese superoxide dismutase knockout mouse. Arch Biochem Biophys. 1999;363:91–97. doi: 10.1006/abbi.1998.1060. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Fang F, Miriyala S, Crooks PA, Oberley TD, Chaiswing L, et al. KEAP1 is a redox sensitive target that arbitrates the opposing radiosensitive effects of parthenolide in normal and cancer cells. Cancer Res. 2013;73:4406–4417. doi: 10.1158/0008-5472.CAN-12-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.