Abstract
The fundamental pathophysiology of sickle cell disease is predicated by the polymerization of deoxygenated (T-state) sickle hemoglobin (Hb S) into fibers that distort red blood cells into the characteristic sickle shape. The crystal structure of deoxygenated Hb S (DeoxyHb S) and other studies suggest that the polymer is initiated by a primary interaction between the mutation βVal6 from one Hb S molecule, and a hydrophobic acceptor pocket formed by the residues βAla70, βPhe85 and βLeu88 of an adjacent located Hb S molecule. On the contrary, oxygenated or liganded Hb S does not polymerize or incorporate in the polymer. In this paper we present the crystal structure of carbonmonoxy-ligated sickle Hb (COHb S) in the quaternary classical R-state at 1.76 Å. The overall structure and the pathological donor and acceptor environments of COHb S are similar to those of the isomorphous CO-ligated R-state normal Hb (COHb A), but differ significantly from DeoxyHb S as expected. More importantly, the packing of COHb S molecules does not show the typical pathological interaction between βVal6 and the βAla70, βPhe85 and βLeu88 hydrophobic acceptor pocket observed in DeoxyHb S crystal. The structural analysis of COHb S, COHb A and DeoxyHb S provides atomic level insight into why liganded hemoglobin does not form a polymer.
Keywords: Crystal structure, Hemoglobin, R-state, Allosteric, Sickle cell disease, Mutation
1. Introduction
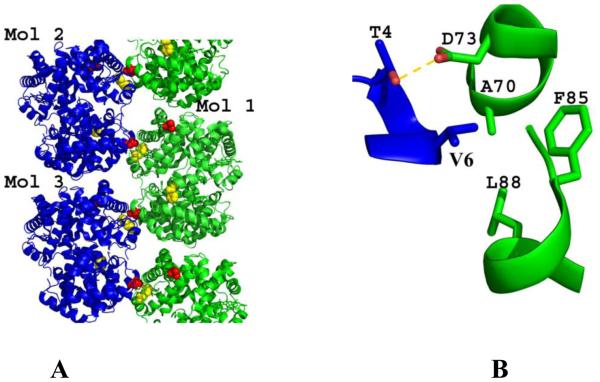
Sickle cell disease occurs as a result of a single point mutation of βGlu6 in normal hemoglobin (Hb A) to βVal6 in sickle hemoglobin (Hb S) (Pauling & Itano, 1949; Ingram, 1959). When Hb S is in its deoxygenated T-state form, it assembles into long, rigid, insoluble fibers that lead to sickling of red blood cells (RBCs) and several cascading adverse events, including, but not limited to oxidative stress, hemolysis of RBCs, inflammation, vaso-occlusion and impaired microvascular blood flow, painful crises, chronic organ damage, morbidity and mortality (Habara & Steinberg, 2016; Kim, 2014; Akinsheye & Klings, 2010; De Franceschi, 2009; Turhan et al, 2002;). The crystal structure of deoxygenated Hb S (DeoxyHb S) has been elucidated (Harrington et al, 1997), and in conjunction with other biochemical studies show the fundamental structure of deoxygenated Hb S fiber as double strand fibrils of Hb S (sometimes referred to as Wishner-Love double strand) arranged into a 14-filament strand fiber (Fig. 1A) (Eaton & Hofrichter, 1990; Carragher et al, 1988; Lewis et al, 1994; Nagel et al, 1980; Benesch et al, 1982; Bluemke et al, 1987). Individual fibrils are stabilized by numerous intra-strand (axial) and inter-strand (lateral) contacts (Harrington et al, 1997; Cretegny & Edelstein, 1993; Watowich et al, 1993; Edelstein, 1981), including the primary interaction between the pathological β2Val6 from one Hb S molecule and a hydrophobic acceptor pocket in the region of β1Ala70, β1Phe85 and β1Leu88 (hereafter the βPhe85 pocket) of a laterally-located heterotetramer (Fig. 1). This hydrophobic contact initiates the polymerization process, and is augmented by an adjacent hydrogen-bond interaction between β2Thr4 and β1Asp73 (Fig. 1B).
Fig. 1. Structure of DeoxyHb S (PDB code 2HBS).
(A) Ribbon figure of the crystal packing of deoxyHb S. (B) The pathological β2Val6 in one strand (blue) interacts with a hydrophobic pocket formed by β1Ala70, β1Phe85, and β1Leu88 from the β1 subunit of a heterotetramer positioned in the adjacent polymeric strand (green). This interaction is stabilized by a hydrogen-bond contact between β2Thr4 and β1Asp73.
Oxygenated Hb S, unlike DeoxyHb S, does not form a polymer. Additionally, polymeric DeoxyHb S excludes sickle hemoglobins that form quaternary R-state from the polymer following oxygen binding (Nagel et al, 1980; Nagel et al, 1979; Bookchin et al, 1975; Bunn & Forget, 1986; Goldberg et al, 1977; Benesch et al, 1980). Liganded Hb exists as multi relaxed states, including the classical R, R2, R3, etc., each with a distinct quaternary conformation (Safo & Abraham, 2005; Jenkins et al, 2009; Lukin et al, 2003, Gong et al, 2006) . Detailed analyses of the different quaternary relaxed Hb states have been recently reviewed extensively (Safo et al, 2011; Jenkins et al, 2009). While the structure of liganded sickle Hb (Hb S) in the R2 state conformation (Patskovska et al, 2005) or liganded normal Hb (Hb A) in the classical R-state conformation (Perutz et al, 1968) have been elucidated that of Hb S in the classical R-state--to the best of our knowledge--has not been reported. The current study reports liganded Hb S structure in the classical R-state conformation, and compares it with the known structures of liganded Hb S in the R2-state, liganded Hb A in the R-state, anddeoxygenated T-state Hb S.
2. Materials and methods
2.1. Source of Hb for studies
Normal Hb was purified (Safo & Abraham, 2003) from de-identified and discarded normal RBCs obtained from the American Red Cross Mid-Atlantic Blood Services Region. Sickle Hb was purified (Safo & Abraham, 2003) from de-identified and discarded sickle RBCs from patients who visited the Virginia Commonwealth University (VCU) Hospital for apheresis. The use of these samples does not require IRB.
2.2. Crystallization, data collection and structure determination of COHb S
Sickle Hb (40 mg/mL) was deoxygenated with a few pellets of sodium dithionite, followed by saturation with CO to generate the CO-bound Hb S form, which was then crystallized with 3.0-3.4 M phosphate buffer, pH 6.4, and one drop of toluene (Safo & Abraham, 2003; Safo & Abraham, 2005). The crystal was washed with mother liquor containing 15% glycerol, then flash cooled prior to diffraction data collection at 100 K with a Rigaku IV ++ image plate detector (CuKα X-rays; λ = 1.54 Å), and a MicroMax-007 source fitted with Varimax Confocal optics (Rigaku, The Woodlands, TX). The dataset was processed with the d*trek software (Rigaku) and the CCP4 suite of programs (Winn et al, 2011). The isomorphous native human CO-bound hemoglobin (COHb A) structure (PDB code 1LJW; α1β1 dimer) (Safo et al, 2002) was used as the starting model to refine the structure, using both Phenix and CNS refinement programs (Adams et al, 2010; Brunger et al, 1998). Prior to the refinement, βGlu6 in 1LJW was mutated to alanine. Model building and correction were carried out using COOT (Emsley & Cowtan, 2004). A round of refinement expectedly identified the sixth residue of the β-chain as valine and was included in the refinement. Also, included in the final model are two molecules of toluene, a molecule of phosphate ions, and 442 water molecules. The structure refined to a final Rfactor/Rfree of 19.3/24.3%. The atomic coordinate and structure factor files have been deposited in the RCSB Protein Data Bank with accession codes 5E6E. Detailed crystallographic and structural analysis parameters are reported in Table 1.
Table 1.
Data collection and refinement statistics of COHb S. Numbers in parentheses are for the highest resolution shell.
| Data Collection | |
| Space group | P41212 |
| Unit-cell a, b, c (Å) | 53.35, 53.35, 191.07 |
| Resolution (Å) | 53.35-1.76 (1.82-1.76) |
| Unique reflections | 27374 (2014) |
| Redundancy | 4.76 (1.81) |
| Completeness (%) | 95.9 (72.9) |
| Average I/σ(I) | 14.7 (3.9) |
| Rmerge (%)a | 6.2 (17.5) |
| Refinementb | |
| Resolution (Å) | 29.61-1.76 (1.82-1.76) |
| No. of reflections | 27308 (2013) |
| Rwork (%) | 19.3 (29.8) |
| Rfree (%) | 24.3 (35.2) |
| R.m.s.d. bonds (Å) | 0.011 |
| R.m.s.d. angles (°) | 1.9 |
| Dihedral angles | |
| Most favored (%) | 96.1 |
| Allowed (%) | 3.9 |
| Average B (Å2) / atoms | |
| All atoms | 27.7 |
| Protein | 25.4 |
| Hemes | 22.6 |
| CO | 16.5 |
| Toluene | 40.2 |
| Phosphate | 48.9 |
| Water | 39.2 |
| PDB ID code |
Rmerge = ΣhklΣi|Ii(hkl) – <I(hkl)>|/ΣhklΣiIi(hkl).
Rfree was calculated from 5% randomly selected reflection for cross-validation. All other measured reflections were used during refinement.
2.3. Dynamic light scattering studies
Dynamic light scattering (DLS) studies were conducted to determine the aggregation state of COHb S, COHb A, DeoxyHb A and DeoxyHb S in 0.01M ammonium phosphate buffer, pH7.0 at 25°C using Malvern, Zetasizer Nano-S DLS instrument (Chen et al, 2004). 4 molar excess of sodium dithionite were added to 150 μM of Hb A and Hb S to make DeoxyHb A and Deoxy Hb S, respectively. Aliquots of the DeoxyHb A and DeoxyHb S solutions were saturated with CO gas to obtain COHb A and COHb S, respectively. 500 μL of each sample was taken in a disposable polystyrene visible cuvette and used for the DLS experiment (Chen et al, 2004). The final reported data are average of 4 measurements as reported in Table 2.
Table 2.
Average effective diameter of Hb A and Hb S
| Hb solution | D (nm) |
|---|---|
| COHb A | 6.9 ±1.4 |
| COHb S | 7.4 ± 0.17 |
| DeoxyHb A | 6.9 ± 1.5 |
| DeoxyHb S | 20.0 ± 0.95 |
3. Results and Discussion
We have determined the crystal structure of carbonmonoxy Hb S (COHb S) at 1.76 Å resolution, which, the first such reported liganded classical R-state Hb S structure. The crystal structures of normal Hb (COHb A; PDB code 1LJW) in the R-state (Safo et al, 2002) and sickle Hb (COHb S; PDB code 1NEJ) in the R2-state (Patskovska et al, 2005) have been reported. Crystals of COHb S and R-state COHb A are isomorphous with space group P41212 and typical cell parameters of 53, 53 and 192 Å. Initial and final electron density maps of COHb S suggested valine at the 6th position of the β-chain (Fig. 2). The final structure contains a dimer (α1β1) in the asymmetric unit, comprising 141 residues in the α-subunit, 146 residues in the β-subunit, 2 heme groups, 2 CO-ligated heme ligands, 2 toluene molecules, a phosphate molecule, and 442 water molecules. Toluene, which is routinely used to aid in the crystallization of liganded Hb was observed bound in two positions at the hydrophobic pocket formed by αTrp14, αVal17, αTyr24, αLue105, αLeu109, αLeu125, αPhe128, αVal10, αVal70 and αLeu66. Similar toluene binding has been reported in COHb A structure (Safo & Abraham, 2005). The phosphate also binds to the surface of the protein at a cavity formed by αHis20, αTyr24, αHis112, and βLys120. The two R-state structures, COHb S and COHb A are indistinguishable with root mean square deviation (rmsd) of~ 0.2 Å using the dimers (α1β1) or tetramers (α1β1α2β2) for comparison (Table 3). Likewise, the heme environment, the α1β2 dimer interface, and the A helix where the mutation βVal6 occurs in Hb S are very similar. The dimeric structures of R-state COHb S and R2-state COHb S are also similar (~0.6 Å), but differ in their tetrameric structures (rmsd of ~1.6 Å), which is the result of a rotation of the α1β1 dimer relative to the α2β2 dimer by ~11° during the R↔R2 transition (Table 3). Similar quaternary structural differences are also observed between R-state COHb A and R2-state COHb A structures (Safo & Abraham, 2005). Despite the quaternary structure differences, the heme environment is similar in the liganded structures.
Fig. 2. Crystal structure of COHb S.
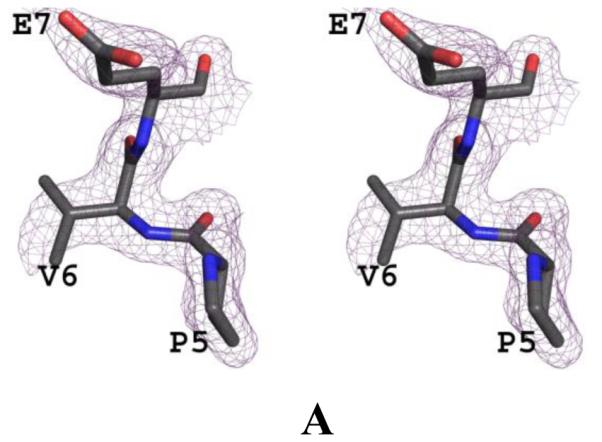
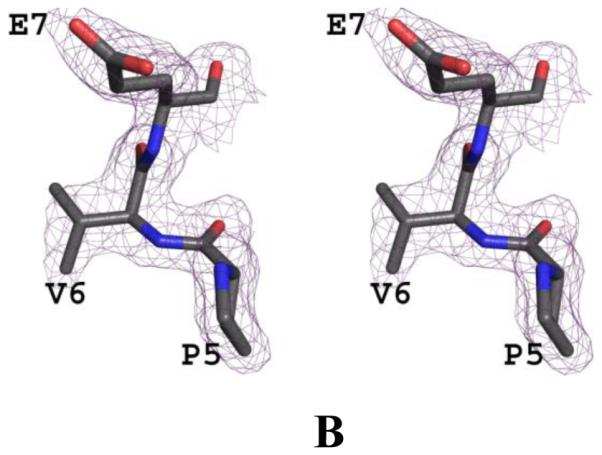
(A) Stereo-view of the initial electron density (2Fo-Fc) map with alanine at the 6th position of the β-subunit during the refinement, contoured at 1.0σ. (B) Stereo-view of the final 2Fo-Fc map with valine at the 6th position of the β-subunit during the refinement, contoured at 1.0σ. The two maps are superimposed with the final refined model.
Table 3.
Structural differences between T, R and R2 states of COHb A and COHb S. The least-squares calculations involved only Cα atoms.
| Transition | Rmsd(Å) | Screw- rotation angle(°) |
||||||
|---|---|---|---|---|---|---|---|---|
| α1 | α2 | β1 | β2 | α1β1 | α2β2 | α1β1α2β2 | ||
| COHb S(R)-COHb A(R) | 0.17 | 0.17 | 0.19 | 0.19 | 0.21 | 0.21 | 0.24 | 0.9 |
| COHb S(R)-COHb S(R2) | 0.69 | 0.69 | 0.45 | 0.43 | 0.61 | 0.61 | 1.61 | 11.2 |
| COHb A(R)-COHb S(R2) | 0.63 | 0.64 | 0.42 | 0.42 | 0.55 | 0.56 | 1.64 | 12.1 |
| COHb S(R)-DxHb S(T) | 0.48 | 0.48 | 0.75 | 0.89 | 0.81 | 0.75 | 2.37 | 14.2 |
| COHb A(R)-DxHb S(T) | 0.48 | 0.49 | 0.79 | 0.91 | 0.91 | 0.81 | 2.37 | 13.9 |
| COHb S(R2)-DxHb S(T) | 0.70 | 0.72 | 0.82 | 1.01 | 1.02 | 1.01 | 2.59 | 21.6 |
The dimeric structures of the R-state COHb S and T-state DeoxyHb S (PDB code 2HBS) (Harrington et al, 1997) when superposed gives rmsd of ~0.8 Å, while the tetramers, as expected differ significantly with rmsd of ~2.4 Å. Similar structural differences are also observed between R-state COHb A and DeoxyHb A (PDB accession 2DN2) (Park et al., 2006) or between R-state COHb A and DeoxyHb S (Table 3) (Safo & Abraham, 2005). The large quaternary structural differences between the T and R structures is due to rotation of the α1β1 dimer relative to the α2β2 dimer by ~14° during the T↔R transition (Table 3) (Safo et al, 2011; Jenkins et al, 2009; Baldwin & Chothia, 1979). It is notable that the T↔R2 transition, either sickle or normal Hb, also results in ~22° rotation between the two non-superposed dimers (Table 3) (Jenkins et al, 2009; Safo & Abraham, 2005; Silva et al, 1992).
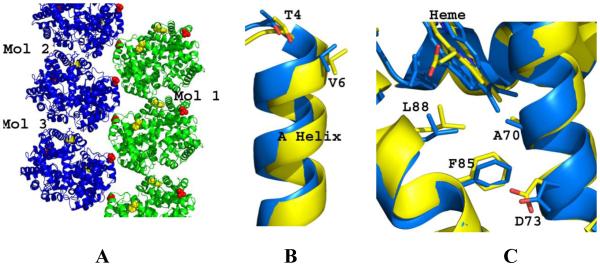
The building block of the Wishner-Love double strand could be described as two axially- and one laterally-located heterotetramers (Fig. 1A; Molecules 1-3). The laterally-located molecules (with the pathologic interaction between β2Val6 and β1Phe85 pocket) are related by a 2-fold screw axis. Interestingly, like the DeoxyHb S crystal, the crystal lattice of COHb S in the R-state shows packing of Hb tetramers in double layers, with the laterally-located molecules also related by a 2-fold screw axis (Fig. 3A). However, there are significant differences between the two crystal packings, notwithstanding the fact that even though the mutated residue βVal6 is located at the interface of two laterally-located Hb molecules in COHb S, it does not make any interaction with the βPhe85 pocket. In fact, βVal6 in COHb S is not involved in any crystal contact. The relative stabilities of DeoxyHb S and COHb S crystal packings, quantified by calculating the buried solvent accessible surface area for the three consecutive molecules that form the two lateral contacts and one axial contact (Molecules 1-3, Figs. 1A and 3A) are different. While the buried surface for the axial contact (between molecules 2 and 3) is similar in COHb S (768 Å2) and DeoxyHb S (748 Å2), the total buried surface for the two lateral contacts (between molecules 1 and 2, and between molecules 1 and 3) in COHb S (889 Å2) pales in comparison to that of DeoxyHb S (3292 Å2). Note that the primary contact between β2Val6 and the β1Phe85 pocket in DeoxyHb S, augmented by the hydrogen-bond interaction between β2Thr4 and β1Asp73 is a major contributor to the large buried surface at the lateral interface (Fig. 1B). Like R-state COHb S, the mutated β2Val6 in R2-state COHb S is not involved in any crystal interaction in the lattice packing (Patskovska et al., 2005). It is obvious from the large buried surface why DeoxyHb S molecules aggregate into a double strand in solution to form a fiber. In contrast, it appears that the crystal packing in COHb S (either in the R or R2 quaternary form) is only tenuous and non-specific; making clear that ligated Hb S cannot form a fiber like DeoxyHb S. Consistently, dynamic light scattering experiment showed DeoxyHb S, even at low protein concentration of 150 μM and phosphate buffer (0.01M) to aggregate while COHb S, COHb A or DeoxyHb A showed no aggregation (Table 2). Similar results have been reported by Cheng et al. (2004) who also showed DeoxyHb S to aggregate at very low phosphate buffer concentration (0.05 M) while Hb S or Hb A did not, although at high phosphate buffer concentration (>1.5 M) all the Hbs aggregated. It is important to note that while individual double-stranded fibrils are initiated and stabilized by the primary interaction between β2Val6 and β1Phe85, several secondary interactions between the molecules in the strands are also of critical importance to the Hb S polymer stability (Harrington et al, 1997; Cretegny & Edelstein, 1993; Watowich et al, 1993; Edelstein, 1981). This is evidenced by the large number of naturally occurring mutations that reduce polymer formation and disease severity by attenuating these polymer-stabilizing contacts. (Nagel et al, 1980; Bunn & Forget, 1986; Benesch et al, 1982; Benesch et al, 1976; Rhoda et al, 1983).
Fig. 3. Structure analysis of COHb S and DeoxyHb S.
(A) Ribbon figure of the crystal packing of COHb S. (B) The β-subunit A helix (with βVal6 and βThr4) of COHb S (yellow) and DeoxyHb S (blue) after superposing the β2-subunits (3-138 residues) of the two structures. (C) The hydrophobic acceptor pocket formed by βAla70, βPhe85, and βLeu88 of COHb S (yellow) and DeoxyHb S (blue) after superposing the β1-subunits (3-138 residues) of the two structures.
The intriguing question is why does βVal6 in COHb S not make a hydrophobic interaction with the β1Phe85 pocket that is at least required to initiate polymer formation? We compared the donor and acceptor regions of DeoxyHb S with those of R-state COHb S. The β2-subunit A helix (residues 4-12), with the pathological β2Val6 and β2Thr4 exhibits a displacement of 1.5 Å, almost twice the rmsd value of 0.8 Å calculated for all the β2-subunit residues (residues 3-138) (Fig. 3B). The positions of the three β1-subunit residues β1Ala70, β1Phe85, and β1Leu88 that make up the hydrophobic acceptor pocket, as well as β1Asp73 that makes hydrogen-bond interaction with β2Thr4 also show an average displacement of 1.0 Å compared to the rmsd value of 0.7 Å for all the β1-subunit residues (Fig. 3C). These differential positions of the donor and acceptor residues, especially the former could result in inefficient alignment between β2Val6 and the β2Phe85 pocket, and may in part explain why the interaction between β2Val6 and the β2Phe85 pocket and consequently the Wishner-Love double strand packing does not occur in liganded Hb S. Likewise, significant positional shift of β2Thr4 (1.6 Å) suggests that the hydrogen-bond interaction between β2Thr4 and β1Asp73 is likely to be abrogated or weakened. The positional importance of these two residues is consistent with studies that show the natural substitutions βAsp73→Asn as in Hb Korle Bu or βAsp73→Val as in Hb Mobile increases the solubility of DeoxyHb S mixtures by eliminating the critical β2Thr4-β1Asp73 hydrogen-bond interaction (Adachi et al, 1987; Bookchin et al, 1970; Converse et al, 1985). It is also important to point out that because the T↔R transition results in 14° rotation of the α1β1 dimer relative to the α2β2 dimer, several polymer secondary contacts that are known to stabilize the polymer in DeoxyHb S cannot occur in COHb S, and may also account for the inability of COHb S to form the Wishner-Love strand. Similar argument can also be made between COHb S in the R2 quaternary state and DeoxyHb S, where the T↔R2 transition involves ~22° and the position of β2Val6 in the two structures differ by ~1.8 Å.
Conclusion
We have determined the crystal structure of COHb S in the quaternary R-state form. Despite the pathologic βVal6 mutation, the structure is indistinguishable from liganded normal human Hb structure, even at the A helix where the mutation occurs in Hb S. Structural analysis of COHb S and DeoxyHb S provides atomic level insight into why liganded hemoglobin does not form a polymer.
Acknowledgement
This work was supported by NIH/NIMHD grant MD009124 (MKS), NIH/NHLBI grant K01HL103186 (OA), the NSTIP strategic technologies program in the Kingdom of Saudi Arabia, Project No. 10-BIO1253-03. Structural biology resources were provided in part by NIH/NCI grant P30CA016059 to the VCU Massey Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi K, Kim J, Kinney TR, Asakura T. Effect of the beta 73 amino acid on the hydrophobicity, solubility, and the kinetics of polymerization of deoxyhemoglobin S. The Journal of Biological Chemistry. 1987;262:10470–10474. [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinsheye I, Klings ES. Sickle cell anemia and vascular dysfunction: The nitric oxide connection. Journal of Cellular Physiology. 2010;224:620–625. doi: 10.1002/jcp.22195. [DOI] [PubMed] [Google Scholar]
- Baldwin J, Chothia C. Haemoglobin: The structural changes related to ligand binding and its allosteric mechanism. Journal of Molecular Biology. 1979;129:175–220. doi: 10.1016/0022-2836(79)90277-8. [DOI] [PubMed] [Google Scholar]
- Benesch RE, Yung S, Benesch R, Mack J, Schneider RG. Alpha-chain contacts in the polymerisation of sickle haemogloblin. Nature. 1976;260:219–221. doi: 10.1038/260219a0. [DOI] [PubMed] [Google Scholar]
- Benesch RE, Kwong S, Benesch R. The effects of alpha chain mutations cis and trans to the beta6 mutation on the polymerization of sickle cell haemoglobin. Nature. 1982;299:231–234. doi: 10.1038/299231a0. [DOI] [PubMed] [Google Scholar]
- Benesch RE, Edalji R, Benesch R, Kwong S. Solubilization of hemoglobin S by other hemoglobins. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:5130–5134. doi: 10.1073/pnas.77.9.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemke DA, Carragher B, Potel MJ, Josephs R. The three-dimensional structure of sickle hemoglobin macrofibers. Progress in Clinical and Biological Research. 1987;240:31–46. [PubMed] [Google Scholar]
- Bookchin RM, Nagel RL, Balazs T. Role of hybrid tetramer formation in gelation of haemoglobin S. Nature. 1975;256:667–668. doi: 10.1038/256667a0. [DOI] [PubMed] [Google Scholar]
- Bookchin RM, Nagel RL, Ranney HM. The effect of beta 73 asn on the interactions of sickling hemoglobins. Biochimica et Biophysica Acta. 1970;221:373–375. doi: 10.1016/0005-2795(70)90279-5. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D, Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bunn HF, Forget BG. Hemoglobin: Molecular, Genetic and Clinical Aspects. W. B. Saunders; Philadelphia: 1986. pp. 502–564. [Google Scholar]
- Carragher B, Bluemke DA, Gabriel B, Potel MJ, Josephs R. Structural analysis of polymers of sickle cell hemoglobin. I. sickle hemoglobin fibers. Journal of Molecular Biology. 1988;199:315–331. doi: 10.1016/0022-2836(88)90316-6. [DOI] [PubMed] [Google Scholar]
- Cheng K, Ballas SK, Hantgan RR, Kim-Shapiro DB. Aggregation of Normal and Sickle Hemoglobin in High Concentration Phosphate Buffer. Biophysical Journal. 2004;87:4113–4121. doi: 10.1529/biophysj.104.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse JL, Sharma V, Reiss-Rosenberg G, Ranney HM, Danish E, Bowman LS, Harris JW. Some properties of hemoglobin mobile (alpha 2 beta 2 73 Asp----Val) Hemoglobin. 1985;9:33–45. doi: 10.3109/03630268508996980. [DOI] [PubMed] [Google Scholar]
- Cretegny I, Edelstein SJ. Double strand packing in hemoglobin S fibers. Journal of Molecular Biology. 1993;230:733–738. doi: 10.1006/jmbi.1993.1195. [DOI] [PubMed] [Google Scholar]
- De Franceschi L. Pathophisiology of sickle cell disease and new drugs for the treatment. Mediterranean Journal of Hematology and Infectious Diseases. 2009;1:e2009024. doi: 10.4084/MJHID.2009.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton WA, Hofrichter J. Sickle cell hemoglobin polymerization. Advances in Protein Chemistry. 1990;40:63–279. doi: 10.1016/s0065-3233(08)60287-9. [DOI] [PubMed] [Google Scholar]
- Edelstein SJ. Molecular topology in crystals and fibers of hemoglobin S. Journal of Molecular Biology. 1981;150:557–575. doi: 10.1016/0022-2836(81)90381-8. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallographica Section D, Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goldberg MA, Husson MA, Bunn HF. Participation of hemoglobins A and F in polymerization of sickle hemoglobin. The Journal of Biological Chemistry. 1977;252:3414–3421. [PubMed] [Google Scholar]
- Habara A, Steinberg MH. Genetic basis of heterogeneity and severity in sickle cell disease. Experimental Biology and Medicine (Maywood) 2016 doi: 10.1177/1535370216636726. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington DJ, Adachi K, Royer WE., Jr The high resolution crystal structure of deoxyhemoglobin S. Journal of Molecular Biology. 1997;272:398–407. doi: 10.1006/jmbi.1997.1253. [DOI] [PubMed] [Google Scholar]
- Rhoda MD, Martin J, Blouquit Y, Garel MC, Edelstein SJ, Rosa J. Sickle cell hemoglobin fiber formation strongly inhibited by the Stanleyville II mutation (alpha 78 asn leads to lys) Biochem. Biophys. Res. Commun. 1983;111:8–13. doi: 10.1016/s0006-291x(83)80109-0. [DOI] [PubMed] [Google Scholar]
- Ingram VM. Abnormal human haemoglobins. III. the chemical difference between normal and sickle cell haemoglobins. Biochimica et Biophysica Acta. 1959;36:402–411. doi: 10.1016/0006-3002(59)90183-0. [DOI] [PubMed] [Google Scholar]
- Jenkins JD, Musayev FN, Danso-Danquah R, Abraham DJ, Safo MK. Structure of relaxed-state human hemoglobin: Insight into ligand uptake, transport and release. Acta Crystallographica Section D, Biological Crystallography. 2009;65:41–48. doi: 10.1107/S0907444908037256. [DOI] [PubMed] [Google Scholar]
- Kim HC. Red cell exchange: special focus on sickle cell disease. Hematology Am Soc Hematol Educ Program. 2014;2014(1):450–456. doi: 10.1182/asheducation-2014.1.450. [DOI] [PubMed] [Google Scholar]
- Lewis MR, Gross LJ, Josephs R. Cryo-electron microscopy of deoxy-sickle hemoglobin fibers. Microscopy Research and Technique. 1994;27:459–467. doi: 10.1002/jemt.1070270512. [DOI] [PubMed] [Google Scholar]
- Lukin JA, Kontaxis G, Simplaceanu V, Yuan Y, Bax A, Ho C. Quaternary structure of hemoglobin in solution. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:517–520. doi: 10.1073/pnas.232715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Simplaceanu V, Lukin JA, Giovannelli JL, Ho NT, Ho C. Quaternary structure of carbonmonoxyhemoglobins in solution: structural changes induced by the allosteric effector inositol hexaphosphate. Biochemistry. 2006;45:5140–5148. doi: 10.1021/bi052424h. [DOI] [PubMed] [Google Scholar]
- Nagel RL, Johnson J, Bookchin RM, Garel MC, Rosa J, Schiliro G, Wajcman H, Labie D, Moo-Penn W, Castro O. Beta-chain contact sites in the haemoglobin S polymer. Nature. 1980;283:832–834. doi: 10.1038/283832a0. [DOI] [PubMed] [Google Scholar]
- Nagel RL, Bookchin RM, Johnson J, Labie D, Wajcman H, Isaac-Sodeye WA, Honig GR, Schiliro G, Crookston JH, Matsutomo K. Structural bases of the inhibitory effects of hemoglobin F and hemoglobin A2 on the polymerization of hemoglobin S. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:670–672. doi: 10.1073/pnas.76.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patskovska LN, Patskovsky YV, Almo SC, Hirsch RE. COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000. Acta Crystallographica Section D, Biological Crystallography. 2005;61:566–573. doi: 10.1107/S0907444905004622. [DOI] [PubMed] [Google Scholar]
- Pauling L, Itano HA. Sickle cell anemia a molecular disease. Science (New York, N.Y.) 1949;110:543–548. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Yokoyama T, Shibayama N, Shiro Y, Tame JR. 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J. Mol. Biol. 2006;360:690–701. doi: 10.1016/j.jmb.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Muirhead H, Cox JM, Goaman LC. Three-dimensional fourier synthesis of horse oxyhaemoglobin at 2.8 A resolution: The atomic model. Nature. 1968;219:131–139. doi: 10.1038/219131a0. [DOI] [PubMed] [Google Scholar]
- Safo MK, Burnett JC, Musayev FN, Nokuri SS, Abraham DJ. Crystal structure of human carbonmonoxy hemoglobin at 2.16 Å: A snapshot of the allosteric transition. Acta Crystallogr. 2002;58,:2031–2037. doi: 10.1107/s0907444902015809. [DOI] [PubMed] [Google Scholar]
- Safo MK, Abraham DJ. The enigma of the liganded hemoglobin end state: a novel quaternary structure of human carbonmonoxy hemoglobin. Biochemistry. 2005;44:8347–8359. doi: 10.1021/bi050412q. [DOI] [PubMed] [Google Scholar]
- Safo MK, Abraham DJ. X-ray crystallography of hemoglobins. Methods in Molecular Medicine. 2003;82:1–19. doi: 10.1385/1-59259-373-9:001. [DOI] [PubMed] [Google Scholar]
- Safo MK, Ahmed MH, Ghatge MS, Boyiri T. Hemoglobin-ligand binding: Understanding hb function and allostery on atomic level. Biochimica et Biophysica Acta. 2011;1814:797–809. doi: 10.1016/j.bbapap.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Silva MM, Rogers PH, Arnone A. A third quaternary structure of human hemoglobin A at 1.7-A resolution. The Journal of Biological Chemistry. 1992;267:17248–17256. [PubMed] [Google Scholar]
- Turhan A, Weiss LA, Mohandas N, Coller BS, Frenette PS. Primary role for adherent leukocytes in sickle cell vascular occlusion: A new paradigm. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3047–3051. doi: 10.1073/pnas.052522799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watowich SJ, Gross LJ, Josephs R. Analysis of the intermolecular contacts within sickle hemoglobin fibers: Effect of site-specific substitutions, fiber pitch, and double-strand disorder. Journal of Structural Biology. 1993;111:161–179. doi: 10.1006/jsbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D. Biological Crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]