SUMMARY
Systems approaches have been used to describe molecular signatures driving immunity to influenza vaccination in humans. Whether such signatures are similar across multiple seasons and in diverse populations is unknown. We applied systems approaches to study immune responses in young, elderly, and diabetic subjects vaccinated with the seasonal influenza vaccine across five consecutive seasons. Signatures of innate immunity and plasmablasts correlated with and predicted influenza antibody titers at 1 month after vaccination with >80% accuracy across multiple seasons but were not associated with the longevity of the response. Baseline signatures of lymphocyte and monocyte inflammation were positively and negatively correlated, respectively, with antibody responses at 1 month. Finally, integrative analysis of microRNAs and transcriptomic profiling revealed potential regulators of vaccine immunity. These results identify shared vaccine-induced signatures across multiple seasons and in diverse populations and might help guide the development of next-generation vaccines that provide persistent immunity against influenza.
INTRODUCTION
Seasonal influenza infection kills several hundred thousand people every year, with the majority of deaths occurring among the elderly (Pica and Palese, 2013; Simonsen et al., 2005). Although vaccination is considered the most effective method for preventing influenza, it shows limited efficacy in the elderly (Sasaki et al., 2011). The vaccine recommended for this age group is the inactivated influenza vaccine that contains virus hemagglutinin (HA) proteins from three (trivalent, TIV) or four of the circulating influenza H1N1, H3N2, and B strains. The lower efficacy of the influenza vaccine in elderly compared to young adults has been associated with immunosenescence (Duraisingham et al., 2013), such as impaired generation of antibody-secreting cells (ASCs) (Sasaki et al., 2011) and memory CD8+ T cells (Wagar et al., 2011) and CD4+ T cells (Kang et al., 2004). However, the molecular mechanisms underlying the decreased vaccine efficacy remain unexplored.
Systems vaccinology is an emerging field that applies systems biology approaches and predictive modeling to vaccinology and provides a powerful tool for unraveling the molecular mechanisms of vaccine immunity (Pulendran, 2014; Pulendran et al., 2010). Recently, systems vaccinology has been successfully used to study the immune response to the influenza vaccine in young adults (Bucasas et al., 2011; Cao et al., 2014; Franco et al., 2013; Furman et al., 2013; Nakaya et al., 2011; Tsang et al., 2014) as well as to other vaccines such as the yellow fever (Gaucher et al., 2008; Querec et al., 2009) and meningococcus (Li et al., 2013) vaccines. These studies were able to identify gene signatures (Nakaya et al., 2011), as well as cellular compositions and gene modules (Furman et al., 2013), post-vaccination that are predictive of the later antibody response vaccination. In addition, such approaches provided important insights into the pathways driving immune responses to vaccination (Oh et al., 2014; Ravindran et al., 2014).
However, several fundamental issues in the field remain unaddressed. First, there is still no comprehensive analysis of the similarity of signatures to influenza vaccination across multiple seasons (Pica and Palese, 2013). This is a crucial issue because the virus strains in the vaccine can change from year to year and the impact of these variations on transcriptional signatures is unknown. Second, there is limited information about the gene regulatory networks and cellular responses that underlie the sub-optimal immunity observed in the elderly population. Third, because all previous studies have focused on signatures that predict the antibody response at 4 weeks after vaccination, the extent to which transcriptional signatures are associated with the longevity of the antibody response has not been examined. This analysis is important for understanding how innate immunity can generate long-lasting antibody responses to influenza vaccination. Finally, little is known about the role of microRNAs (miRNAs) in fine-tuning the transcriptional responses of immune cells after influenza vaccination.
To address these issues, we used systems vaccinology approaches to examine >400 young and elderly adults, including diabetics, vaccinated with seasonal TIV during five consecutive influenza seasons (2007–2011). These subjects included 212 individuals from our study and 218 individuals from a previously published study (Franco et al. 2013). Our systems analysis identified universal signatures of immunity to vaccination spanning multiple years in diverse human populations including the young, elderly, and diabetic. Integrative analysis of microRNAs and transcriptomic profiling revealed potential regulators of vaccine immunity. These results can help guide the development of next-generation vaccines that provide persistent immunity against influenza.
RESULTS
Antibody Responses to Influenza Vaccination Correlate with Age but Not Gender, Race, or Diabetic Status
We vaccinated with TIV a total of 212 subjects across 5 influenza vaccine seasons from 2007 to 2011, among whom 54 were elderly (>65 years old in 2010 and 2011 cohorts) (Figures S1A and S1B). The 2011 cohort also included 17 subjects diagnosed with type 2 diabetes (T2D). Overall, there was a 65:35 ratio of females to males, and the majority of subjects were of European descent (Figure S1B). Blood samples were collected at baseline (day of vaccination) and at the time points indicated (Figure 1A). We performed microarray analyses on the peripheral blood mononuclear cells (PBMCs) as indicated (Figure 1A). For the 2008 and 2009 seasons, we included in our analyses published data from a previous study by Franco et al. (2013) containing 218 additional subjects. For a subset of the 2010 season cohort, fluorescence-activated cell sorting (FACS) measurements as well as miRNA profiling were also performed (Figure 1A). The entire dataset is publicly available online at Immport (Bhattacharya et al., 2014) (http://immport.niaid.nih.gov/).
Figure 1. Experimental Approach and Humoral Immunity to Influenza Vaccination in Young and Elderly.
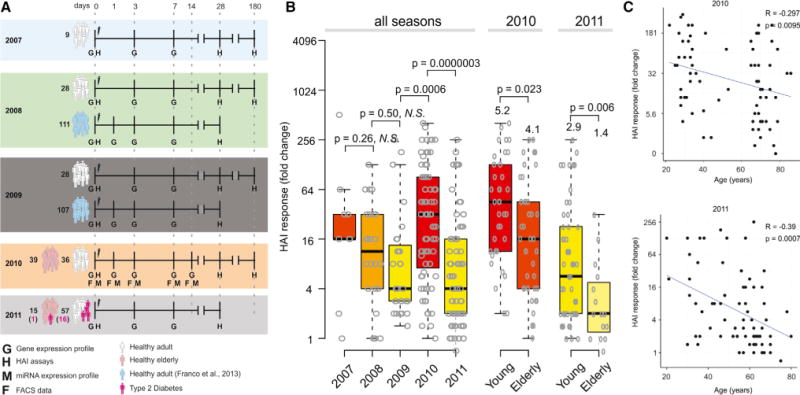
(A) Experimental approach used to study five consecutive influenza vaccination seasons. Microarray experiments and HAI assays were used to obtain the gene expression profiles and antibody responses of 413 TIV vaccinees. Flow cytometry and miRNA expression data were obtained for vaccinees from 2010 season. For 2008 and 2009 seasons, publicly available data (Franco et al., 2013) were included.
(B) HAI responses by season. The maximum HAI response (highest day 28-day 0 fold-induction among all three strains) is shown for all 212 subjects separated by flu season, along with box plots indicating the first and third quartiles and median. For 2010 and 2011, subjects are also separated into young (<65 years old) and elderly (65 or older). p values represent results of independent two-sample t test between responses of young and elderly.
(C) Correlation of HAI responses with age in the 2010 season and 2011 season. R and p represent the Pearson correlation coefficient and associated p value, respectively.
See also Figure S1.
We evaluated the plasma antibody responses of all vaccinated individuals via hemagglutination-inhibition (HAI) antibody titer assays. The individual antibody response for each of the three influenza strains included in the vaccine was calculated as the fold change between the HAI titer at day 28 relative to the baseline titer. We then defined the magnitude of the HAI response as the maximum fold change among the three influenza strains (Figure 1B; Nakaya et al., 2011). Subjects were classified as “high responders” if (1) their HAI response had at least a 4-fold increase at 28 days (Sullivan et al., 2010) and (2) the day 28 antibody titer was 1:40 or more for at least one strain; subjects were classified as “low responders” otherwise. In the 2010 and 2011 seasons, in which elderly subjects were included, antibody responses showed a significant decrease with age (Figures 1B, 1C, and S1C), consistent with previous studies (Seidman et al., 2012). Vaccines from different seasons induced varying levels of HAI responses, and there was a significant decrease in the response with age (Figure 1C). Subjects vaccinated during the 2010 season, the year after the H1N1 2009 pandemic, generated the strongest HAI responses. There was no significant difference in antibody response based on gender (Figure S1D), diabetic status (Figure S1E), or race (data not shown). There was no significant difference in the response between males and females in any of the seasons examined (data not shown).
Gene Signatures of Antibody Responses across Multiple Seasons in the Young and the Elderly
In order to identify transcriptional pathways associated with the antibody response to vaccination, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005) on genes correlated with the HAI response in each influenza season (Figure 2A). For this, we used a set of blood transcriptional modules (BTMs) previously identified by our group through large-scale network integration of publicly available human blood transcriptomes (Li et al., 2014). BTMs related to the induction of interferons as well as the activation of dendritic cells (Figures 2A and 2B) were enriched on days 1 and 3 after TIV vaccination, whereas modules related to T cells at these time points were negatively associated with the antibody response. These results were validated by analysis of both seasons of the Franco et al. (2013) dataset. On day 7, there was a robust enrichment of ASC and cell cycle-related modules, consistent with our original study (Nakaya et al., 2011).
Figure 2. Signatures Associated with the Antibody Response Induced by TIV.
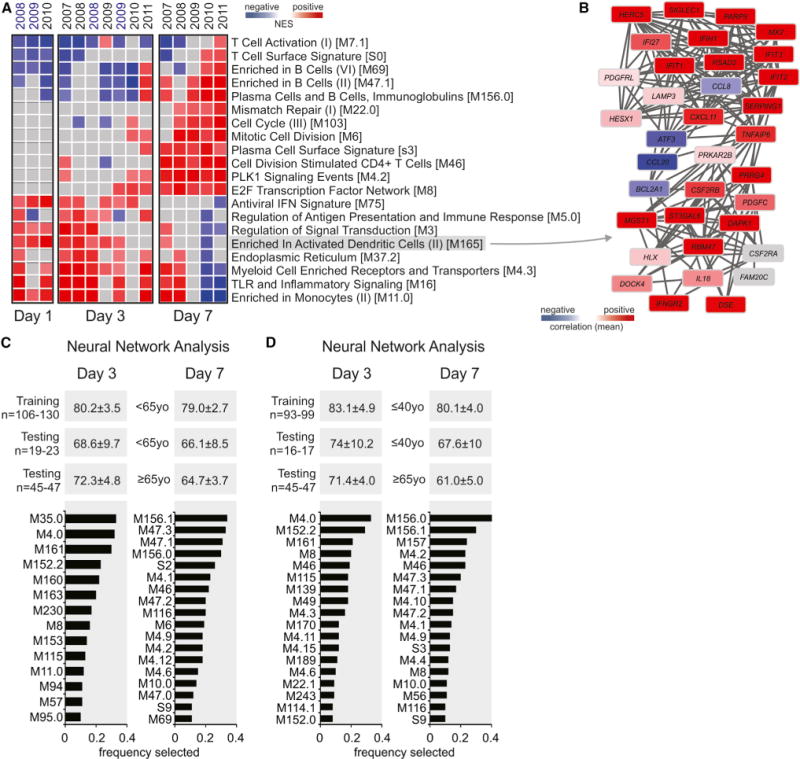
(A) Heat map of blood transcription modules (BTMs, rows) and TIV seasons (columns) whose activity at days 1, 3, or 7 after vaccination is associated with HAI response at day 28 after vaccination. Gene set enrichment analysis (GSEA, nominal p < 0.05; 1,000 permutations) was used to identify positive (red), negative (blue), or no (gray) enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and HAI response. Seasons labeled in blue are from Franco et al. (2013) dataset. Modules shown are those consistently enriched in at least 70% of seasons on a given day. Abbreviation is as follows: NES, normalized enrichment score.
(B) Genes in BTM M165; each “edge” (gray line) represents a coexpression relationship, as described in Li et al. (2014); colors represent the mean correlation for seven TIV seasons between baseline-normalized gene expression at day 3 and HAI response at day 28 after vaccination.
(C and D) Identification of BTMs that predict antibody responses via neural network nalysis. Single sample GSEA (Barbie et al., 2009) enrichment scores were generated for each BTM on day 3 and day 7 after vaccination in 85% of the young subjects (training set) and used as inputs to an artificial neural network classifier to predict the day 28 antibody responses, in the remaining 15% of the young (young test set) or the elderly (elderly test set) subjects (see Supplemental Experimental Procedures for details). The mean accuracies and standard deviations out of 100 randomized trials are shown, along with the frequency with which each module was selected by the algorithm as an input to the classifier. In (C) “young” is <65 years and “elderly” >65 years and in (D) “young” is <40 years and “elderly >65 years.
See also Figures S2–S4.
Additionally, we compared transcriptional responses based on gender, age, race, or diabetic status. We first performed differential expression analysis between these groups (male versus female, type-2-diabetes-positive versus -negative, and European versus African and/or Asian) using fold change expression data from day 3 and day 7. There were very few differentially expressed genes based on gender (Figure S2A) or race (data not shown), indicating similar responses after vaccination. Comparison of gene expression between type-2-diabetes-positive and -negative subjects revealed a modest number of differentially expressed genes (Figure S2B), but GSEA on genes ranked by correlation with the day 28 HAI response in these two groups showed a high degree of overlap in the modules associated with a high antibody response (Figure S2C). Based on the similarity of these transcriptional responses, we decided not to segregate subjects based on gender, race, or diabetic status in subsequent analyses.
Given that we could identify pathways that consistently correlated with the magnitude of HAI response (Figure 2A), we sought to identify transcriptional signatures of immunogenicity across multiple influenza seasons. In our previous study, we have used DAMIP (discriminant analysis via mixed integer programming) (Brooks and Lee, 2010) to find sets of 3–5 discriminatory genes that could predict the antibody response of subjects from the 2007–2009 seasons (Nakaya et al., 2011). Here we applied DAMIP to a larger number of subjects across a greater number of influenza vaccine seasons (Figure S3), as described in the Experimental Procedures and Supplemental Experimental Procedures. Here DAMIP was able to correctly predict the HAI response in the blind testing group with 67.6%–70.0% accuracy (Tables S1 and S2). Consistent with our previous work, the most frequently selected genes included a large number of ASC genes (Table S1).
Given the robustness of the module-level responses across different TIV seasons, we wanted to investigate whether module-level features were capable of accurately predicting vaccine-induced immunity in a multi-year dataset and in the elderly. Therefore, we first generated BTM normalized enrichment scores for each subject using single-sample GSEA (ssGSEA) (Barbie et al., 2009) and then used these scores as inputs to an artificial neural network classifier (Dreiseitl and Ohno-Machado, 2002) (see Supplemental Experimental Procedures for details). In brief, the young subjects from all seasons were randomly divided into training and testing groups in an 85%/15% ratio. For each bootstrapping cycle, we split the young subjects (from all years) as 85% (young training set) and 15% (young testing set) and put all elderly aside (from both 2010 and 2011 years, the elderly testing set). Then, we used the young training set to select the features (in this case the BTMs) and to check their performance in the same young training set. Next, we checked the performance of these “predictive BTMs” in the young testing set and elderly testing set. This process was repeated for 100 trials in order to ensure that the performance was robust across many random divisions of the data. For each of the 100 times that this 85%/15% split in young subjects was done, we tested the prediction on the same elderly testing set. With this approach, we identified BTMs that predicted HAI responses in both the young and elderly groups with accuracies ranging between 79.0% and 80.2% and between 64.7% and 72.3%, respectively, using both day 3 and day 7 signatures (Figure 2C). Examination of the modules most frequently selected by the algorithm in the 100 randomized trials revealed pathways related to innate immune cell responses as well as leukocyte differentiation and antigen presentation on day 3 and B cell/immunoglobulin production and cell cycle pathways on day 7 (Table S3).
Interestingly, we observed similar classification accuracy in both the young and elderly test sets when using the predictive signatures trained on the young dataset (Figure 2C). This suggests that although the elderly had lower antibody responses to vaccination, the molecular signatures that were predictive of high HAI response remained the same. One potential caveat to this was that in the analysis in Figure 2C we had defined young as <65 years and elderly as >65 years, a narrow chronological distinction that could hamper quantitative analysis of age-related effects. To address this issue, we performed additional analysis by removing the intermediate-age subjects (between 40 and 65 years) from our analysis (Figure 2D). Thus here we defined young as subjects younger than 40, and elderly as subjects older than 65. The results are similar to our original work (Figure 2C versus Figure 2D). In addition, as the 2010 cohort has a sharper distinction between the young and the elderly (Figure S1A), we performed GSEA on genes ranked by correlation with HAI response (similar to the analysis of Figure 2A) for young and elderly subjects in the 2010 cohort (Figure S4). There were a number of consistently enriched modules in both the young and elderly, in particular those relating to the interferon response and activation of dendritic cells on day 1 and the plasmablast response on day 7. These results suggest that there were consistent transcriptional signatures associated with the day 28 antibody response in both groups. Together these results demonstrate that module-level features are able to successfully predict vaccine immunogenicity in TIV seasons in both young and elderly vaccinees.
Baseline Signatures Associated with Antibody Responses
We also examined whether there were pre-vaccination transcriptional signatures that were associated with antibody responses. Although there has been previous work aimed at identifying baseline cellular and molecular predictors of influenza vaccine response (Frasca et al., 2010; Furman et al., 2013; Tsang et al., 2014), these studies examined subjects within only a single influenza season, and the robustness of these signatures in predicting immunity to influenza vaccination across multiple seasons has not been determined. First, we performed GSEA (Subramanian et al., 2005) in each season by using genes ranked by correlation between baseline expression and the HAI response. However, this approach resulted in little overlap between seasons, with no module consistently enriched in more than three out of five seasons (Figure S5). This might have been due to a reduced signal-to-noise ratio and increased batch effects owing to the inability to use fold-change expression at baseline. Although we attempted to adjust for batch effects in the day 0 expression data across seasons by using the ComBat software (Johnson et al., 2007), these effects might have not been completely removed.
To increase the power of this analysis so as to detect the more subtle transcriptional signatures at baseline, we repeated the procedure with all five seasons combined. In addition, publicly available data from previously published influenza vaccine studies by Franco et al. (2013) and Furman et al. (2013) were included as independent validation sets (see Supplemental Experimental Procedures for details). This approach revealed several B-cell- and T-cell-related modules whose expression pre-vaccination was positively correlated with an increased antibody response to vaccination in all studies (Figure 3A). In contrast, modules related to monocytes were negatively correlated with antibody responses (Figures 3B–3G). Interestingly, contained within the monocyte-related modules were genes such as TLR4, TLR8, NOD2, and ASGR2, which encode proteins involved in innate sensing, and genes encoding IFN-γR, IL-13R, TIMP2, LYN, SYK, and other molecules involved in inflammation. This supports the concept that inflammatory responses at baseline might be detrimental to the induction of vaccine-induced antibody responses (Haq and McElhaney, 2014; Pawelec et al., 2014). Investigation of the correlation with antibody response at a gene level (Figures 3B–3G) highlights the strength of pathway-level analyses: although the module enrichment was consistent across datasets, the individual genes contributing to this enrichment varied from study to study.
Figure 3. Baseline Signatures Are Associated with the Antibody Response.
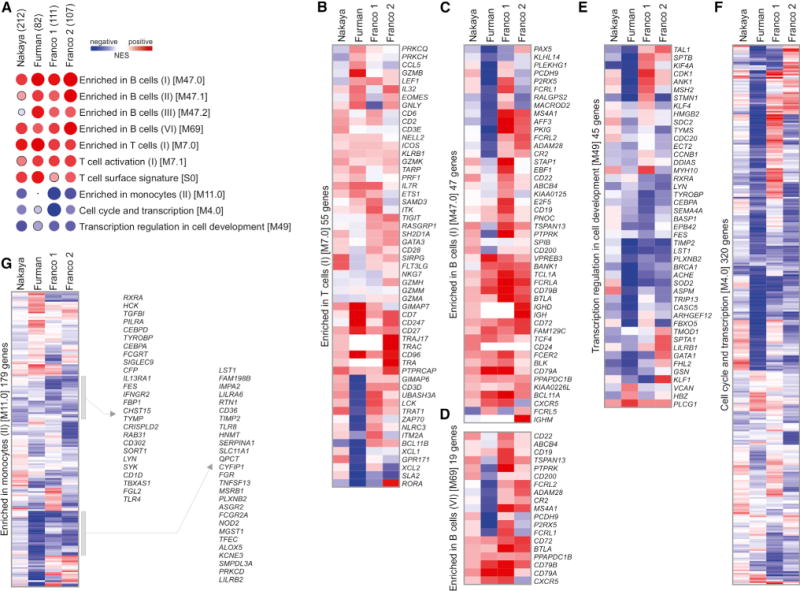
(A) Heat map of BTMs (rows) and TIV studies (columns) whose activity before vaccination is associated with HAI response at day 28 after vaccination. GSEA (nominal p < 0.05; 1,000 permutations) was used to identify positive (red) or negative (blue) enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and HAI response. Circle size is proportional to the normalized enrichment score (NES). Numbers in parentheses next to each study represent number of subjects in the study. Modules shown are those consistently enriched in at least three out of four studies.
(B–G) Heat maps of genes within BTMs from (A); colors represent the mean correlation in each study between baseline gene expression and HAI response at day 28 after vaccination.
See also Figure S5.
Signatures Induced by Vaccination with TIV in Young and Elderly
We next investigated the influence of age on the vaccine-induced transcriptional signatures. To this end, we first identified differentially expressed genes after vaccination in the young versus elderly subjects from the 2010 season (Figure 4A) (in which there is a sharp chronological separation between the young and the elderly [Figure S1A]). By far the largest difference in overall expression between the two groups occurred on day 1, where the young exhibited a much greater number of both up- and downregulated genes. Weighted correlation network analysis (WGCNA) (Langfelder and Horvath, 2008) was used to find clusters of highly correlated genes among the TIV-regulated genes in young (clusters Y1–Y6) and elderly (clusters E1–E5) (Figure 4B). This method compares the correlation in expression patterns among genes across all young or elderly subjects and utilizes hierarchical clustering with dynamic tree cut to define modules of genes that are temporally co-expressed within each group. Genes in common to clusters Y4 and E3 were associated with antibody-secreting cells (ASCs), whereas the overlap between clusters Y1 and E1 contained several interferon-related genes (Figure 4C). Although the genes shared by clusters from young and elderly vaccinated individuals had similar temporal expression profiles, the magnitude of the expression of interferon-related genes differed, being higher in young individuals (Figure 4C).
Figure 4. Molecular Signatures Induced by Vaccination with TIV in Young Adults and in Elderly.
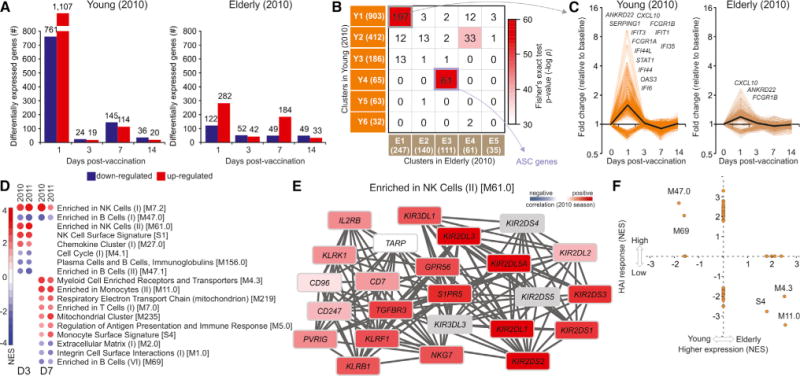
(A) Number of genes differentially expressed (log2 fold-change > 0.2 and t test p value < 0.01) in young (<65 years) (left) and elderly (≥65 years) (right) vaccinees on days 1, 3, 7, and 14 after vaccination (2010 season).
(B) Heat map of highly correlated gene modules within the differentially expressed genes in (A) for young (rows, modules Y1–Y6) and elderly (columns, modules E1–E5), generated by weighted correlation network analysis (Langfelder and Horvath, 2008). The number of genes in each module is shown in parentheses and the number of genes in common between two modules is shown inside the squares. Colors represent the Fisher’s exact test p value of the overlap between clusters. The 61 genes in common between Y4 and E3 are associated with ASCs.
(C) Temporal expression patterns of 197 interferon-related genes in common between modules Y1 and E1 from (B). Black line represents the mean fold change of all genes.
(D) BTMs (rows) whose activity at days 3 or 7 after vaccination (columns) is associated with the age of vaccinees from 2010 and 2011 seasons. GSEA (nominal p < 0.05; 1,000 permutations) was used to identify positive (red) or negative (blue) enrichment of BTMs (gene sets) within pre-ranked gene lists, where genes were ranked according to their correlation between expression and increasing age. The intensity of the color and the size of the circles represent the normalized enrichment score (NES) of GSEA. In this analysis we used age as a quantitative variable, rather than arbitrarily splitting the cohorts young versus elderly. Modules shown are those consistently enriched in both seasons.
(E) Genes in BTM M61.0; each “edge” (gray line) represents a coexpression relationship, as described in Li et al. (2014); colors represent the correlation for 2010 season between baseline-normalized gene expression at day 3 after vaccination and the age of vaccinees.
(F) BTMs whose activity at day 7 after vaccination is correlated with the age of vaccinees (x axis) and/or is correlated with HAI response (y axis) in both 2010 and 2011 seasons. Values represent the mean of the NES obtained independently for each season. NES receives a value of zero if the BTM is not significantly associated with age or HAI response (nominal p < 0.05; 1,000 permutations) in either season.
See also Figures S6 and S7.
We then performed GSEA using genes ranked by their correlation with age to identify modules with enriched expression in either young or elderly vaccinees (Figure 4D). It is important to note that in this analysis we used age as a quantitative variable, rather than arbitrarily splitting the cohorts into young versus elderly. In order to ensure the robustness of our results, we performed this analysis separately on the two trials containing elderly subjects (2010 and 2011) and identified modules that were consistently enriched in both seasons. Natural killer (NK) cell-related modules showed increased expression with age on both day 3 and day 7 after vaccination, and several monocyte modules showed increased expression with age on day 7. In contrast, many B cell modules showed decreased expression in older subjects on both day 3 and day 7. These results indicate that elderly subjects might be mounting a significant innate response, but that their adaptive B cell response is diminished. One of the modules showing increased expression in the elderly was BTM M61.0 (Figure 4E), which contains many killer cell immunoglobulin-like receptor (KIR) and lectin-like receptor (KLR) genes. These are inhibitory receptors that suppress the cytotoxic activity of NK cells when bound to MHC I molecules (KIRs) and cadherins and other ligands (KLRs) (Long et al., 2013).
Next, we examined whether or not any of the age-related transcriptional differences were also associated with impaired antibody responses. By plotting the age-based enrichment score of each module against its antibody response-based enrichment score on day 7, we were able to identify five modules whose expression varied with age and were associated with either a high or low antibody response (Figure 4F). Two B cell modules, M47.0 and M69, showed increased expression in younger subjects and in subjects with high antibody responses, whereas three monocyte- and myeloid-cell-related modules, S4, M4.3, and M11.0, were enriched in the elderly and were also associated with a decreased antibody response in the 2010 and 2011 seasons. Interestingly, two of these modules, M11.0 and M4.3, showed association with an increased antibody response in the 2007 and 2008 seasons (Figure 2A), indicating that this association is not consistent across all five seasons on day 7. These results demonstrate that TIV induces distinct but overlapping transcriptional responses in the young versus elderly. The early innate response at day 1, comprised of antiviral and type IFN-related genes, seems to be impaired in the elderly. In contrast, several transcriptional modules related to NK cells and monocytes appear to have enhanced expression at baseline and after vaccination, in the elderly relative to the young.
Cellular Responses Induced by Vaccination with TIV in the Young and Elderly
The aforementioned transcriptional signatures of NK cells and monocytes in the elderly subjects could have been caused by de novo transcriptional induction of genes expressed in these cell types or by changing representations of these cell types within the PBMC compartment. In order to distinguish between these two possibilities, we performed FACS analysis on PBMCs in a subset of subjects from the 2010 cohort. Our analysis showed that proportions of total NK cells in elderly subjects were higher than those of young subjects at baseline as well as at all the time points studied (days 0–14). Of note, vaccination induced an increase in the percentage of NK cells in the elderly subjects; in contrast, in the young there was a reduction in the percentage of NK cells on day 1 after vaccination (Figure 5A). Similar trends were observed in the absolute numbers of NK cells (data not shown). To examine this trend in the 2011 cohort, where FACS data were unavailable, we used a deconvolution method (Abbas et al., 2009; Gaujoux and Seoighe, 2013; Shen-Orr and Gaujoux, 2013) to estimate cell frequencies based on gene expression of a mixed cell population. This approach revealed increased estimated NK cell frequencies in the elderly on day 3 after vaccination when compared to young, agreeing with the 2010 FACS data (data not shown). These results confirm our findings at a molecular level (Figure 4D) and show that the increase in NK-cell-related expression in the elderly is due in part to enhanced representation of this cell population after vaccination.
Figure 5. Flow Cytometry Analysis of NK Cells in Young and Elderly after TIV Vaccination.
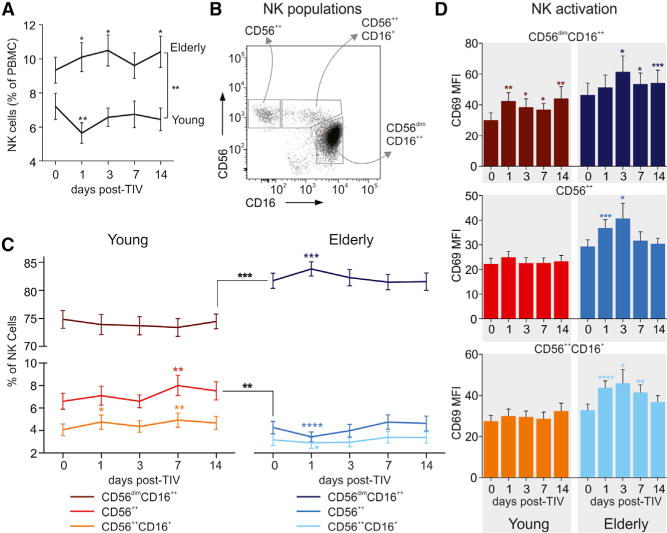
(A) Changes in total NK cell population after vaccination represented as percent within all PBMCs for young and elderly. Mean ± SEM.
(B) Blood NK cells were defined within the CD3−CD4−CD19−CD14− PBMCs. Dot blot represents three distinct NK cell populations defined by CD56 and CD16 markers: CD56++ NK, CD56++CD16+ NK, and CD56dimCD16++ NK.
(C) Kinetics of magnitudes of CD56++ NK, CD56++CD16+ NK, and CD56dimCD16++ NK cell subsets in young (left) and elderly (right) after vaccination. Mean ± SEM.
Areas under curve (AUC) in (A) and (C) were calculated to compare magnitudes of total NK cells and NK cells subsets between young and elderly cohorts throughout the study duration (days 0–14) and compared by t test. Changes of each of the NK subset on the indicated time points after vaccination were compared to the day 0 (baseline) time point by t test.
(D) Activation of each of the NK cell subsets were assessed by CD69 staining and compared with day 0 (baseline) by t test. Data are represented as the geometric mean fluorescence intensity (MFI) for young (left) and elderly (right) at each time point, mean ± SEM.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We also explored the distribution of the NK compartment into three subpopulations: CD56++ NK (CD56bright), CD56++CD16+ NK, and CD56dimCD16++ NK (Figure 5B). These NK cell subsets have distinct functions: CD56dimCD16++ cells, the most abundant population of NK cells in the blood, have significantly higher cytotoxic activity, whereas CD56++ cells are characterized by increased cytokine production and can exhibit immunoregulatory properties under certain conditions (Poli et al., 2009). We observed a higher frequency of CD56dimCD16++ cells coincident with a lower frequency of CD56++ cells in the elderly at baseline, consistent with previous studies (Figure 5C; Solana et al., 2014). The distribution of these subpopulations was relatively stable during the response, with CD56dimCD16++ cells showing a small but significant increase in the elderly on day 1 after vaccination and CD56++ cells increasing in the young on day 7. Additionally, we examined the activation of these subsets through immunostaining for CD69, a marker associated with increased cytotoxicity and IFN-γ production (Figure 5D; Clausen et al., 2003; Gorski et al., 2006). Elderly subjects showed increasing activation after vaccination in all three subsets, with highest expression on day 3. In contrast, the young exhibited activation of the CD56dimCD16++ subset as measured by CD69 expression only on day 1 and at a lower level than in the elderly.
In addition to NK cells, we also observed higher proportions of monocytes among the elderly at baseline and during the entire duration of the study compared to young subjects (Figure S6A). In both groups, the percentage of total monocytes in PBMCs increased substantially on day 1 after vaccination, then returned to or below baseline levels. We further examined the distribution of monocytes into three subsets: “classical” (CD14+CD16−), “intermediate” (CD14+CD16+), and “non-classical” (CD14dimCD16++) (Figure S6B). The distribution of monocyte subsets was similar in both groups, with the elderly having a slight increase in intermediate monocytes compared to young (Figure S6C). The activation of these subsets was also analyzed by immunostaining for CCR5, a receptor for a number of inflammatory cytokines, and CD86, a costimulatory molecule involved in T cell activation (Figure S6D). In both the young and elderly, CCR5 expression peaked in classical and intermediate monocytes on day 1 after vaccination, corresponding with the inflammation associated with the innate immune response. Elderly subjects showed increased CCR5 expression in classical and intermediate monocytes compared to the young, whereas young subjects had higher CD86 expression in these subsets. These results are consistent with the transcriptional changes observed (Figure 4) and support the concept that persistent and excessive inflammatory responses might be detrimental to the induction of vaccine-induced antibody responses (Frasca et al., 2014; Haq and McElhaney, 2014; Pawelec et al., 2014).
Molecular Signatures Associated with the Persistence of Antibody Responses
A desirable feature of a good vaccine is the ability to induce long-lasting protection from infection. To investigate how effectively the influenza vaccine generates a long-lived response and identify the mechanisms responsible for this response, we measured antibody titers in a subset of subjects at 180 days (D180) after vaccination. The antibody responses peaked at day 28 (D28), with a significant decline in most subjects by D180 (Figure 6A). By applying the same criteria for seroconversion used for the D28 high- versus low-responder classification to the D180 antibody responses, we saw that just over half of D28 high responders maintained their responder status on day 180 (“persistent” responders), whereas the remainder no longer met the criteria for seroconversion on day 180 (“temporary” responders). Because the D180 antibody titer shows significant dependence on the original antibody response generated on day 28 (Figure 6B), examining the molecular profile associated with the D180 response would identify many of the mechanisms responsible for generating the D28 response. In order to study the relative persistence of the response, we calculated residuals from a linear fit between the D28 and D180 responses, thereby removing the effect of the D28 response on the D180 response. We observed that subjects with a higher D28 titer had a greater decrease in HAI response between D28 and D180, indicating increased difficulty to maintain higher levels of antibodies. The linear fit therefore represents the expected D180 response given a particular initial D28 response, and the residual can be considered an “adjusted” D180 response taking into account the original D28 response. Subjects with positive residuals have a more persistent response, whereas a negative residual represents a waning response. As expected, correlation analysis of plasmablast module BTM S3 shows a positive association with the D180 antibody response due to the effect of D28 antibody responses, generated through plasmablast expansion (Figure 6C). However, when we performed correlation between the plasmablast module activity and the D180/D28 residual, we no longer saw an association with the plasmablast expression (Figure 6D). This result suggests that factors other than the expansion of plasmablasts are responsible for maintaining a persistent antibody response.
Figure 6. Signatures Associated with the Persistence of TIV-Induced Antibody Response.
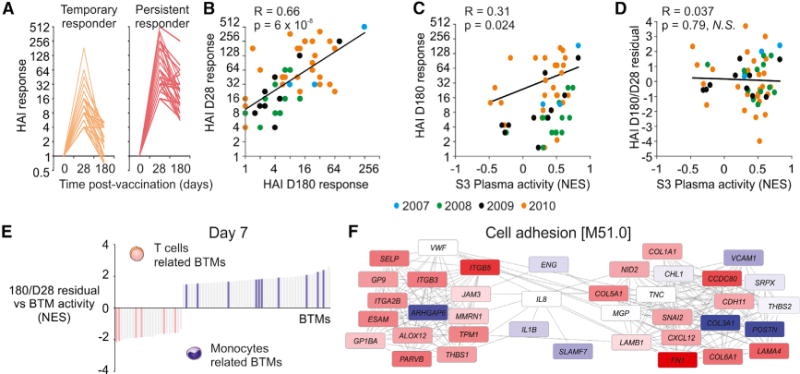
(A) HAI response (fold change of HAI titer relative to baseline) through 180 days for temporary (n = 28) and persistent (n = 34) responders. Temporary responders met the FDA criteria for seroconversion (minimum 1:40 titer and 4-fold increase after vaccination) on day 28 but not on day 180, whereas persistent responders met the criteria on both days.
(B) Comparison between day 28 and day 180 HAI responses. Each symbol represents a single vaccinee and the color represents the season that they were vaccinated (n = 62). Black lines represent the regression line (Pearson) for all vaccinees combined. Day 180/day 28 residual is computed as the (vertical) distance from each sample to the regression line.
(C) Comparison between “S3 Plasma” BTM activity and the HAI responses at day 28 after vaccination. Each symbol represents a single vaccinee and the color represents the season that they were vaccinated (n = 62). Black lines represent the regression line (Pearson) for all vaccinees combined.
(D) Comparison between “S3 Plasma” BTM activity and the HAI D180/D28 residual. Each symbol represents a single vaccinee and the color represents the season that they were vaccinated (n = 62). Black lines represent the regression line (Pearson) for all vaccinees combined.
(E) Genes in BTM M51.0; each “edge” (gray line) represents a coexpression relationship, as described in Li et al. (2014); colors represent the correlation between baseline-normalized gene expression at day 3 after vaccination and the D180/D28 residual.
(F) BTMs (bars) whose activity at day 7 after vaccination is significantly associated with the HAI D180/D28 residual (GSEA; nominal p < 0.05; 1,000 permutations). Vaccinees from 2007 to 2010 seasons were combined. BTMs related to T cell functions (pink bars) or monocyte functions (purple bars) are shown.
To identify the pathways associated with a persistent or waning response, we performed GSEA on genes correlated with the D180 versus D28 residual. Among the BTMs most enriched in subjects with a persistent response on both days 3 and 7 were several modules associated with cell movement and adhesion, such as BTM 51 (Figure 6E). In fact, one of the genes in module M51 that was highly correlated with the response persistence was P-selectin (SELP), which plays an important role in the adhesion and extravasation of leukocytes out of the blood circulation and into organs or tissues (Wang et al., 2007). In addition, the day 7 expression of a number of T-cell-related modules was negatively associated with the D180/D28 residual, indicating that increased T cell responses might potentially result in more transient antibody responses (Figure 6F). The plasmablast signature at day 7, which was predictive of the HAI titers at day 28, did not correlate with the D180/D28 residual (Figure 6D).
Post-transcriptional Gene Regulation of the Immune Response to Vaccination
MicroRNAs (miRNAs) have been identified as key regulators of gene expression at a post-transcriptional level (Filipowicz et al., 2008). Although there is some knowledge about miRNA regulation of immune pathways (Lodish et al., 2008), their role in responses to vaccination remains unaddressed. To assess whether or not miRNAs contribute to the differences in response we saw between young and elderly vaccinees, we measured the miRNA expression profiles of 672 human miRNAs on days 1, 3, and 7 after vaccination from a subset of subjects in the 2010 cohort (Figure 7A). We saw a significant difference in the miRNA profiles between these two groups. Whereas a majority of the differentially expressed miRNA in the elderly were upregulated after vaccination, particularly on days 1 and 7, the young exhibited predominantly downregulation of miRNA. We also identified miRNA whose expression correlated with the day 28 antibody response (Figure 7B). We again saw differences between the young and elderly subjects, with the elderly having a larger number of miRNA negatively correlating with the antibody response when compared with the young. These results suggest that miRNA might indeed be important regulators of the immune response to influenza vaccination. miRNA expression reduces the translation of its target genes through mRNA degradation or silencing via the RNA-induced silencing complex, so the increase in miRNA expression in the elderly after vaccination suggests a possible mechanism for the impaired responses in this population that merits further exploration.
Figure 7. MicroRNA Expression Profiling of Young and Elderly TIV Vaccinees.
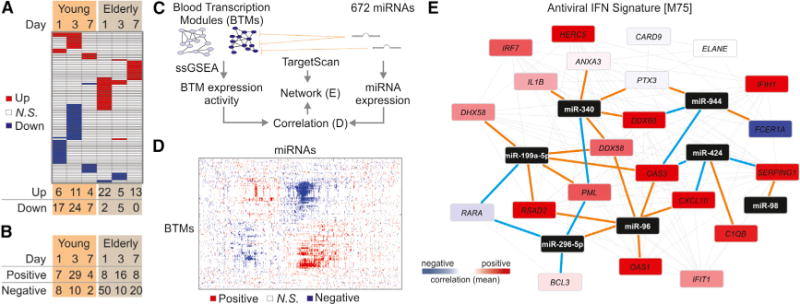
(A) Heat map of miRNAs (rows) up- (red) or downregulated (blue) at days 1, 3, and 7 after vaccination in young and elderly (columns); paired t test (p < 0.05); total number of differentially expressed miRNAs are shown at the bottom.
(B) MicroRNAs whose expression is positively or negatively correlated with HAI response in young and elderly; Pearson correlation (p < 0.05).
(C) Identification of networks potentially regulated by miRNAs. Activity of BTMs was determined by single-sample GSEA (Barbie et al., 2009) and correlated with the expression of miRNAs. TargetScan database (Garcia et al., 2011) was used to identify the potential target genes of miRNAs.
(D) Heat map of BTMs (rows) whose activity at day 1 after vaccination correlated with the baseline-normalized expression of miRNAs (columns) at the same time point. Positive and negative correlations are shown in red and blue, respectively.
(E) Genes in BTM M75; each gray line represents a coexpression relationship, as described in Li et al. (2014); each brown line connects a miRNA and its potential target gene; each blue line represents a negative correlation (Pearson, p < 0.15) between the expression of miRNA and the expression of the potential target gene; colors represent the mean correlation between baseline-normalized gene expression at day 1 and HAI response at day 28 after vaccination in the 2010 TIV season.
To identify the miRNA that regulate particular immune pathways during vaccine response, we implemented a strategy to integrate the mRNA and miRNA omics-level measurements (Figure 7C). Because miRNA often have multiple related target genes, we first generated normalized enrichment scores for 252 BTMs on a per subject basis by performing ssGSEA on the transcriptomic data. We then performed correlation analysis between all miRNA-BTM pairs, revealing groups of modules that appear to be regulated together by several miRNA (Figure 7D) on day 1 after vaccination.
Finally, we used the TargetScan miRNA database (Garcia et al., 2011) to identify the genes in a given module predicted to be targets of negatively correlating miRNAs. Of particular interest was the regulation of module M75-antiviral interferon signature (Figure 7E) on day 1 after vaccination, because the interferon pathway plays a key role during the innate immune response. This miRNA-mRNA network suggests potential novel regulators of the interferon response after vaccination, such as miR-424. This miRNA was predicted to target OAS3, a 2′–5′-oligoadenylate synthase involved in viral RNA degradation (Samuel, 2001), as well as CXCL10, an important chemokine induced by IFN-γ that serves as a chemoattractant for lymphocytes (Dufour et al., 2002).
DISCUSSION
In this paper we were able to identify shared and consistent molecular signatures of immunogenicity to TIV, across five influenza vaccine seasons. At day 1 after vaccination, the pathways positively associated with the later antibody response revealed a strong innate response marked by expression of interferons and activation of dendritic cells. This response was also observed on day 3, with enrichment of TLR signaling and antigen presentation. By day 7, there was a strong signature from the expansion of plasmablasts. Using the same modules in a single sample approach as inputs to an artificial neural network classifier allowed us to successfully predict antibody responses to vaccination across all seasons included in our study. Using pathway-level features as predictors of immune response is a promising approach to not only reduce the variability in gene expression across influenza seasons but also to provide improved biological context to the predictive signatures.
In addition, we were able to establish baseline signatures related to B and T cell expression that were associated with an increased antibody response at day 28 after vaccination. These signatures were validated in two independent datasets, confirming that they are not specific to strain or study and are consistent with previous findings that showed that the frequency of B cell and T cell subsets at baseline was predictive of day 28 antibody responses (Tsang et al., 2014). Therefore, the increased statistical power of our dataset allowed us to identify relatively weak baseline transcriptional differences that were detectable only using flow cytometry in previous studies.
Our analysis of responses in young and elderly subjects revealed important aspects about the relationship between immunosenescence and vaccine response. We observed that although the elderly exhibit diminished B cell responses after vaccination, they also have enhanced NK cell and monocyte numbers compared to young. Furthermore, the elderly displayed increased frequency and activation of cytotoxic NK cells, as well as increased CCR5 but diminished CD86 on proinflammatory monocytes. Previous studies have shown that although the NK cell population increases with age, there are concurrent changes in the receptors of these cells, including decreases in the activating receptors NKp30 and NKp46 (Almeida-Oliveira et al., 2011). NKp30 is involved in crosstalk with dendritic cells (Walzer et al., 2005), and NKp46 has been shown to bind to influenza hemagglutinin to allow NK-cell-mediated recognition of influenza-virus-infected cells (Mandelboim et al., 2001), suggesting potential mechanisms by which NK cells might regulate adaptive immunity. Most importantly, monocytes were also increased in the elderly before vaccination, and our baseline analysis revealed a negative association between day 0 monocyte expression and magnitude of antibody response. This indicates a potential connection between the baseline state of the immune system in the elderly and reduced responsiveness to vaccination. Together these results suggest potential mechanisms by which changes to the innate response in the elderly might result in diminished antibody responses to vaccination.
In addition, we also investigated the signatures associated with the longevity of antibody responses. Although the goal of all vaccines is to induce lasting immunity, waning immunity to vaccination is a major issue in vaccinology (Pichichero, 2009). Indeed, we observed a significant drop in antibody titers in a majority of subjects within 6 months of vaccination. By comparing the relative persistence of the day 180 antibody responses with BTM-normalized enrichment scores, we saw that the expansion of plasmablasts, which plays an important role in the development of the day 28 antibody response, had little effect on response longevity. Instead, we observed a potential role for cell movement and adhesion in maintaining a persistent antibody response. This transcriptional signature might be an indicator of migration of plasmablasts into the bone marrow, a crucial step in the generation of long-lived plasma cells (Radbruch et al., 2006). This analysis demonstrates the role that initial immune processes, as early as 3 days after vaccination, can have in shaping the antibody response as much as 6 months later.
Finally, we sought to explore how the previously examined transcriptional responses to vaccination might be regulated, in particular through the expression of miRNAs. miRNAs have been shown to play an important role in modulating the signaling pathways of the immune system (Lodish et al., 2008), but their role in vaccine response has not been studied. By integrating transcriptomic and miRNA expression data, we were able to identify several potential miRNA regulators of the interferon response after vaccination, such as miR-424. Previous studies have shown that absence of regulation of interferon signaling by miRNA can result in impaired CD8 T cell responses (Gracias et al., 2013). These results demonstrate how the balance between positive and negative signals controlling the innate response is necessary for a successful adaptive response and underscore the importance of understanding the way in which miRNAs help achieve this balance.
In summary, this longitudinal study across five influenza seasons provided the opportunity to identify conserved molecular elements present in immune response to TIV vaccination. Our pathway-level analyses revealed previously unknown mechanisms that contribute to impaired responses to influenza vaccination in the elderly. These systems approaches can help delineate the molecular responses to vaccination in other immunocompromised populations, such as infants and HIV-positive individuals. They might also offer insights into the pathways by which adjuvants improve vaccine response. By integrating this knowledge, we will be able to provide a more complete picture of how the immune system responds to vaccination and help guide the development of the next generation of vaccines that provide long-lasting immunogenicity and better protection of at-risk populations.
EXPERIMENTAL PROCEDURES
Human Subjects and Specimen Collection
A total of 212 subjects were vaccinated with TIV during the 2007–2011 influenza seasons. Written informed consent was obtained from each subject with institutional review and approval from the Emory University Institutional Review Board. Blood samples were collected at baseline and on days 3, 7, 28, and 180 days after vaccination. Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood and stored in liquid nitrogen (−210°C). Detailed vaccination and sample collection procedures can be found in the Supplemental Experimental Procedures. In general, the health of all subjects was under control with no severe symptoms. Presence of relevant co-morbidities was recorded and T2D patients did not have cancer, or major infectious or autoimmune diseases, for at least 12 months prior to enrollment. Diabetic individuals were 20 years or older and with an established diagnosis of T2D for more than 6 months. Individuals had to be free of influenza and of any symptoms associated with respiratory infections at the time of enrollment. All T2D patients were under metformin treatment. Pregnancy and documented current substance and/or alcohol abuse were also exclusion criteria, as well as use of medicines known to alter the immune response, such as high-dose corticosteroids, within 6 months prior to enrollment. As “co-morbidities,” a few patients had hypertension, hyperlipidemia, pain (joint, back), or hypothyroidism.
RNA Isolation and Microarray Analysis
Total RNA from PBMCs (~1.5 × 106 cells) was purified using Trizol (Invitrogen, Life Technologies Corporation) according to the manufacturer’s instructions. Samples were checked for purity and hybridized on Human U133 Plus 2.0 Arrays (Affymetrix). Microarray intensity data were normalized by RMA (Irizarry et al., 2003) for each cohort separately. Pathway analyses were performed using GSEA (Subramanian et al., 2005). In this 5-year analysis, we utilized a multi-step DAMIP strategy as described in the Supplemental Experimental Procedures (Figure S3). Classification rules or signatures associated with three to five discriminatory genes were identified. We reported the classification rules in which the predictive accuracies of HAI response in all other seasons are at least 70%. This resulted in a total of 175 predictive rules, with accuracy ranging between 80.0% and 87.3% (Table S1, Figure S3). For each of these rules, a second layer of blind prediction was then performed on the remaining blind test set (15% of subjects from each season) (Table S2). Detailed description of analyses can be found in the Supplemental Experimental Procedures.
Flow Cytometry Analysis
2–3 × 106 PBMCs were thawed and stained with live/dead marker (Alexa Fluor 430, Life Technologies) to exclude dead cells. Cells were then stained with an appropriate antibody cocktail. Cells were washed, fixed, and analyzed on the LSRII flow cytometer (BD). All flow cytometry analysis was done with FlowJo (Treestar). Blood NK cells were defined within the live singlets gate CD3−CD4−CD19−CD14− cells as the CD56++ NK, CD56++CD16+ NK, and CD56dimCD16++ NK. The CD56++CD16+ subset is considered an intermediate population in transition from the CD56++ to the CD56dimCD16++ subset (Béziat et al., 2011). Monocytes were defined within the live singlet CD3−CD19−PBMC as the CD14+CD16−, CD14+CD16+, and CD14dimCD16++ monocytes. The detailed staining procedures and list of antibodies are presented in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Systems analysis of vaccine immunity to influenza across seasons and populations
Signatures of innate immunity and plasmablasts predicted influenza antibody titers
Certain vaccine-induced signatures were shared among all analyzed populations
Defined baseline signatures of immunogenicity that might help guide vaccine design
Acknowledgments
We are grateful to Mary Bower, Srilatha Edugupanti, and the Hope Clinic Staff who assisted with the clinical work. We also thank Emory Immunology/Flow Cytometry Core for technical assistance. H.I.N. receives a CNPq (Brazilian Research Council) research fellowship. This work was supported by NIH grants U19AI090023 (B.P.), U19AI057266 (R.A. and B.P.), HHSN272201400004C (B.P.), U54AI057157 (B.P.), R37AI48638 (B.P.), and R37DK057665 (B.P.) and NSF Grant STC-0939370 (S.S.).
Footnotes
ACCESSION NUMBERS
The GEO accession numbers for the microarray data reported in this paper are GSE: GSE74817 (influenza seasons 2009–2012) and GSE29619 (influenza seasons 2007–2009).
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, four tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2015.11.012.
AUTHOR CONTRIBUTIONS
H.I.N., S.S.D., M.K., N.R., D.F., A.K.M., G.-M.L., R.A., M.J.M., and B.B.B. collected, processed, and prepared samples and assisted with the analyses. H.I.N. and T.H. performed most of the microarray analyses. E.K.L. performed the DAMIP model analyses. T.H. performed the neural network predictive analyses. S.S.D. and M.K. performed the FACS analyses. R.G. and S.S.-O. performed the deconvolution analyses. S.S. and B.P. supervised the analyses. M.G. and S.G. assisted with analysis of microarray data. B.P., R.A., N.R., and M.J.M. conceived the study and designed the experiments. H.I.N., T.H., S.S., and B.P. wrote the paper.
References
- Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS ONE. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Oliveira A, Smith-Carvalho M, Porto LC, Cardoso-Oliveira J, Ribeiro AdosS, Falcão RR, Abdelhay E, Bouzas LF, Thuler LCS, Ornellas MH, Diamond HR. Age-related changes in natural killer cell receptors from childhood through old age. Hum Immunol. 2011;72:319–329. doi: 10.1016/j.humimm.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat V, Duffy D, Quoc SN, Le Garff-Tavernier M, Decocq J, Combadière B, Debré P, Vieillard V. CD56brightCD16+ NK cells: a functional intermediate stage of NK cell differentiation. J Immunol. 2011;186:6753–6761. doi: 10.4049/jimmunol.1100330. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, Berger P, Desborough V, Smith T, Campbell J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- Brooks JP, Lee EK. Analysis of the consistency of a mixed integer programming-based multi-category constrained discriminant model. Ann Oper Res. 2010;174:147–168. [Google Scholar]
- Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao RG, Suarez NM, Obermoser G, Lopez SM, Flano E, Mertz SE, Albrecht RA, García-Sastre A, Mejias A, Xu H, et al. Differences in antibody responses between trivalent inactivated influenza vaccine and live attenuated influenza vaccine correlate with the kinetics and magnitude of interferon signaling in children. J Infect Dis. 2014;210:224–233. doi: 10.1093/infdis/jiu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J, Vergeiner B, Enk M, Petzer AL, Gastl G, Gunsilius E. Functional significance of the activation-associated receptors CD25 and CD69 on human NK-cells and NK-like T-cells. Immunobiology. 2003;207:85–93. doi: 10.1078/0171-2985-00219. [DOI] [PubMed] [Google Scholar]
- Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35:352–359. doi: 10.1016/s1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Duraisingham SS, Rouphael N, Cavanagh MM, Nakaya HI, Goronzy JJ, Pulendran B. Systems biology of vaccination in the elderly. Curr Top Microbiol Immunol. 2013;363:117–142. doi: 10.1007/82_2012_250. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Franco LM, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, Arden N, Renwick A, Yu P, Quarles JM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D, Diaz A, Romero M, Landin AM, Phillips M, Lechner SC, Ryan JG, Blomberg BB. Intrinsic defects in B cell response to seasonal influenza vaccination in elderly humans. Vaccine. 2010;28:8077–8084. doi: 10.1016/j.vaccine.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D, Diaz A, Romero M, Landin AM, Blomberg BB. High TNF-α levels in resting B cells negatively correlate with their response. Exp Gerontol. 2014;54:116–122. doi: 10.1016/j.exger.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29:2211–2212. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- Gorski KS, Waller EL, Bjornton-Severson J, Hanten JA, Riter CL, Kieper WC, Gorden KB, Miller JS, Vasilakos JP, Tomai MA, Alkan SS. Distinct indirect pathways govern human NK-cell activation by TLR-7 and TLR-8 agonists. Int Immunol. 2006;18:1115–1126. doi: 10.1093/intimm/dxl046. [DOI] [PubMed] [Google Scholar]
- Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry EJ, Turner M, Katsikis PD. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq K, McElhaney JE. Immunosenescence: Influenza vaccination and the elderly. Curr Opin Immunol. 2014;29:38–42. doi: 10.1016/j.coi.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kang I, Hong MS, Nolasco H, Park SH, Dan JM, Choi JY, Craft J. Age-associated change in the frequency of memory CD4+ T cells impairs long term CD4+ T cell responses to influenza vaccine. J Immunol. 2004;173:673–681. doi: 10.4049/jimmunol.173.1.673. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Nakaya HI, Kazmin DA, Oh JZ, Pulendran B. Systems biological approaches to measure and understand vaccine immunity in humans. Semin Immunol. 2013;25:209–218. doi: 10.1016/j.smim.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G, Goldeck D, Derhovanessian E. Inflammation, ageing and chronic disease. Curr Opin Immunol. 2014;29:23–28. doi: 10.1016/j.coi.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- Pichichero ME. Booster vaccinations: can immunologic memory outpace disease pathogenesis? Pediatrics. 2009;124:1633–1641. doi: 10.1542/peds.2008-3645. [DOI] [PubMed] [Google Scholar]
- Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126:458–465. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B. Systems vaccinology: probing humanity’s diverse immune systems with vaccines. Proc Natl Acad Sci USA. 2014;111:12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbruch A, Muehlinghaus G, Luger EO, Inamine A, Smith KG, Dörner T, Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, Park Y, Jones DP, Chappert P, Davoust J, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, Nishtala M, Wrammert J, Smith K, James JA, et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest. 2011;121:3109–3119. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JC, Richard SA, Viboud C, Miller MA. Quantitative review of antibody response to inactivated seasonal influenza vaccines. Influenza Other Respi Viruses. 2012;6:52–62. doi: 10.1111/j.1750-2659.2011.00268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165:265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- Solana R, Campos C, Pera A, Tarazona R. Shaping of NK cell subsets by aging. Curr Opin Immunol. 2014;29:56–61. doi: 10.1016/j.coi.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SJ, Jacobson R, Poland GA. Advances in the vaccination of the elderly against influenza: role of a high-dose vaccine. Expert Rev Vaccines. 2010;9:1127–1133. doi: 10.1586/erv.10.117. [DOI] [PubMed] [Google Scholar]
- Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagar LE, Gentleman B, Pircher H, McElhaney JE, Watts TH. Influenza-specific T cells from older people are enriched in the late effector subset and their presence inversely correlates with vaccine response. PLoS ONE. 2011;6:e23698. doi: 10.1371/journal.pone.0023698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: “l’union fait la force”. Blood. 2005;106:2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- Wang HB, Wang JT, Zhang L, Geng ZH, Xu WL, Xu T, Huo Y, Zhu X, Plow EF, Chen M, Geng JG. P-selectin primes leukocyte integrin activation during inflammation. Nat Immunol. 2007;8:882–892. doi: 10.1038/ni1491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


