Abstract
Autophagy is a conserved biological process for digestion and recycling of cytoplasmic constituents in eukaryotic cells. Autophagy may trigger cell death or promote cell survival following various forms of stress. The emerging roles of autophagy in megakaryopoiesis, thrombopoiesis, and platelet function have been uncovered using not only in vitro and in vivo genetic models, but also in clinical observations of autophagic structure in patients with thrombocytopenic disorders. Inhibition of autophagy in early stage of megakaryocyte differentiation appears to impede megakaryocyte maturation, reduce platelet formation, and affect platelet function, whereas autophagic deficiency in mature megakaryocytes gives rise to abnormal platelet activation and function without changing platelet size and number. On the other hand, induction of autophagy by rapamycin in megakaryocytes exhibited substantial therapeutic benefits in patients with immune thrombocytopenic purpura (ITP). This mini-review is to highlight recent progresses in understanding the regulation of autophagy in megakaryopoiesis, thrombopoiesis and platelet function to bridge the gap between autophagy and megakaryocyte/platelet pathophysiology.
Keywords: Autophagy, megakaryopoiesis, thrombopoiesis, platelets
Introduction
Autophagy is a conserved catabolic process in eukaryotic cells. During autophagy, targeted cytoplasmic components undergo sequestration by double membrane vesicles called autophagosomes, which then deliver their contents to lysosomes for degradation or recycle [1]. Since its discovery in 1960s, autophagy has been flourishingly investigated and comprehensively linked to various biological processes and pathological conditions. According to different transportation and substrates, autophagy can be divided into three distinct forms termed macroautophagy, microautophagy and chaperone-mediated autophagy, of which macroautophagy consists of pexophagy, mitophagy and non-selective autophagy [2].
The crucial roles of autophagy have been implicated in nutrient starvation, infection, cell death and repair [3-5]. Autophagy is considered to play a mainly protective role in cellular stress response by removing aggregated protein and recycling degraded products [6]. On the flip side of the coin, autophagy may trigger programmed cell death under certain conditions [7]. The complete process of autophagy, which includes activation, cargo identification, autophagosome formation, lysosome fusion and degradation, is mediated by a series of autophagy-related genes (ATGs) [8]. Over the past decades, the pleiotropic roles of autophagy in the development of hemocytes have been characterized in erythropoiesis, as well as in blood cancers [9,10]. Recently, autophagy has been demonstrated to be indispensable for normal megakaryopoiesis and platelet function using animal models with lineage specific deletion of ATG [11,12]. In addition, accumulating evidence of autophagy has hitherto been noted in ITP, myelodysplastic syndromes, and chronic myelogenous leukemia since the initial discovery of putative autophagic vacuoles from megakaryocytes in 1970s [13-15]. In the present review, we will first discuss the molecular basis of the autophagic machinery briefly and then move on to more specific regulation of megakaryopoiesis and thrombopoiesis by autophagic modulators, followed by touching upon the clinical relevance of autophagic regulation and thrombocytopenic disorders.
Autophagic signaling
The process of autophagy was initially discovered by Porter et al in 1962 from glucagon treated liver cells, in which lysosomes containing other organelles were observed [16]. Further discoveries in injury response and recycling/degradation of cellular constituents led to the invention of the term “autophagy” by de Duve [17]. Amongst all the three types of autophagy identified, macroautophagy is the canonical pathway that has been most extensively studied [18]. Damaged organelles are subjected to sequestration by double layered autophagosomes that subsequently deliver their contents for acidic hydrolysis and degradation by fusing with lysosomes [2]. Microautophagy, to some extent, resembles macroautophagy in spite of direct engulfment of intracellular constituents by lysosomes [19]. On the other hand, chaperone-mediated autophagy operated in a quite different manner involving the hsc70-containing complex with high selectivity for substrates [20].
Autophagy-related genes (ATGs) are the key regulators of autophagic signaling. Originally cloned from the yeast Saccharomyces cerevisiae, the homologues of ATGs in mammalian cells have been identified, and their functions unveiled [21,22]. In mammals, nutritional deprivation, growth factors, and oxidative stress can regulate autophagy through AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) [23,24]. In the setting of starvation, increased ratio of AMP/ATP activates AMPK, which then induces cellular protective autophagy through inhibition of mTORC1 [25]. UNC-51 like kinase 1 (ULK1), focal adhesion kinase family interacting protein of 200 kD (FIP200), ATG101 and ATG13 form a protein complex with mTORC1 that is an inhibitor of autophagy [26]. Increased formation of this complex following dephosphorylation of ATG13 induced by metabolic stress or rapamycin promotes autophagy [27]. Similarly, activated UKL1 phosphorylates Beclin-1, which forms a complex with ATG14L, P150, and phosphatidylinositol 3-kinase VPS34 (Vsp34), to initiate the formation of autophagosomes [28]. Once Vsp34 is activated, the generation of phosphatidylinositol 3-phosphate (PtdInS(3)P) will recruit WD repeat domain phosphoinositide-interacting protein 2 (WIPI2) to the surface of the phagophore through binding with ATG16L [29]. Concurrently, ATG12 and ATG5 bind to ATG16L thus forming an E3-like complex, which then binds to ATG3 and promotes autophagosome nucleation. On the other hand, activated ATG3 covalently binds to LC3, which is lipidated by ATG16L and conjugates with PE on the membrane of autophagosomes [30]. During this step, p62 pinpoints specific organelles as a cargo docking receptor, as well as guiding LC3 into autophagosomes [31]. Eventually, the autophagosomes fuses with the acidic lysosomes to form autolysosome, from which the outside LC3 molecules are conversed to the cleaved form by ATG4, while the inner LC3 and cargos undergo degradation [32].
Recently, the roles of autophagy in hematopoiesis have been recognized. Several studies demonstrate that autophagy is involved in both megakaryopoiesis and erythropoiesis, for instance, in differentiation of reticulocytes to red blood cells, as well as in thrombopoiesis [11,33,34]. Induction of autophagy is associated with cell death and differentiation in chronic myeloid leukemia cell line, when the efficacy of induction of autophagy has been tested in thrombocytopenic disorders [35,36].
Megakaryopoiesis and thrombopoiesis
Megakaryopoiesis is a complicated process mediated by different hematopoietic cells and extracellular factors. Hematopoietic stem cells are committed to megakaryocyte lineage, including mixed progenitor colony (CFU-Mix), or common myeloid progenitor (CMP), colony-forming unit-granulocyte-erythrocyte-monocyte-megakaryocyte (CFU-GEMM), mixed MK/erythroid progenitor cell (MEP), colony-forming unit-megakaryocyte (CFU-MK), burst-forming unit-megakaryocyte (BFU-MK), and eventually differentiated mature megakaryocytes [37]. The maturation of MKs is characterized by endomitosis, cytoplasmic maturation, and assembling of all constituents required for the production of functional platelets. Mature megakaryocytes can be identified by specific cell surface markers including CD41, CD61 (integrin αIIbβ3), CD42 (glycoprotein Ib) and glycoprotein V [38]. The engagement of transcription factors, cytokines and extracellular stress synergically promotes the maturation of megakaryocytes [39]. Transcription factors, such as SCL, GATA1, GATA2, NF-E2, allows the development of megakaryocyte/erythroid progenitor cells [40]. FOG1 (ZFPM1) regulates the transcriptional activity of GATA-1 and contributes to early megakaryocyte differentiation, when NFE2 and SCL-1 regulate the later differentiation of megakaryocytes and production of platelets [41,42]. Moreover, PU.1 and Fli-1 also guide the differentiation of megakaryocytes from progenitor cells [43]. In contrast, C-myb (MYB) balances with GATA1 by playing an inhibitory role in megakaryopoiesis [44]. Besides transcription factors, the differentiation of megakaryocytes is also fine-tuned by hormones, especially thrombopoietin. By interaction with its receptor c-MPL, thrombopoietin acts as the canonical potentiator of differentiation of megakaryocytes and thrombopoiesis [45]. Activation of downstream signals consisting of MAPK, PI3K, and STATs, works in concert to promote megakaryopoiesis [46]. Bone marrow niches provide platforms for megakaryocytes and platelet development by supplying gradients of oxygen, chemokines, and infrastructures for megakaryocyte migration [47]. Additionally, megakaryocyte migration is regulated by stromal cell-derived factor-1α (SDF-1), angiopoietin 1 (Ang-1), and TPO, etc. SDF-1α navigates terminal megakaryocytes towards the vascular endothelium, when activation of VEGFR1 promotes SDF-1 mediated migration of megakaryocytes to the vascular niches and increases platelet production [48]. Recently, the gradients of reactive oxygen species are also implicated in megakaryocyte maturation [49].
Upon maturation, megakaryocytes acquire all necessary cellular constituents for thrombopoiesis, which are demarcated into micro anucleated particles prior to shedding [50]. Meanwhile, the cell membrane and cell skeleton including tubulin and actin undergo extensive rearrangement, which allows the protrusion of massive pseudopodial projections termed proplatelets [51,52]. Eventually, these proplatelets dispatch from megakaryocytes with their constituents, thus giving rise to circulating platelets. The dispatching of platelets from mature megakaryocytes is termed thrombopoiesis. Of note, apoptosis has been shown to be involved in the eventual stage of platelet release [53].
Roles of autophagy in megakaryocyte maturation and platelet function
There is substantial evidence supporting the roles of autophagy in megakaryopoiesis. In 1970s, TEM (Transmission Electron Microscope) studies from patients with carcinoid syndrome identified autophagosome-like structures inside platelets but not megakaryocytes, indicating the possible involvement of increased autophagy [13]. A later study in dogs with burn injury revealed the presence of autophagocytosis in megakaryocytes, which was considered as potential cell recycling process [54]. Evidence of activated autophagy is also noticed in ITP, which exhibits extensive cytoplasmic vacuoles representing programmed cell death. Nevertheless, these vacuoles observed appear to be with single membrane rather than with double membranes resembling autophagosomes [15].
The inhibitor of autophagy, mTORC1, has been reported to regulate both early and late steps of megakaryocyte development [55]. Additionally, inhibition of mTORC1 with rapamycin induces autophagy, decreases the size and ploidy of megakaryocytes, and impedes the maturation of megakaryocytes through a p21 and cyclin D3 dependent manner [56]. These findings suggest an important role of autophagy in the regulation of megakaryocyte development. Of note, both mTORC1 containing rapamycin-associated TOR protein (Raptor) and mTORC2 containing rapamycin-insensitive companion of Raptor (Rictor) are involved in the regulation of cell cycle [57]. However, it can be difficult to conclude that autophagy directly regulates megakaryopoiesis due to the non-specificity of pharmacological approach such as rapamycin. In that case, genetic approaches using gene knockout mice were developed to underscore the role of autophagy. Consequently, a recent study demonstrated that abrogation of autophagy from stem cell stage by hematopoietic knockout of ATG7 lead to impaired megakaryopoiesis, thrombopoiesis and hemostasis, producing larger but fewer dysfunctional platelets [11]. However, deletion of ATG7 in mature megakaryocytes and platelets using PF4-driven crew method only result in abnormal hemostasis while platelet number and size remain unchanged. Further investigation demonstrated abnormal aggregation and cargo granule packing in these platelets [12]. In light of these findings, autophagy is likely to be indispensable for the early stage of megakaryocyte development, and is required for normal platelet function as well (Figure 1).
Figure 1.
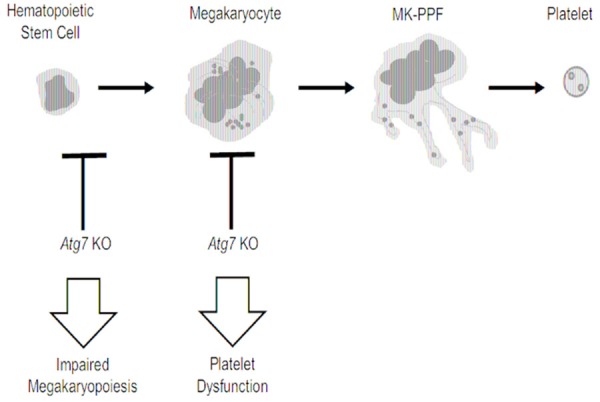
Inhibition of autophagy in hematopoietic stem cell impairs megakaryopoiesis and platelet function. Inhibition of autophagy by genetic deletion of Atg7 in mice at different stages of lineage development impairs megakaryocyte differentiation, as well as platelet function. MK-PPF, Proplatelet-forming megakaryocyte. Atg7 KO, Atg7 knockout.
Being a chronic myelogenous leukemia (CML) cell line, K562 retains the capability of megakaryocytic differentiation, thus providing a favorable tool to study megakaryopoiesis. One group reported that inhibition of autophagy in K562 cells by knockdown of autophagic genes substantially impedes megakaryopoiesis [58]. Consistently, Lapatinib treatment induces autophagic cell death and megakaryocytic differentiation in K562 cells, both of which can be inhibited by knockdown of ATG7 or application of 3-MA [35]. In contrast, another group showed that although autophagy was readily observed during induction of megakaryocytic differentiation by 12-O-tetradecanoyl-phorbol-13-acetate (TPA), it does not seem to be required for cell differentiation [59]. Further studies in MO7e cells showed that cycle progression and nuclear division are regulated by mTORC1, whereas cell size and cell death were controlled by mTORC2 [57]. The relationship between autophagy and megakaryopoiesis should be interpreted cautiously since rapamycin used in this study may also suppress P70S6K and 4E-BP pathways. In addition to megakaryocytes, further studies in platelets confirmed the presence of autophagic proteins and showed that class III PtdIns3K dependent autophagy was required for normal platelet function [60].
Closing remarks
Autophagy, as a conserved biological process, has been well studied and associated with cancer, metabolic disorders, autoimmune disease, and radiation damage. Altered autophagy is implicated in hematopoietic and blood cells indicated by morphological studies. Given that most evidence of autophagy hitherto is from cancer cells, the explicit function of autophagy in megakaryocytes and platelets remains to be elucidated. Fortunately, recent genetic studies uncovered the indispensable role of autophagy in both megakaryopoiesis and platelet function. In addition, results from a clinical trial on ITP suggest that rapamycin is effective for treating immune-induced thrombocytopenia. Thus, targeting autophagy may yield a promising approach for thrombocytopenic disease, for example, in MDS or secondary to chemo-/radiation-therapy.
Acknowledgements
This work was supported by the grants from Natural Science Foundation of China [grant 81370373, 81170132, 91439112 to LZ]; the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) of China [to LZ].
Disclosure of conflict of interest
None.
References
- 1.Kobayashi S. Choose Delicately and Reuse Adequately: The Newly Revealed Process of Autophagy. Biol Pharm Bull. 2015;38:1098–1103. doi: 10.1248/bpb.b15-00096. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- 3.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 4.Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, Rosas IO, Morse D. Autophagy in idiopathic pulmonary fibrosis. PLoS One. 2012;7:e41394. doi: 10.1371/journal.pone.0041394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 6.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datan E, Shirazian A, Benjamin S, Matassov D, Tinari A, Malorni W, Lockshin RA, Garcia-Sastre A, Zakeri Z. mTOR/p70S6K signaling distinguishes routine, maintenance-level autophagy from autophagic cell death during influenza A infection. Virology. 2014;452-453:175–190. doi: 10.1016/j.virol.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nencioni A, Cea M, Montecucco F, Longo VD, Patrone F, Carella AM, Holyoake TL, Helgason GV. Autophagy in blood cancers: biological role and therapeutic implications. Haematologica. 2013;98:1335–1343. doi: 10.3324/haematol.2012.079061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Wu K, Xiao X, Liao J, Hu Q, Chen H, Liu J, An X. Autophagy as a regulatory component of erythropoiesis. Int J Mol Sci. 2015;16:4083–4094. doi: 10.3390/ijms16024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Cai J, Zhang S, Yuan N, Li X, Fang Y, Song L, Shang M, Liu S, Zhao W, Hu S, Wang J. Loss of autophagy leads to failure in megakaryopoiesis, megakaryocyte differentiation, and thrombopoiesis in mice. Exp Hematol. 2015;43:488–494. doi: 10.1016/j.exphem.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Ouseph MM, Huang Y, Banerjee M, Joshi S, MacDonald L, Zhong Y, Liu H, Li X, Xiang B, Zhang G, Komatsu M, Yue Z, Li Z, Storrie B, Whiteheart SW, Wang QJ. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood. 2015;126:1224–1233. doi: 10.1182/blood-2014-09-598722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JC, Maldonado JE, Mann KG, Moertel CG. Ultrastructural cytochemistry of platelets and megakaryocytes in the carcinoid syndrome. Mayo Clin Proc. 1976;51:585–593. [PubMed] [Google Scholar]
- 14.Ortega Aramburu JJ. [Idiopathic thrombocytopenic purpura--autoimmune thrombocytopenia in children] . An Esp Pediatr. 1985;23:145–151. [PubMed] [Google Scholar]
- 15.Houwerzijl EJ, Blom NR, van der Want JJ, Vellenga E, de Wolf JT. Megakaryocytic dysfunction in myelodysplastic syndromes and idiopathic thrombocytopenic purpura is in part due to different forms of cell death. Leukemia. 2006;20:1937–1942. doi: 10.1038/sj.leu.2404385. [DOI] [PubMed] [Google Scholar]
- 16.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santambrogio L, Cuervo AM. Chasing the elusive mammalian microautophagy. Autophagy. 2011;7:652–654. doi: 10.4161/auto.7.6.15287. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 23.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 24.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S, Choi SG, Yoo SM, Son JH, Jung YK. Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy. 2014;10:1906–1920. doi: 10.4161/auto.32177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortensen M, Simon AK. Nonredundant role of Atg7 in mitochondrial clearance during erythroid development. Autophagy. 2010;6:423–425. doi: 10.4161/auto.6.3.11528. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- 35.Huang HL, Chen YC, Huang YC, Yang KC, Pan H, Shih SP, Chen YJ. Lapatinib induces autophagy, apoptosis and megakaryocytic differentiation in chronic myelogenous leukemia K562 cells. PLoS One. 2011;6:e29014. doi: 10.1371/journal.pone.0029014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Wang Z, Dai L, Cao L, Su J, Zhu M, Yu Z, Bai X, Ruan C. Effects of rapamycin combined with low dose prednisone in patients with chronic immune thrombocytopenia. Clin Dev Immunol. 2013;2013:548085. doi: 10.1155/2013/548085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquet JM, Gross BS, Gratacap MP, Quek L, Pasquet S, Payrastre B, van Willigen G, Mountford JC, Watson SP. Thrombopoietin potentiates collagen receptor signaling in platelets through a phosphatidylinositol 3-kinase-dependent pathway. Blood. 2000;95:3429–3434. [PubMed] [Google Scholar]
- 38.Hodohara K, Fujii N, Yamamoto N, Kaushansky K. Stromal cell-derived factor-1 (SDF-1) acts together with thrombopoietin to enhance the development of megakaryocytic progenitor cells (CFU-MK) Blood. 2000;95:769–775. [PubMed] [Google Scholar]
- 39.Deutsch VR, Tomer A. Megakaryocyte development and platelet production. Br J Haematol. 2006;134:453–466. doi: 10.1111/j.1365-2141.2006.06215.x. [DOI] [PubMed] [Google Scholar]
- 40.McDonald TP, Sullivan PS. Megakaryocytic and erythrocytic cell lines share a common precursor cell. Exp Hematol. 1993;21:1316–1320. [PubMed] [Google Scholar]
- 41.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU. 1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Takayama M, Fujita R, Suzuki M, Okuyama R, Aiba S, Motohashi H, Yamamoto M. Genetic analysis of hierarchical regulation for Gata1 and NF-E2 p45 gene expression in megakaryopoiesis. Mol Cell Biol. 2010;30:2668–2680. doi: 10.1128/MCB.01304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood. 2012;119:3724–3733. doi: 10.1182/blood-2011-09-380634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di Rago L, Hilton AA, Willson TA, Roberts AW, Ramsay RG, Nicola NA, Alexander WS. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc Natl Acad Sci U S A. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaushansky K, Fox N, Lin NL, Liles WC. Lineage-specific growth factors can compensate for stem and progenitor cell deficiencies at the postprogenitor cell level: an analysis of doubly TPO- and G-CSF receptor-deficient mice. Blood. 2002;99:3573–3578. doi: 10.1182/blood.v99.10.3573. [DOI] [PubMed] [Google Scholar]
- 46.Kaushansky K. Determinants of platelet number and regulation of thrombopoiesis. Hematology Am Soc Hematol Educ Program. 2009:147–152. doi: 10.1182/asheducation-2009.1.147. [DOI] [PubMed] [Google Scholar]
- 47.Avecilla ST, Hattori K, Heissig B, Tejada R, Liao F, Shido K, Jin DK, Dias S, Zhang F, Hartman TE, Hackett NR, Crystal RG, Witte L, Hicklin DJ, Bohlen P, Eaton D, Lyden D, de Sauvage F, Rafii S. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10:64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- 48.Mazharian A. Assessment of megakaryocyte migration and chemotaxis. Methods Mol Biol. 2012;788:275–288. doi: 10.1007/978-1-61779-307-3_19. [DOI] [PubMed] [Google Scholar]
- 49.Shinohara A, Imai Y, Nakagawa M, Takahashi T, Ichikawa M, Kurokawa M. Intracellular reactive oxygen species mark and influence the megakaryocyte-erythrocyte progenitor fate of common myeloid progenitors. Stem Cells. 2014;32:548–557. doi: 10.1002/stem.1588. [DOI] [PubMed] [Google Scholar]
- 50.Schulze H, Korpal M, Hurov J, Kim SW, Zhang J, Cantley LC, Graf T, Shivdasani RA. Characterization of the megakaryocyte demarcation membrane system and its role in thrombopoiesis. Blood. 2006;107:3868–3875. doi: 10.1182/blood-2005-07-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Italiano JE Jr, Lecine P, Shivdasani RA, Hartwig JH. Blood platelets are assembled principally at the ends of proplatelet processes produced by differentiated megakaryocytes. J Cell Biol. 1999;147:1299–1312. doi: 10.1083/jcb.147.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel SR, Richardson JL, Schulze H, Kahle E, Galjart N, Drabek K, Shivdasani RA, Hartwig JH, Italiano JE Jr. Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood. 2005;106:4076–4085. doi: 10.1182/blood-2005-06-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Josefsson EC, James C, Henley KJ, Debrincat MA, Rogers KL, Dowling MR, White MJ, Kruse EA, Lane RM, Ellis S, Nurden P, Mason KD, O’Reilly LA, Roberts AW, Metcalf D, Huang DC, Kile BT. Megakaryocytes possess a functional intrinsic apoptosis pathway that must be restrained to survive and produce platelets. J Exp Med. 2011;208:2017–2031. doi: 10.1084/jem.20110750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng TM, Lin Y, Gu DQ, Luo CK, Zheng HE. Ultrastructural changes of bone marrow megakaryocytes in several types of injury. Burns Incl Therm Inj. 1984;10:282–289. doi: 10.1016/0305-4179(84)90007-x. [DOI] [PubMed] [Google Scholar]
- 55.Drayer AL, Olthof SG, Vellenga E. Mammalian target of rapamycin is required for thrombopoietin-induced proliferation of megakaryocyte progenitors. Stem Cells. 2006;24:105–114. doi: 10.1634/stemcells.2005-0062. [DOI] [PubMed] [Google Scholar]
- 56.Raslova H, Baccini V, Loussaief L, Comba B, Larghero J, Debili N, Vainchenker W. Mammalian target of rapamycin (mTOR) regulates both proliferation of megakaryocyte progenitors and late stages of megakaryocyte differentiation. Blood. 2006;107:2303–2310. doi: 10.1182/blood-2005-07-3005. [DOI] [PubMed] [Google Scholar]
- 57.Fuhler GM, Tyl MR, Olthof SG, Lyndsay Drayer A, Blom N, Vellenga E. Distinct roles of the mTOR components Rictor and Raptor in MO7e megakaryocytic cells. Eur J Haematol. 2009;83:235–245. doi: 10.1111/j.1600-0609.2009.01263.x. [DOI] [PubMed] [Google Scholar]
- 58.Colosetti P, Puissant A, Robert G, Luciano F, Jacquel A, Gounon P, Cassuto JP, Auberger P. Autophagy is an important event for megakaryocytic differentiation of the chronic myelogenous leukemia K562 cell line. Autophagy. 2009;5:1092–1098. doi: 10.4161/auto.5.8.9889. [DOI] [PubMed] [Google Scholar]
- 59.Mishima Y, Terui Y, Mishima Y, Taniyama A, Kuniyoshi R, Takizawa T, Kimura S, Ozawa K, Hatake K. Autophagy and autophagic cell death are next targets for elimination of the resistance to tyrosine kinase inhibitors. Cancer Sci. 2008;99:2200–2208. doi: 10.1111/j.1349-7006.2008.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng W, Chang C, Luo D, Su H, Yu S, Hua W, Chen Z, Hu H, Liu W. Dissection of autophagy in human platelets. Autophagy. 2014;10:642–651. doi: 10.4161/auto.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]


