Abstract
Lung cancer is the most common type of cancer-related death in developed countries. MicroRNAs (miRNAs) are small non-coding RNAs, which regulates gene expression in cancer. Recent studies demonstrate that the microRNA-293-3p (miR-293-3p) may play as an oncogene or a tumor suppressor. However, its expression and roles in non-small cell lung cancer (NSCLC) is not known. In this study, our purpose is to investigate the expression and roles of miR-296-3p in NSCLC. The findings indicated that miR296-3p inhibited NSCLC cell proliferation, enhance the drug resistance, and apoptosis. Data of luciferase reporter assays demonstrated that the CX3CR1 gene was a direct regulator of tumorsuppressive miR296-3p. Moreover, overexpressed CX3CR1 was confirmed in NSCLC clinical specimens. Inhibition of CX3CR1 could inhibit cancer cellular survival and increase chemotherapy sensitivity. There was a negative relationship between miR296-3p and CX3CR1 expression in NSCLC tissues. Our study elucidates that miR296-3p plays a suppressive role in NSCLC by inhibiting CX3CR1 expression.
Keywords: Lung cancer, miR-296-3p, CX3CR1
Introduction
Lung cancer is the most common type of cancer in the world, which is a leading cause of cancer related death [1,2]. Operation, chemotherapy and radiotherapy are the most common ways for lung cancer therapy. But, the results of clinical therapy are not good. With the development of the molecular mechanism of basic theory on lung carcinogenesis, there are many new molecules discovered and may be used as potential good targets for therapy.
MicroRNAs (miRNAs) are endogenous small non-coding RNA molecules with around 20-29nt which bind to the 3’-UTRs of target mRNAs. miRNAs regulate gene expression at the posttranscriptional level [3-6]. More and more studies show that miRNAs involve in various cellular processes such as proliferation, apoptosis, metastasis, differentiation, stem cell, autophagy, metabolism and therapy response of non-small cell lung cancer (NSCLC) [7-10]. Recent studies showed that the microRNA-293-3p (miR-296-3p) may play as an oncogene or a tumor suppressor [11-13]. However, its expression and roles in NSCLC is not known.
In this study, our purpose is to investigate the expression and roles of miR-296-3p in NSCLC cells and explore its mechanism. The expression and cellular function of miR296-3p NSCLC cells was studied. CX3CR1 was verified as a direct target gene of miR-296-3p. CX3CR1 expression and its relationship to miR-29-3p were also studied. The study elucidated that miR296-3p played a suppressive role in NSCLC by suppressing CX3CR1 expression.
Materials and methods
Cell culture
Lung cancer cell lines A549, H157, NIH-H358, Calu-3, LAX, HCC827, LTEP-2, D6, SPCA1 and normal lung epithelial cells (BEAS-2B) were primarily obtained from ATCC. BEAS-2B cells were cultured according to the instructions. The NSCLC cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin and streptomycin and maintained at normal conditions.
RNA isolation and quantitative real-time-PCR
Total RNAs of cells and tissues were extracted using Trizol (Invitrogen) according to the manufacturer’s protocol. The reverse-transcription reaction and quantitative real-time PCR (qRT-PCR) were performed: 95°C for 10 min, 40 cycles of 95°C (15 s) and annealed/extended at 60°C for 1 min. The ΔΔCt was calculated by subtracting the Ct of U6. Fold change was calculated using the equation 2-ΔΔCt.
Cell survival assay
A549 and H157 cells were transfected with miR-296-3p or anti-miR-296-3p or CX3CR1 siRNA overnight. For cell proliferation, 103 cells/well was seeded in 96-well plates and measured by CCK-8 assay (Dojindo, Japan) after 24, 48, 72 hours according to the manufacturer’s instructions. For drug sensitivity assay, cells were treated with 5-FU, DDP and paclitaxel at a concentration. Cell viability was measured at the day 3.
Target gene prediction
The candidate targets of miR-296-3p were predicted by the following applications: TargetScan (http://www.targetscan.org) and database (www.mirbase.org).
Cell transfection and luciferase assay
A549 and H157 cells were transfected with the miR-296-3p, pGL-WT and pGL-MT using Lipofectamine 2000 according to the manufacturer’s instructions. Twenty-four hours after transient transfection, the cells were harvested and luciferase assays were performed. The relative luciferase activities (ratios of firefly and renilla luciferase activity) of lysates were measured by the dual luciferase reporter assay system (Promega, Madison, WI, USA).
Luciferase activity assay
The CX3CR1 3’-UTR sequence was amplified from human cDNAs by PCR. The wildtype and mutated 3’-UTR regions of CX3CR1 were cloned into pMiRREPORT vector (Ambion). These constructs were validated by DNA sequencing. The reporter plasmids were co-transfected with miR-296-3p mimics or the control into lung cancer cells using Lipofectamine 2000 (Invitrogen) in 24-well plates for luciferase activity testing using luciferase assay system (Promega) after 48 hours.
Tissue samples of NSCLC patients and cell lines
Samples were obtained from patients with 56 specimens of lung cancer in the First Affiliated Hospital of Harbin Medical University (Harbin, China) between 2008 and 2014. Tumor samples were collected after obtaining the written informed consents from the patients. Primary tumor cultures were done using the method described by Brassesco et al. [18]. All cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin-streptomycin (Gibco) at 37°C in a humidified incubator with 5% CO2. The Ethics Review Board of the hospital approved this study.
Western blotting
Total cellular protein were electrophoresed through a 10% denaturing polyacrylamide gel and transferred to PVDF membranes. The blots were probed with the anti-CX3CR1 or anti-β-actin (Abcam, Cambridge, MA). The bands were detected using an ECL Plus kit according to the manufacturer’s protocol.
Statistical analysis
SPSS15.0 was used for data analysis. The correlation between miR-296-3p and CX3CR1 mRNA expression was evaluated using Pearson’s chi-square test. All data were represented as mean ± SEM from triplicate experiments. Results were considered statistically significant at p<0.05.
Results
miR-296-3p attenuates lung cancer cell proliferation in lung cancer cells
To investigate the role of miR-296-3p in lung cancer, firstly, we used 10 lung cancer cell lines to examine miR-296-3p expression by real time RT-PCR. It was found that miR-296-3p levels was very low in most of the cell lines (Figure 1A). Next, A549 and H157 cells were selected for their lack of miR-296-3p expression. A549 and H157 cells were transfected with miR-296-3p mimics and miR-296-3p was up-regulated (Figure 1B). Next, cell proliferation of A549 and H157 cell with miR-296-3p overexpression was assayed and miR-296-3p could inhibit cell proliferation (Figure 1C and 1D). When miR-296-3p expression was inhibited in A549 and H157 cells (Figure 1E), the cell proliferation was increased (Figure 1F and 1G).
Figure 1.
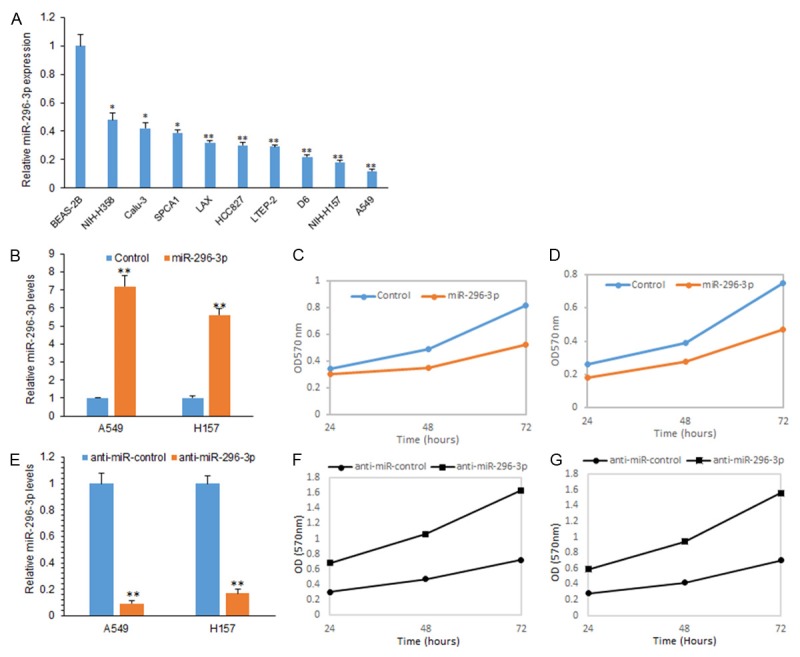
miR-296-3p attenuates lung cancer cell proliferation in lung cancer cells. A. Real time RT-PCR analysis of miR-296-3p expression in lung cancer cells including A549, H157, NIH-H358, Calu-3, LAX, HCC827, LTEP-2, D6 and SPCA1 and normal lung epithelial cells (BEAS-2B). B. Real time RT-PCR analysis of miR-296-3p expression in A549, H157 cells transfected with miR-296-3p mimics and miRNA controls. C. CCK8 assay was used to analyze cell proliferation of A549 cells with miR-296-3p mimics transfection. D. CCK8 assay was used to analyze cell proliferation of H157 cells with miR-296-3p mimics transfection. E. Real time RT-PCR analysis of miR-296-3p expression in A549, H157 cells transfected with anti-miR-296-3p and anti-miRNA controls. F. CCK8 assay was used to analyze cell proliferation of A549 cells transfected with anti-miR-296-3p and anti-miRNA controls. G. CCK8 assay was used to analyze cell proliferation of H157 cells transfected with anti-miR-296-3p and anti-miRNA controls. **p<0.01; *p<0.05.
miR-296-3p increases drug sensitivity to 5-FU, DDP and paclitaxel lung cancer cells
Drug resistance in lung cancer chemotherapy is a very frustrated question. Here, we want to know whether miR-296-3p is related to chemotherapy responses. Whether miR-296-3p enhances the drug sensitivity in lung cancer cells? A549 and H157 cells were transfected with miR-296-3p mimics and then exposed to the drug which was common used in lung cancer therapy, and CCK8 assay and colony formation were used to examine cell proliferation. We found that when the A549 cells were exposed to different dose of DDP and paclitaxel for 3 days and the survival rate significantly decreased (Figure 2A and 2B), and there were similar results in H157 cells (Figure 2C and 2D).
Figure 2.
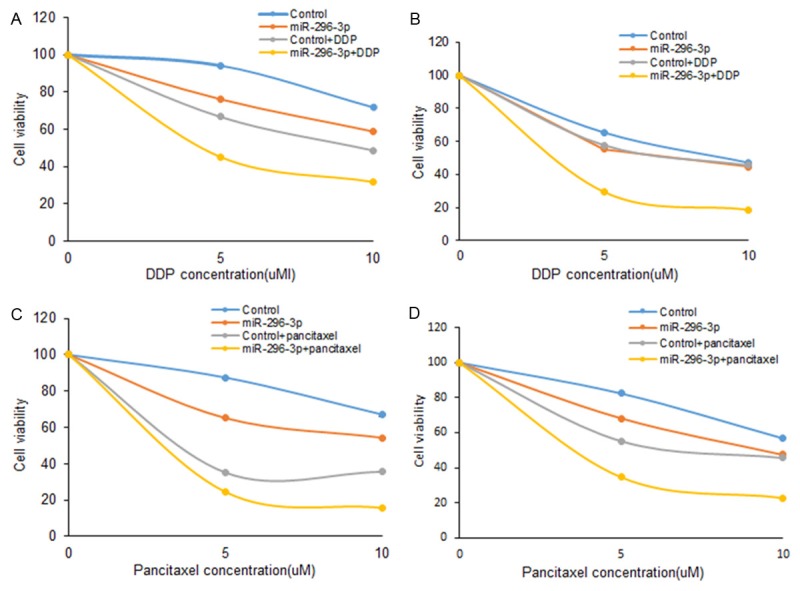
miR-296-3p increases drug sensitivity to 5-FU, DDP and paclitaxel lung cancer cells. A, B. CCK8 assay was used to analyze cell survival of A549 cells. The cells were transfected with miR-296-3p mimics or anti-miR-296-3p transfection and then exposed to DDP and paclitaxel for 3 days. C, D. CCK8 assay was used to analyze cell survival of H157 cells. The cells were transfected with miR-296-3p mimics or anti-miR-296-3p transfection and then exposed to DDP and paclitaxel for 3 days.
miR-296-3p regulates CX3CR1 expression in lung cancer cells
CX3CR1 is one of the potential target genes of miR-296-3p using TargetScan and miRBase (Figure 3A). Dual reporter assays revealed that introduction of miR-296-3p in A549 cells suppressed the activity of a luciferase reporter fused to the wild-type (WT) 3’-UTR of CX3CR1, but did not suppress that of a reporter fused to a mutant (MUT) version of the 3’-UTR (Figure 3B). Introduction of miR-296-3p in A549 and H157 cells reduced CX3CR1 expression at the mRNA and protein levels (Figure 3C and 3D). These results suggest that CX3CR1 is a direct target of miR-296-3p in lung cancer cells.
Figure 3.
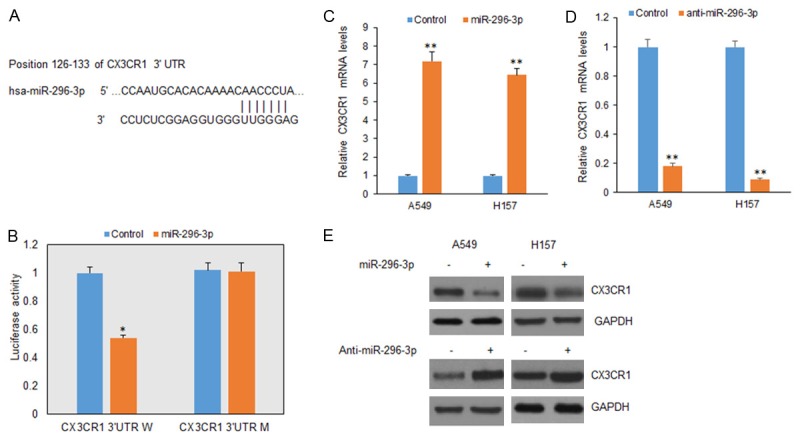
CX3CR1 is a direct target of miR-296-3p in lung cancer cells. A. Wild typed and mutant target sites for miR-296-3p sequences in 3’UTR of CX3CR1 were shown. B. Luciferase reporter assays were performed to verify the binding of miR-296-3p in 3’-UTR of CX3CR1. W: wild type; M: mutation. C. qRT-PCR assay was performed to detect the mRNA level of CX3CR1 in A549 and H157 cells treated with miR-296-3p mimics. miR-296-3p was measured by real-time RT-PCR in A549 cells after miR-296-3p mimic transfection. D. qRT-PCR assay was performed to detect the mRNA level of CX3CR1 in A549 and H157 cells treated with anti-miR-296-3p. miR-296-3p was measured by real-time RT-PCR in A549 cells after miR-296-3p mimic transfection. E. Western blotting analysis was used to measure CX3CR1 protein in A549 and NCI-H157 cells treated with miR-296-3p mimics or anti-miR-296-3p. *p<0.05, **p<0.01 vs control.
Knocking down of CX3CR1 inhibits cell proliferation and promotes apoptosis in NSCLC cells
To verify whether CX3CR1 promotes cell proliferation and promotes apoptosis in NSCLC cells, A549 and H157 cells were transfected with CX3CR1 siRNA and total protein was extracted for western blotting. CX3CR1 protein was down-regulated in the two cells lines (Figure 4A). Analysis of cell proliferation showed that cell survival rate decreased in the cells after CX3CR1 knocking down (Figure 4B). We then analyzed CX3CR1 expression in NSCLC tissues. The result showed that CX3CR1 mRNA levels in NSCLC samples was aberrantly over-expressed (Figure 4C), associated with metastasis in clinical features (Figure 4D).
Figure 4.
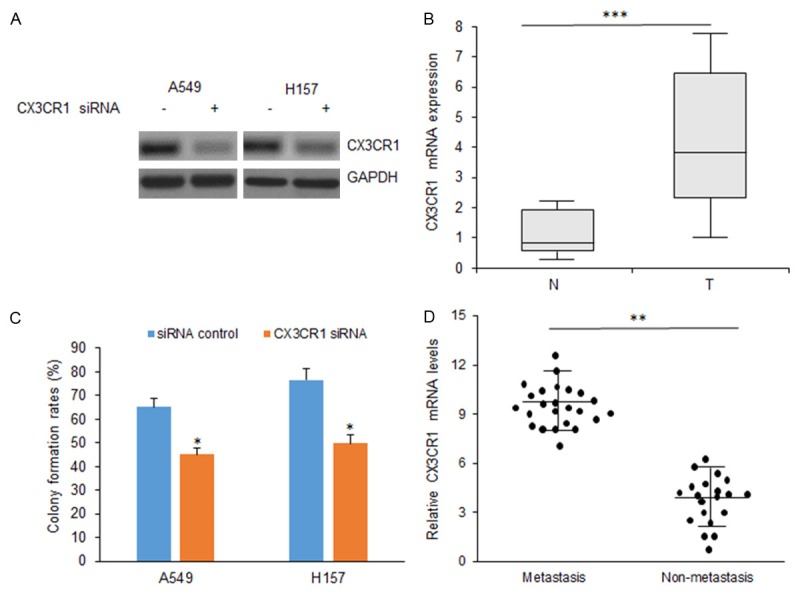
Knocking down of CX3CR1 inhibits cell proliferation and promotes apoptosis in NSCLC cells. A. CX3CR1 expression in NSCLC cells with CX3CR1 siRNA transfection by western blotting. B. Cellular proliferation was assayed in A549 and H157 cells with CX3CR1 siRNA transfection by colony formation. C. CX3CR1 mRNA levels in NSCLC samples. D. Relationship between CX3CR1 mRNA and NSCLC metastasis. **p<0.01; *p<0.05.
miR-296-3p suppresses lung cancer cell proliferation and invasion by down-regulation of CX3CR1
CX3CR1 was identified as a target gene of miR-296-3p, but the functional relationship between miR-296-3p and CX3CR1 is not known. A549 cells were transfected with CX3CR1 siRNA and miR-296-3p and cell proliferation was assayed by CCK8. It was shown that cells with CX3CR1 knocking down grew slower, and miR-296-3p combining with inhibition of CX3CR1 suppressed cell growth significantly (Figure 5A). In H157 cells, there was a similar result (Figure 5B). A549 cells were transfected with CX3CR1 siRNA and miR-296-3p and cell apoptosis was assayed by flow cytometry. Apoptosis rate was higher in the cells with CX3CR1 siRNA and miR-296-3p than the ones with only CX3CR1 siRNA or miR-296-3p transfection (Figure 5C), so was in H157 cells (Figure 5D). Taken together, the results demonstrated that the repression of cell progression by miR-296-3p was typically a consequence of decreased CX3CR1 expression in lung cancer cells.
Figure 5.
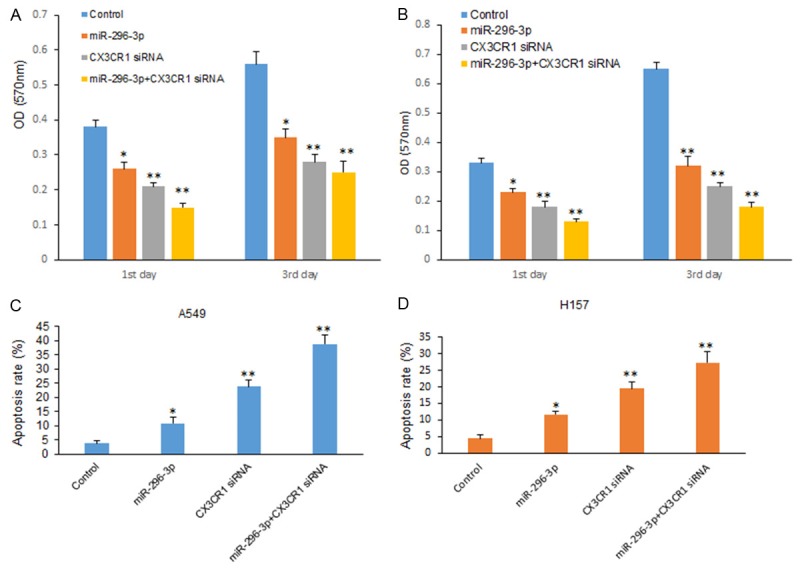
miR-296-3p suppresses lung cancer cell proliferation and invasion by down-regulation of CX3CR1. A. Cell proliferation of A549 cells transfected with miR-296-3p and CX3CR1 siRNA was assayed by CCK8. B. Cell proliferation of H157 cells transfected with miR-296-3p and CX3CR1 siRNA was assayed by CCK8. C. Cell apoptosis of A549 cells transfected with miR-296-3p and CX3CR1 siRNA was assayed by flow cytometry. D. Cell apoptosis of H157 cells transfected with miR-296-3p and CX3CR1 siRNA was assayed by flow cytometry. **p<0.01; *p<0.05.
Low levels of miR-296-3p is associated with clinical features of NSCLC
miR-296-3p expression levels in lung cancer tissues were examined by qRT-PCR analysis and significantly lower in 42 human Lung cancer tissues than their normal adjacent normal lung tissues (Figure 6A). Furthermore, miR-296-3p expression levels were compared between metastatic tissues and non-metastatic lung ones and the former was greatly lower than the latter (Figure 6B). Further analysis showed that miR-296-3p expression was negatively associated with the histological grade of lung cancer (Figure 6C). CX3CR1 was found to be negatively related to miR-296-3p expression in lung cancer tissues (Figure 6D). The results demonstrated that lack of miR-296-3p expression in lung cancer is positively related to the advanced stage, metastasis, and poor prognosis.
Figure 6.
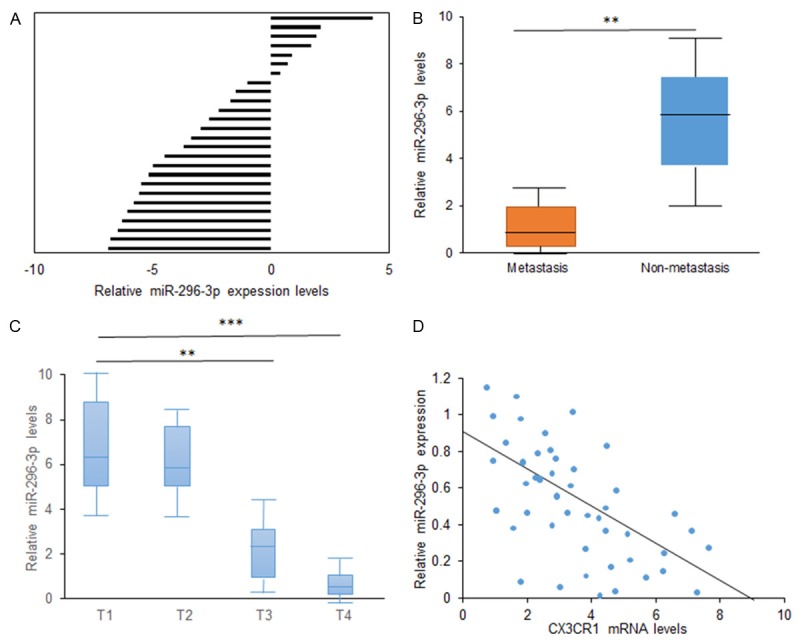
Low levels of miR-296-3p is associated with clinical features of NSCLC. A. Real time RT-PCR analysis of miR-296-3p expression in NSCLC tissues. N: normal adjacent tissues; T: NSCLC tissues. B. Data analysis of miR-296-3p expression in NSCLC tissues with low metastasis and high metastasis. C. Data analysis of miR-296-3p expression in NSCLC tissues with different staging. D. Data analysis of relationship between miR-296-3p and CX3CR1 expression in NSCLC tissues (r=-0.538). **p<0.01; *p<0.05.
Discussion
Previous study showed that miR-296-3p is down-regulated in tumor but its biological function and expression in NSCLC is still not known. In the current study, the expression level of miR-296-3p in NSCLC cell lines was lower than the normal lung epithelial cells. Cellular functional studies showed miR-296-3p inhibited NSCLC cell proliferation and enhanced the sensitivity to the chemotherapy. Inhibition of miR-296-3p in NSCLC cells could reverse the inhibition of cell proliferation. The results suggested that miR-296-3p plays a critical role in non-small cell lung cancer cell proliferation and increases the sensitivity of drug therapy.
There are a few studies showing the expression and role of miR-296-3p in tumor. A report showed that the expression level of miR-296-3p is much lower in the non-metastatic human prostate cancer cell line P69 than that in the highly metastatic cell line M12, which is derived from P69, which suggests miR-296-3p plays roles as an oncogene in prostate cancer [11]. Another study indicates that miR-296-3p was down-regulated in drug resistant glioma U251AR cells and over-expression of miR-296-3p enhanced the sensitivity of glioblastoma (GBM) cells to anticancer drugs [12]. So, the role of miR-296-3p in cancer depends on the tumor background. In our study, miR-296-3p expression was down-regulated in NSCLC tissues compared with their normal tissues. It suggests that miR-296-3p plays as a tumor suppressor in NSCLC.
There are other target genes of miR-296-3p such as EAG1, ICAM1. In our study, CX3CR1 was predicted and verified as a direct target of miR-296-3p in human lung cancer. The results showed that CX3CR1 was significantly up-regulated in human NSCLC tissues. Knockdown of CX3CR1 decreased A549 and H157 cell proliferation. CX3CR1 is the receptor of the ligand for chemokine CX3CL1. CX3CL1 is defined as a membrane as well as a soluble chemokine expressed by neurons and activated endothelial cells. Recent evidence has shown that the CX3CL1/CX3CR1 pair plays a major role in adhesion, migration and survival of tumor cells including pancreatic cancer cells [13-15]. CX3CR1 is normally over-expressed in lung cancer, breast cancer, esophageal carcinoma, prostate cancer, oral cancer and pancreatic cancer and so on [16-18].
In conclusion, we demonstrated that the expression of miR-296-3p was usually low in NSCLC including the samples from the patients and cell lines compared with their controls. It was demonstrated that miR-296-3p could significantly inhibited the cell proliferation, increased cell apoptosis and enhanced drug sensitivity through by directly down-regulating CX3CR1. The study gives us an evidence that miR-296-3p may be a new therapeutic way for NSCLC therapy. However, there needs to find new target genes of miR-296-3p in cancer.
References
- 1.Patel JN, Ersek JL, Kim ES. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015;4:503–514. doi: 10.3978/j.issn.2218-6751.2015.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda M, Okamoto I, Nakagawa K. Clinical development of nintedanib for advanced non-small-cell lung cancer. Ther Clin Risk Manag. 2015;11:1701–1706. doi: 10.2147/TCRM.S76646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naidu S, Garofalo M. microRNAs: An Emerging Paradigm in Lung Cancer Chemoresistance. Front Med (Lausanne) 2015;2:77. doi: 10.3389/fmed.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusek AM, Abba M, Eljaszewicz A, Moniuszko M, Niklinski J, Allgayer H. MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol Cancer. 2015;14:34. doi: 10.1186/s12943-015-0302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonagh L, Gray SG, Finn SP, Cuffe S, O’Byrne KJ, Barr MP. The emerging role of microRNAs in resistance to lung cancer treatments. Cancer Treat Rev. 2015;41:160–169. doi: 10.1016/j.ctrv.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Yang Q, Wang S. MicroRNAs: a new key in lung cancer. Cancer Chemother Pharmacol. 2014;74:1105–1111. doi: 10.1007/s00280-014-2559-9. [DOI] [PubMed] [Google Scholar]
- 7.Ulivi P, Zoli W. miRNAs as non-invasive biomarkers for lung cancer diagnosis. Molecules. 2014;19:8220–8237. doi: 10.3390/molecules19068220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi P, Middleton J, Jeon YJ, Garofalo M. MicroRNAs in lung cancer. World J Methodol. 2014;4:59–72. doi: 10.5662/wjm.v4.i2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortunato O, Boeri M, Verri C, Moro M, Sozzi G. Therapeutic use of microRNAs in lung cancer. Biomed Res Int. 2014;2014:756975. doi: 10.1155/2014/756975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guz M, Rivero-Müller A, Okoń E, Stenzel-Bembenek A, Polberg K, Słomka M, Stepulak A. MicroRNAs-role in lung cancer. Dis Markers. 2014;2014:218169. doi: 10.1155/2014/218169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen C, Zhao X, Xu M, Sun Q, Deng R, Zhang H, Qu Y, Huang J, Jiang B, Yu J. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis. 2013;4:e928. doi: 10.1038/cddis.2013.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbagallo D, Piro S, Condorelli AG, Mascali LG, Urbano F, Parrinello N, Monello A, Statello L, Ragusa M, Rabuazzo AM, Di Pietro C, Purrello F, Purrello M. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics. 2013;14:62. doi: 10.1186/1471-2164-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Y, Liao H, Liu T, Zeng X, Xiao F, Luo L, Guo H, Guo L. MiR-296-3p regulates cell growth and multi-drug resistance of human glioblastoma by targeting ether-à-go-go (EAG1) Eur J Cancer. 2013;49:710–724. doi: 10.1016/j.ejca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Wei LM, Cao S, Yu WD, Liu YL, Wang JT. Overexpression of CX3CR1 is associated with cellular metastasis, proliferation and survival in gastric cancer. Oncol Rep. 2015;33:615–624. doi: 10.3892/or.2014.3645. [DOI] [PubMed] [Google Scholar]
- 15.Schmall A, Al-Tamari HM, Herold S, Kampschulte M, Weigert A, Wietelmann A, Vipotnik N, Grimminger F, Seeger W, Pullamsetti SS, Savai R. Macrophage and cancer cell crosstalk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am J Respir Crit Care Med. 2015;191:437–447. doi: 10.1164/rccm.201406-1137OC. [DOI] [PubMed] [Google Scholar]
- 16.Lv CY, Zhou T, Chen W, Yin XD, Yao JH, Zhang YF. Preliminary study correlating CX3CL1/CX3CR1 expression with gastric carcinoma and gastric carcinoma perineural invasion. World J Gastroenterol. 2014;20:4428–4432. doi: 10.3748/wjg.v20.i15.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao T, Gao S, Wang X, Liu J, Duan Y, Yuan Z, Sheng J, Li S, Wang F, Yu M, Ren H, Hao J. Hypoxia-inducible factor-1α regulates chemotactic migration of pancreatic ductal adenocarcinoma cells through directly transactivating the CX3CR1 gene. PLoS One. 2012;7:e43399. doi: 10.1371/journal.pone.0043399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J, Yang M, Shao J, Miao Y, Han J, Du J. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol Cancer. 2013;12:141. doi: 10.1186/1476-4598-12-141. [DOI] [PMC free article] [PubMed] [Google Scholar]


