SUMMARY
Insulin receptor (IR) signaling is critical to controlling nutrient uptake and metabolism. However, only a low-resolution (3.8 Å) structure currently exists for the IR ectodomain, with some segments ill-defined or unmodeled due to disorder. Here, we revise this structure using new diffraction data to 3.3 Å resolution that allow improved modeling of the N-linked glycans, the first and third fibronectin type III domains, and the insert domain. A novel haptic interactive molecular dynamics strategy was used to aid fitting to low-resolution electron density maps. The resulting model provides a foundation for investigation of structural transitions in IR upon ligand binding.
Graphical Abstract
INTRODUCTION
The human insulin receptor (IR) is an (αβ)2 homodimeric receptor tyrosine kinase that plays a key role in glucose homeostasis and regulation of lipid, protein, and carbohydrate metabolism (Saltiel and Kahn, 2001). IR signaling also modulates brain neurotransmitter levels (Adamo et al., 1989). Aberrant IR signaling is implicated in type 2 diabetes (Saltiel and Kahn, 2001), cancer (Frasca et al., 2008), and Alzheimer’s disease (Craft, 2012). The extracellular part of the IR αβ monomer comprises two leucine-rich repeat domains (L1 and L2), a cysteine-rich region (CR), and three fibronectin type III domains (FnIII-1, -2, and -3, respectively) (Figure 1A). An insert domain (ID) lies within FnIII-2 and contains the αβ proteolytic processing site. A single intra-monomer disulfide bond links the α chain to the β chain, with the (αβ)2 homodimer stabilized by disulfide bonds at two locations (Figure 1A). IR is related to the type 1 insulin-like growth factor receptor (IGF-1R); their ectodomains have 57% sequence identity and each binds insulin and the insulin-like growth factors IGF-I and IGF-II, though with varying affinity (Adams et al., 2000). These ligands are understood to interact with two distinct sites (sites 1 and 2) on the ectodomain surface (De Meyts, 2015).
Figure 1. IR Ectodomain Structure.
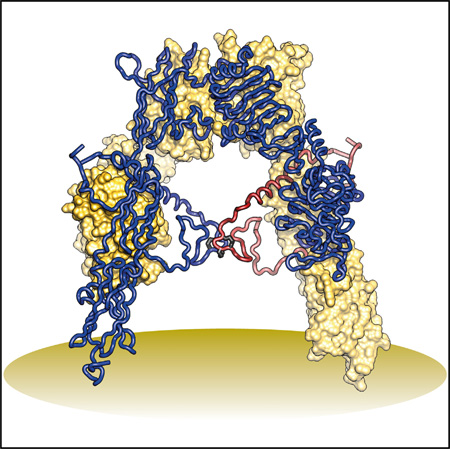
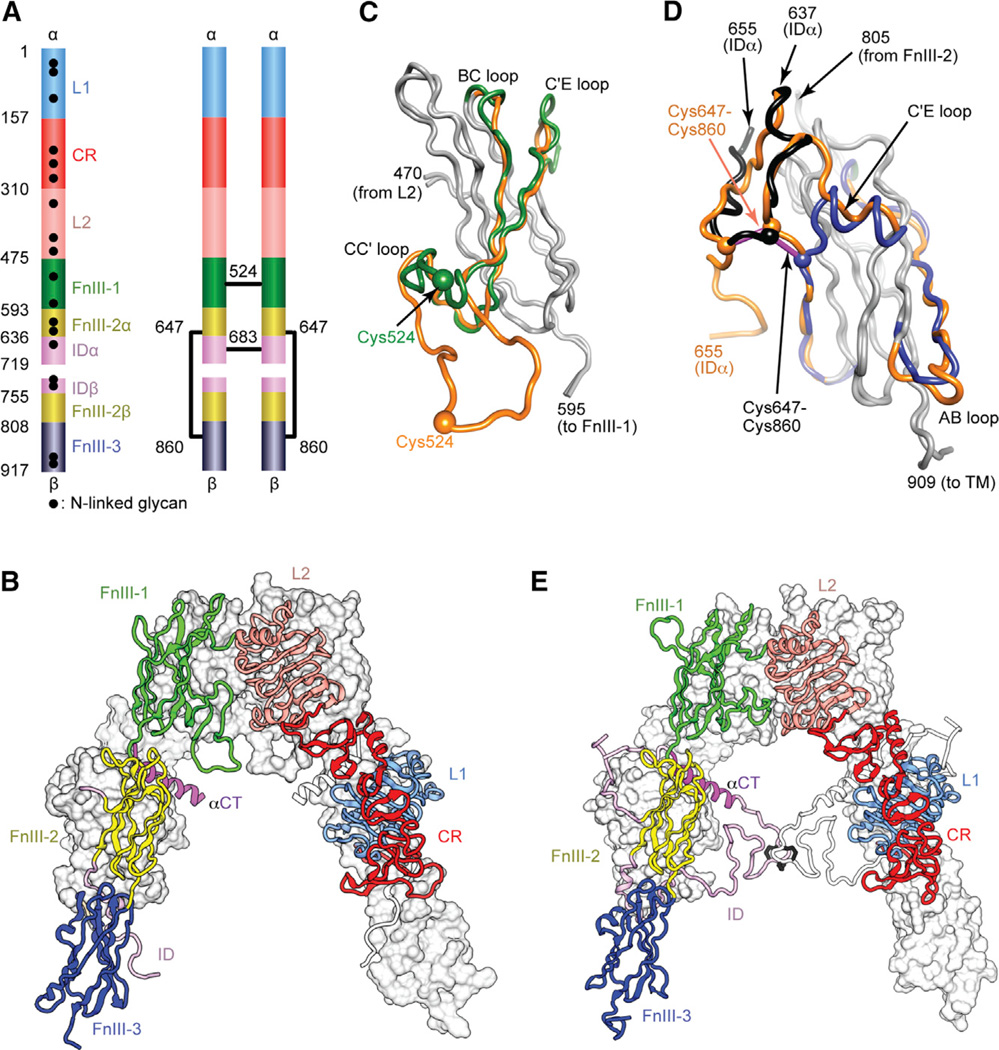
(A) Domain structure of the αβ monomer and of the disulfide-linked (αβ)2 homodimer. Black lines denote inter-chain disulfide bonds. N-linked glycosylation sites are indicated for the monomer. (B) Inverted V-shaped arrangement of the domains within 3LOH. One monomer is in ribbon representation with domains labeled; the other is in molecular surface, apart from its ID which is in ribbon. (C and D) Revisions to FnIII-1 (C) and FnIII-3 (D). Regions of common residue register across 2DTG/ 3LOH and the revised structure are in gray; where the register differs, 2DTG/3LOH residues are in orange and those of the remodeled FnIII-1 in green, remodeled ID in black, and remodeled FnIII-3 in blue. Included in (D) is the ID segment 637–655 with the 647–860 intra-monomer α/β disulfide bond in purple rod representation.
(E) Revised IR ectodomain homodimer, represented as in (B).
The structural biology of the intact IR and IGF-1R ectodomains is incompletely understood. The only structure available is that of the IR ectodomain (αβ)2 homodimer in complex with four antigen-binding fragments (2 × Fab 83-7, 2 × Fab 83-14), determined to 3.8 Å resolution (PDB: 2DTG, McKern et al., 2006, later refined as PDB: 3LOH, Smith et al., 2010; hereafter 2DTG and 3LOH, respectively). Each αβ monomer has an inverted V-shaped conformation, one leg of which comprises L1, CR, and L2 and the other FnIII-1, -2, and -3. The overall (αβ)2 structure is two-fold symmetric (Figure 1B). The ID (residues ~638–757) appeared largely disordered C-terminal to the Cys647 intra-monomer α/β disulfide bridge, apart from residues 693–710, which formed an α helix (αCT; Figure 1B) on the surface of L1 on the opposing monomer (Smith et al., 2010). 2DTG was determined by molecular replacement (using the higher-resolution structures of the IR L1-CR-L2 fragment [Lou et al., 2006] and homologous Fabs) followed by phase combination to 5.5 Å resolution with heavy-atom derivative phases. The FnIII domains were then built directly into solvent- flattened electron density maps, guided by the predicted sequence location of their β strands and distinguishing features in the maps (McKern et al., 2006). However, modeling was complicated by low resolution, absence of structural templates, and the presence of long inter-strand loops. Inspection of crystallographic maps associated with 3LOH has since led us to question the register assignment of certain strands within FnIII-1 and -3.
We present here a revised structure of the Fab-complexed IR ectodomain refined against diffraction data extending to 3.3 Å resolution derived from an alternative crystal. This structure corrects errors in the original modeling of FnIII-1 and -3 and the ID as well as in the underlying sequences. Our revised structure further includes the N-linked glycans (Figure 1A; left unmodeled in 2DTG/3LOH) and tentative modeling of the missing residues of the ID, which together place constraints on the receptor’s conformational flexibility and thus on possible signal transduction mechanisms. Revision was aided by a novel interactive molecular dynamics flexible fitting (iMDFF) strategy that allows haptic, user-directed evaluation of diverse conformational and sequence register alternatives.
RESULTS
Sequencing
Five literature discrepancies exist in the IRΔβ construct cDNA sequence that underpins 2DTG and 3LOH. Re-sequencing revealed that IRΔβ residues 421 and 465 are Ile and Gln (Ullrich et al., 1985), not Thr and Lys (Ebina et al., 1985), whereas 861– 862 are Val-Ser (Ebina et al., 1985), not Asp-Thr (Ullrich et al., 1985). The substitution Y144H present in the IRΔβ construct and in Ebina et al. (1985) is a population variant (The International HapMap Consortium, 2010). Re-sequencing the Fab 83-14 light chain variable region confirms that residues 56 and 79 are Ser and Glu (Boado et al., 2007), not Pro and Lys (McKern et al., 2006) and that residue 104 is Leu (McKern et al., 2006), not Met (Boado et al., 2007).
Protein Production, Crystallization, and Data Collection
Protein production, crystallization, and X-ray data collection (see Experimental Procedures) followed protocols closely similar to those originally reported by McKern et al. (2006). The crystal obtained had similar unit cell packing and a space group identical to that originally described. The improved quality of the diffraction data is evident in Table 1 and Figure S1A. The diffraction limit for the new data was set at 3.3 Å based on a CC1/2 criterion (rather than Rmerge- or <I/σ(I)>-based criteria) to permit maximum exploitation of weak data during maximum-likelihood refinement (Karplus and Diederichs, 2012).
Table 1.
X-Ray Data Processing and Refinement Statistics
| 3.3-Å Dataset | 3.8-Å Dataset (PDB: 3LOH) | |
|---|---|---|
| X-Ray Data Processing | ||
| Space group | C2221 | C2221 |
| Unit cell a, b, c (Å) | 121.87, 321.25, 199.40 | 123.03, 318.65, 204.64 |
| Resolution (Å) | 35.0–3.3 (3.4–3.3)a | 30.6–3.8 (3.9–3.8) |
| No. of reflections | 57,953 (4,329) | 39,268 (2,634) |
| Multiplicity | 7.0 (3.6) | 5.5 (5.4) |
| <I/σ(I)> | 11.39 (0.78) | 11.0 (0.96) |
| CC1/2 | 0.997 (0.243) | |
| Rmerge | 0.145 (1.62) | 0.142 (1.71) |
| Completeness (%) | 97.9 (86.6) | 98.8 (93.4) |
| Refinement | PDB: 4ZXB | Model S1 | 3LOH (As Originally Reported) |
Revision of 3LOH Based on 4ZXB |
Extended Model of 3LOH Based on Model S1 |
|---|---|---|---|---|---|
| No. of reflections | 57,926 | 57,926 | 39,268 | 39,649 | 39,649 |
| Rxpctwork/Rxpctfreeb | 0.205/0.237 | 0.207/0.245 | NA | 0.212/0.256 | 0.195/0.247 |
| Rwork/Rfree | 0.221/0.256 | 0.220/0.263 | 0.225/0.285 | 0.222/0.272 | 0.204/0.260 |
| No. of protein atoms/glycan atoms | 12,604/317 | 13,960/1,492 | 13,090/NAc | 12,815/78 | 13,633/1,358 |
| <B> protein/<B> glycan (Å2) | 133/171 | 131/227 | 185/NA | 164/170 | 164/259 |
| σbonds (Å)/σangles (°) | 0.010/1.3 | 0.010/1.4 | 0.012/1.7 | 0.010/1.4 | 0.010/1.4 |
| Ramachandran plot (%)d | 92.5/5.3/1.7 | 90.1/6.2/3.7 | 70.1/14.6/15.3 | 92.3/4.7/3.0 | 89.9/6.1/4.0 |
Numbers in parentheses refer to the outer shell.
Rxpct: expectation value of the crystallographic R factor (Blanc et al., 2004) as reported by autoBUSTER (Bricogne et al., 2011). Free set is 5% of all reflections.
NA, not applicable.
Preferred/allowed/outliers, calculated using Coot (Emsley and Cowtan, 2004).
Crystallographic Refinement Coupled with iMDFF
Low-resolution crystallographic refinement of protein structures is an underdetermined problem that relies heavily on the incorporation of prior knowledge as “restraints,” with the propensity for trapping the model in a local energy function minimum distant in coordinate space from the putative global minimum. Manual adjustment and/or rebuilding of segments of the model then remain necessary. Most current rebuilding protocols employ tools such as Coot (Emsley and Cowtan, 2004) and begin with rigid-body adjustment of small protein elements (e.g. side chains/short peptide fragments), followed by local minimization to remove unnatural geometry. The weak guidance offered by low-resolution maps often leaves residual error (steric overlap, poor backbone conformation, and/or side-chain outliers) that may not be corrected by reciprocal space refinement. X-Ray molecular dynamics flexible fitting (xMDFF) was devised as a real-space refinement approach that subjects the structure to a force field designed for realistic long-term molecular dynamics simulation (Brooks et al., 2009) within an electron density map co-presented as a three-dimensional (3D) potential energy grid (McGreevy et al., 2014). This approach, while having the advantage that it does not allow steric clashes and leads to improved geometry and R factors in many cases, is unable to overcome the energy barrier of large-scale structural rearrangement.
Here, we proposed that the combination of xMDFF with an interactive molecular dynamics implementation (termed iMDFF) would improve both the speed and accuracy of the IR ectodomain remodeling. We thus extended the existing interactive molecular dynamics plugin autoIMD of the VMD package (Humphrey et al., 1996) to add MDFF functionality along with standard stereochemical restraints, then coupled it to a haptic interface (a Novint Falcon) to allow 3D interaction with real-time force feedback. While any device compatible with the Virtual Reality Peripheral Network (VRPN) libraries (Taylor et al., 2001) may be used in place of the Falcon, mouse interaction currently has limited functionality. The user can thus “pull” on any atom within a running simulation, while it (and its surroundings) responds in a manner akin to a real molecule. In a typical iMDFF calculation, a small “problem” region (~10–20 contiguous residues and their immediate spatial neighbors) is selected as the mobile component of the interactive simulation. A further 8-Å shell of surrounding atoms is included in the simulation to maintain the physical context of the mobile atoms, but remains fixed in space. Pregenerated crystallographic maps are masked to within user-specified distances from the mobile atoms and then converted to potential energy maps to which the simulation is then coupled. Standard stereochemical restraints are included, as are symmetry-related atoms as required.
Conventional X-ray refinement with autoBUSTER coupled with iMDFF allowed us to rebuild the structure of IRΔβ and associated Fabs into the maps derived from the new diffraction data. The details are as follows.
FnIII-1 Remodeling
Prior secondary structure prediction of FnIII-1 (data not shown) suggested that its C′ strand comprises residues ~529–534, not 536–541 as in 2DTG/3LOH. We used multiple rounds of iMDFF and crystallographic refinement to evaluate alternative registers, leading us to assign strand C as residues 502–510 (rather than 499–507) and strand C′ as residues 529–534, implying a shorter CC′ loop and longer C′E loop (Figure 1C). This re-assignment improved interpretability of the map volumes associated with the adjoining BC, C′E, and EF inter-strand loops. No electron density on the two-fold axis could be assigned to the Cys524-Cys524 disulfide, and density corresponding to residues 519–527 remained unclear. Unassigned density near the side chain of L2 residue Tyr374 suggested potential off-axis positioning of the disulfide bond. Residues 516–530 were thus modeled in two tentative alternative configurations consistent with such positioning. All assigned strand registers were then assessed by a post hoc threading procedure that tested, for each strand, the fit of all possible registers within a ten-residue window of the selected register to a B factor-sharpened side-chain omit map (see Experimental Procedures). This procedure ranked both the re-assigned C and C′ strand registers higher than any alternative consistent with the structure of the domain (Table S1).
As a final measure of improvement, we compared the real-space omit-map correlation coefficients (RSCCs) of individual residues within the remodeled residues 496–548 with those derived from a domain-based overlay of 3LOH onto the same omit maps (Figure S2). We saw an overall improvement in RSCC of 0.17 for the segment 496–548 compared with a baseline improvement of 0.09 for the remainder of FnIII-1.
FnIII-3 Remodeling
Inspection of electron density maps and secondary structure predictions (data not shown) and iMDFF modeling led to the following changes. (1) Residues 862–871 (the C′-E loop) are now built as an α helix within density previously assigned to 866–871 in random coil conformation (Figure 1D). (2) Residues 636–646, at the N terminus of the ID and immediately upstream of the Cys647-Cys860 disulfide bond, are now fitted into density vacated by the FnIII-3 C′-E loop, with an α-helical turn at 639– 643. (3) Strand A is now assigned to residues 817–822 rather than 814–819 (Figure 1D), which inter alia we note alleviates the implausibly buried side chain of Asp812 and intra-stand location of Pro817 in 2DTG/3LOH.
The post hoc threading procedure described above was reapplied here and supported the register assigned to strands A, B, E, F, and G (Table S1). In the case of the C-C′ segment, the register of this segment is well determined by the inter-chain disulfide Cys860-Cys647. The segments that were re-assigned had an overall improvement in RSCC of 0.17 compared with 3LOH overlaid as above, versus an improvement of 0.10 for those segments that were not significantly remodeled (Figure S2).
ID Remodeling
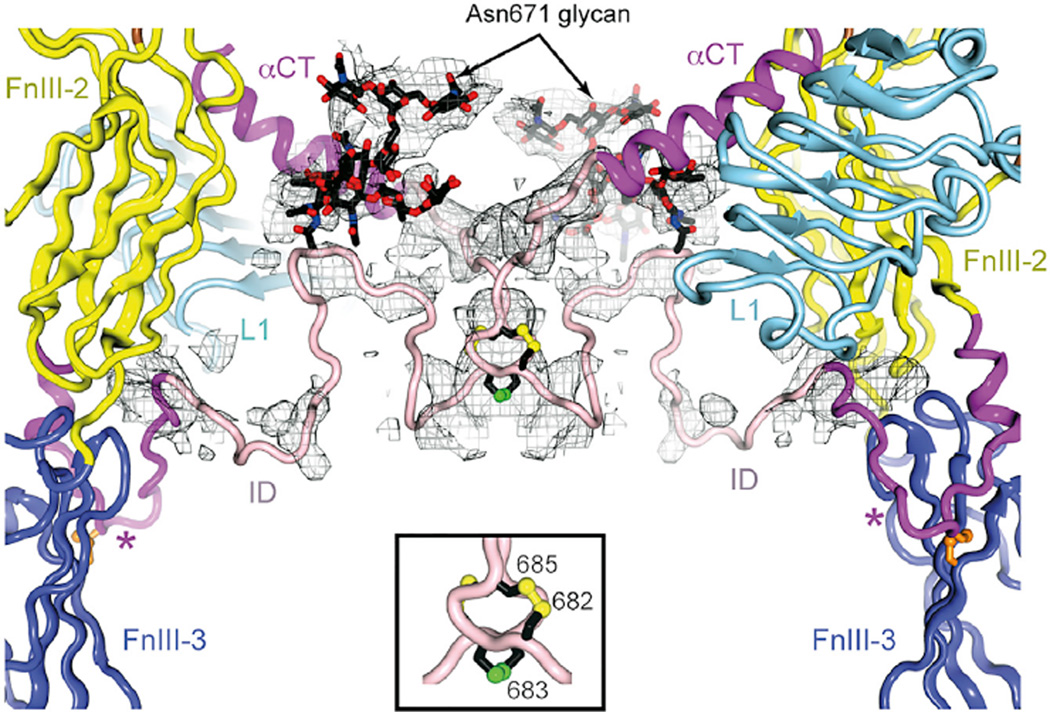
In 2DTG/3LOH, ID residues 638–655 were modeled into weak density extending toward the membrane, with 656–692 left unmodeled. In our revised structure, residues 638–656 are seen to undertake a hairpin-like turn immediately C-terminal to the inter- chain disulfide at Cys647 and direct toward the adjacent domain L1 (Figures 1E and 2). Density C-terminal to Gly650 was increasingly disordered. The final residue assignable was Arg656, modeled into the nexus of domains FnIII-2, FnIII-3, and the opposing domain L1, and potentially interacting with Glu637 and Asp638 (located where the α chain exits FnIII-2), Arg804 (in the FnIII-3 G strand), and Glu154 (in the opposing L1). RSCC calculations similar to those described above indicated that there was an overall RSCC improvement of 0.21 for the remodeled ID segment 637–655 with respect to a 3LOH overlay. We also observed connected density in a “blurred” difference electron density map (Bblur = 60 Å2) similar in shape to the GlcNAc6Man3Fuc glycan expected at Asn671 (Sparrow et al., 2008) and bordered by FnIII-2, the Asn514 glycan and αCT. This volume was adjacent to a second stretch of density broadly consistent with a path of IDα beyond residue 656 to residue 673.
Figure 2. iMDFF Model of Residues Surrounding the Putative Inter-α-Chain Disulfide Bond at Cys683.
Inter-chain disulfide bonds and the Asn671 glycan (in rod representation; labeled) are assigned to a large volume of a σA-weighted (Fobs-Fcalc)-difference electron density (Bblur = 60 Å2), calculated prior to the inclusion of glycan and of the ID residues, in pink. The hairpin-like turn in the ID near Leu648 marked by a star.
The ID includes inter-chain disulfide bond(s) at the 682–685 CCSC motif (Sparrow et al., 1997), unmodeled in 2DTG/3LOH. Although a large electron density feature lay on the two-fold axis proximal to the αCT N terminus (Figure 2), no detailed residue-level fitting was possible. We thus created dimeric (disulfide- linked) models encompassing 674–693 and subjected them to iMDFF against a B factor blurred map (Bblur = 60 Å2) with two-fold symmetry restraint and with Leu693 fixed in its previously refined position. Of these, one with a thioredoxin-like intra-molecular Cys682-Cys685 disulfide and inter-molecular Cys683-Cys683 disulfide was judged the best fit, consistent with receptor monomerization requiring reduction of only two disulfide bonds (Chiacchia, 1991). The resultant dimeric model was then fused to the remainder of the structure using iMDFF.
N-Linked Glycan Modeling
B factor blurred difference electron density maps revealed features at each N-linked site into which glycan could be assembled using iMDFF and guidance on glycosidic bond torsion angles from the GlycoMapsDB (Frank et al., 2007). These features (Table S2) could be reconciled qualitatively with the most common glycoform found by mass spectroscopy (MS) (Sparrow et al., 2008), except in two instances. (1) Although the most common glycoforms at Asn16 are fucosylated (80%), no evidence of fucosylation was present in the maps. Instead, an unusual GlcNAc2Man5 isoform with a single mannose residue on the α-6 arm is suggested, with the glycan antennae located in a highly constrained cleft between two crystallographically related copies of Fab 83-14 and the stem constrained by the poorly resolved β-chain component of the ID. (2) Asn514 shows evidence of fucosylation of the glycan stalk despite no fucosylated species being detected in the MS analysis. These differences may reflect the fact that only a single isoelectric fraction of IRΔβ led to crystal formation (C.W., personal communication).
The Remaining Domains
Inspection of domains L1, CR, L2, and FnIII-2 and the Fabs led to only minor stereochemistry correction. In 3LOH, the constant region of Fab 83-14 was poorly ordered: here disorder is less severe, allowing improvement.
Final Models
Two models are presented. The first comprises only the relatively well-ordered protein and glycan residues and is deposited as PDB: 4ZXB (hereafter 4ZXB), obsoleting 2DTG and 3LOH. The second (Model S1) extends the first to include all remaining residues (Table S3), which have been energy minimized to physically reasonable configurations within B factor-blurred electron density maps. Model S1 provides a starting point for molecular dynamics investigation of the conformational dynamics of ligand binding. Refinement statistics for 4ZXB and Model S1 are shown in Table 1: we note the significant improvement in the Ramachandran plot compared with 3LOH. Table S4 lists residues substantially repositioned in 4ZXB and root-mean-square deviation of their Cα atoms with respect to their 3LOH counterparts.
As a control, we applied the same procedure (iMDFF plus conventional refinement) to 3LOH with respect to the original data guided by the changes made here to the ectodomain structure, again partitioning into two models, one containing the relatively well-ordered protein and glycan residues and the other all residues. These processes led to a small improvement in Rfree but a significant improvement in stereochemical quality (Table 1).
DISCUSSION
iMDFF and the IR Ectodomain
There is a need to increase the speed and fidelity of crystallographic model building at low resolution (Brünger et al., 2009). The non-physical nature of most manual modification methods can unwittingly result in substantial error, leading to a de facto standard wherein stereochemical statistics for low-resolution structures are poorer than those for high-resolution structures. While part of the improvement achieved here in current structure can be attributed to increased data resolution (3.8 to 3.3 Å), the use of iMDFF enabled rapid, highly intuitive testing of hypotheses such as strand register and loop geometry. Critically, iMDFF treats the model as an explicitly physical structure throughout the modeling, with steric clash absent as the surrounding structure adjusts to the interactive modification (a sample of the process is shown in Movie S1). Here, errors in FnIII-1 and -3 have been detected and corrected, N-linked glycans have been modeled, and a tentative path identified for the poorly ordered region of the ID (Figure 1E). The overall stereochemical quality of the ectodomain structure is enhanced, increasing the number of residues in the most favored regions of the Ramachandran plot from ~70% to ~90% (Table 1).
Location of Site 2
Site 2 is proposed (Menting et al., 2013) to reside at the junction of FnIII-1 and -2, as superposition of the structure of insulin in complex with domains L1 and CR, and αCT onto 3LOH directs insulin site 2 binding residues toward that interface. This proposition is supported by alanine scanning mutagenesis of IR residues near the FnIII-1/FnIII-2 junction (Whittaker et al., 2008). Of the overall set scanned, only the FnIII-1 CC′ loop residues 526–530 have revised disposition here, moving from a position adjacent to site 1 in 2DTG/3LOH to one more than 10 Å away. The lack of effect on high-affinity insulin binding upon respective mutation of these residues is thus compatible with their revised location, and we thus see no reason to revise the predicted location of site 2.
Conformational Change upon Hormone Binding
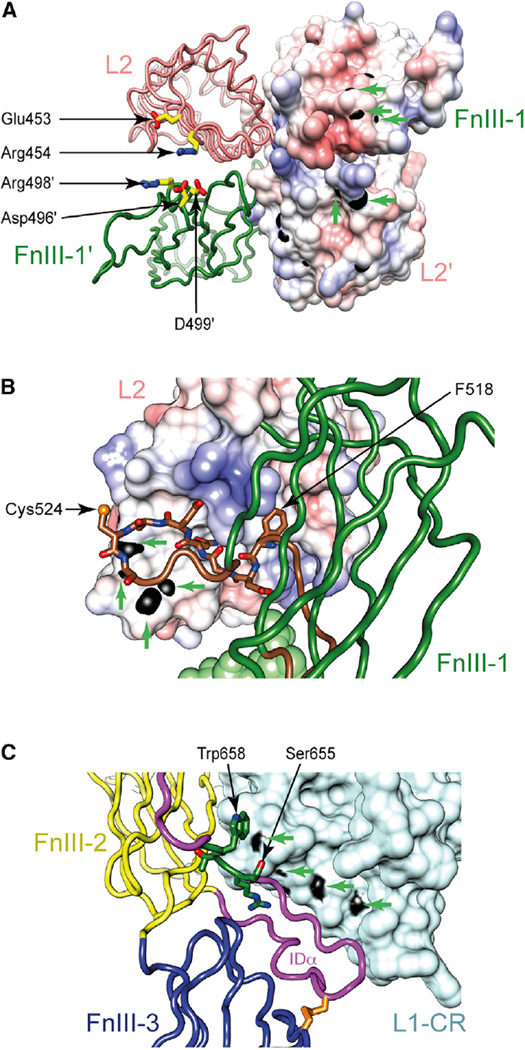
Our revised IR ectodomain structure offers some insight into IR conformational change upon ligand binding. First, the revised structure exhibits enhanced electrostatic complementarity (with respect to 2DTG/3LOH) across the sparsely packed intermonomer L2/FnIII-1 interface. L2 residues Arg454 and Glu453 and FnIII-1 residues Asp496, Arg498, and Asp499 appear capable of forming inter-domain electrostatic interactions upon slight relative domain rotation (Figure 3A). In support of this, we note that peptides containing these residues gain protection from hydrogen/deuterium (H/D) exchange upon ligand binding to both IR and IGF-1R (Houde and Demarest, 2011; Qiang et al., 2014), suggesting that slight reorientation could indeed bring these opposing residues into contact. Second, a previously overlooked feature of 2DTG/3LOH (retained here) is L2 residue Asp464’s juxtaposition with its symmetry-related counterpart in an apparent carboxyl-carboxylate pair. Such pairs are often associated with a pH-dependent conformational switch (Sawyer and James, 1982), and such a role for Asp464 (highly conserved in IR across species) appears consistent with data suggesting that high-affinity insulin binding involves deprotonation of a single IR acid residue (Waelbroeck, 1982). Known mutations of other residues close to Asp464, namely K460E (Kadowaki et al., 1990a, 1988) and N462S (Kadowaki et al., 1990b), are associated with defects in pH-dependent dissociation. Lys460, Asn462, and Asp464 lie in the final turn of L2 and in proximity to the two-fold axis at the apex of the ectodomain, well away from the insulin binding sites (McKern et al., 2006). Third, the revised location of conserved residues 519–529 (within the CC′ loop of FnIII-1, containing the Cys524-Cys524 disulfide) is intriguing. In both IR and IGF-1R, a peptide encompassing L2 residues 344–350 (IR numbering), gains protection from H/D exchange upon ligand binding (Houde and Demarest, 2011; Qiang et al., 2014): solvent-exposed backbone amides of this peptide map to the border of a conserved hydrophobic pocket on the surface of L2. The N-terminal region (residues 519–529) of the remodeled FnIII-1 CC′ loop now lies in immediate proximity to this pocket within the same monomer, with Phe518 (Tyr503 in IGF-1R) positioned ~10 Å away (Figure 3B). We speculate that, upon hormone binding, conformational change within the receptor head may dock the FnIII-1 CC′ loop into this pocket. Taken together, the above three observations suggest only a slight rearrangement within the (L2-FnIII-1)2 head module upon ligand binding.
Figure 3. Polypeptide Segments of IR Showing Altered H/D Exchange Profiles upon Ligand Binding and their Possible Burial upon Ligand Binding.
(A) Asp496, Arg498, and Asp499 (within FnIII-1) lie adjacent to Glu453 and Arg454 (within L2 of the opposite monomer).
(B) Residues 519–525 of the FnIII-1 CC′ loop lie adjacent to a conserved hydrophobic pocket on L2.
(C) Residues 655–658 of the ID lie in proximity to the L1 surface. Residues described are in stick representation, while the molecular surface of amides of residues demonstrating altered H/D exchange profiles is in black and indicated by green arrows.
In our revised structure, N-terminal residues of the ID are directed into electron density at the nexus of FnIII-2, FnIII-3, and the opposing L1 (Figure 3C), a significant alteration with respect to 2DTG/3LOH. We note that IGF-1R residues 641–648 (equivalent to IR 655–662) display an asymmetric pattern of H/D exchange upon IGF-I binding to IGF-1R (Houde and Demarest, 2011). This is intriguing, given that IR residues 657-662, though poorly ordered in the revised model, are anchored by Arg656 in proximity to insulin binding site 1 (Figure 3C). We speculate that their exposure to solvent may thus be altered by ligand-induced change in the relative disposition of domains FnIII-2, FnIII-3, and the opposing L1.
EXPERIMENTAL PROCEDURES
Sequencing
Sequencing of antibody-associated cDNA from 83-7 and 83-14 hybridomas was undertaken by Genscript, and DNA sequencing of IRΔβ by the Australian Genome Research Foundation.
Protein Production, Purification, and Crystallization
Stable expression of IRΔβ in CHO Lec8 cells and its subsequent purification as well as the production and purification of Fabs 83-14 and 83-7 are essentially as described previously (McKern et al., 2006). The complex crystallized was formed by sequential combination of IRΔβ, Fab 83-7, an iodinated form of the undecapeptide J101 (a weak binder to IR [Pillutla et al., 2002]), and Fab 83-14. Crystals were produced at 20°C by vapor diffusion using 1 µl of the complex (2.6 mg/ml) in 10 mM HEPES (pH 7.5), 0.02% sodium azide, and 10% D-trehalose, and 1 µl of well solution (10% polyethylene glycol 8000, 0.1 M MES buffer [pH 6.5], and 0.1 M MgAc2).
Diffraction Data Collection and Processing
A single crystal was placed in well solution plus 20% glycerol prior to cryocooling. Oscillation data (Table 1) were collected at ~100 K (beamline BL5A at the Photon Factory, Japan) and processed with XDS (Kabsch, 2010). The crystal was isomorphic to that of 2DTG/3LOH, allowing initial phasing by rigid-body refinement.
Crystallographic Refinement and Model Building
Tentative rebuilding of FnIII-1 and FnIII-3 was performed initially against maps derived from the 3LOH diffraction data using a combination of manual model building in Coot (Emsley and Cowtan, 2004) and iMDFF, iterated with refinement in autoBUSTER (Bricogne et al., 2011). During this process, iMDFF was developed as an extension of the interactive molecular dynamics (AutoIMD) plugin in VMD (Humphrey et al., 1996) to make use of the existing MDFF functionality of NAMD (McGreevy et al., 2014; Phillips et al., 2005). The source code is available upon request. The partially re-refined structure was then used as a starting model for refinement against the new dataset using iMDFF, Coot, and autoBUSTER. Glycan residues were modeled directly in the electron density maps. Where MS data were available for glycans attached to Lec8-expressed IR ectodomain (Sparrow et al., 2008), coordinates describing the most common glycoform were downloaded from the GLYCAM server (http://www.glycam.org) to initiate model building. Where glycan data were available only for glycans attached to CHO-K1-expressed ectodomain (Sparrow et al., 2008), we assumed that the glycan here would be identical apart from the terminal galactose, given the reasonable agreement of complex glycans across the two cell types (Sparrow et al., 2008). Where no MS data were available, glycan selection was guided by the electron density maps. Glycosidic bond torsion angles were compared against statistical distributions (Frank et al., 2007). No electron density could be assigned to iodinated J101 peptide, suggesting that it was absent or disordered.
Final stages of crystallographic refinement included TLS parameters, restrained individual B factors (except for residues judged to be poorly ordered, which were assigned one B factor per residue), and local structure similarity restraints (IR L1-CR-L2 to PDB: 2HR7 chain A and 2HR7 chain B, 83-7 light chain variable domain to PDB: 3MBX chain L, light chain constant domain to PDB: 1IL1 chain B and PDB: 3MBX chain L and heavy chain to PDB: 1FNS chain H, 83-14 light chain to PDB: 2VXT chain L and heavy chain to PDB: 1MJJ chain B). Refinement statistics are provided in Table 1.
Threading
Sequence register was assessed post hoc by real-space threading. For each FnIII domain in turn, all side-chain atoms were simultaneously assigned zero occupancy and the structure was then re-refined to convergence using the same autoBUSTER protocols as above for the complete refinement. A series of B factor sharpened (Fobs-Fcalc) electron density maps (Bsharp = −150, −160, −170, −180, −190 and −200 Å2) was then calculated—these maps contain features corresponding to the omitted side chains (Figure S1B). The sequence of the FnIII domain was then partitioned into runs of residues (“segments”) judged to be well ordered in the primary electron density maps. The amino acid sequence of the FnIII domain was then “threaded” in one-residue register increments through the volume of the side-chain omit map associated with the segment. Threading employed the decor_guess tool within O (Jones, 2004) and tested a ten-residue window on either side. The sequence register that scored the highest was then tested for agreement with that originally assigned.
RSCC Evaluation
For each of domains FnIII-1 and FnIII-3 in turn, a σA-weighted (2Fobs-Fcalc) omit map was calculated by first setting the occupancy of their remodeled residues to zero and then conducting five large cycles of crystallographic refinement within autoBUSTER to remove bias. An RSCC for each remodeled residue with respect to the above omit map (Bsharp = −100Å) was then calculated using O. The corresponding FnIII domain structure of 3LOH was then overlaid directly onto that of the new model, with the overlay being based solely on residues whose register was unchanged, and the same residue-wise RSCCs calculated for the register-changed residues. The mean of these RSCC values was then compared with that of the same residues in the new structure. As part of the improvement in the RSCC is likely attributable to improved data quality rather than to register re-assignment per se, we also computed as a baseline the increase in RSCC for those residues within the domain whose register was unaltered.
Supplementary Material
Highlights.
The structure of the insulin receptor ectodomain is improved to 3.3 Å resolution
The structure reveals new features within the receptor insert domain
The structure corrects errors in the first and third fibronectin type III domains
A new low-resolution crystallographic model-building strategy, iMDFF, is presented
Acknowledgments
This work was supported by National Health and Medical Research Council Project Grants 1005896 and 1058233 and the Hazel and Pip Appel Fund (to M.C.L.) and was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. J.W. and M.A.W. were supported in part by a grant from the National Institutes of Health (R01 DK040949). We thank CSIRO for making available the X-ray diffraction images, originally collected by Dr. Victor Streltsov (CSIRO) at the Photon Factory during a trip funded by the Australian Synchrotron Research Program, which was in turn funded by the Commonwealth of Australia’s Major National Research Facilities Program. Computational resources used for molecular modeling were provided by the HPC and Research Support Group at QUT. Part of M.C.L.’s laboratory is funded by Sanofi (Germany). J.W. is a consultant to Thermalin Diabetes, LLC (Cleveland, OH and Waban, MA, USA). M.A.W. is a co-founder and shareholder of Thermalin Diabetes, LLC, its Chief Scientific Officer, and a member of its Board of Directors.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structures factors have been deposited with the PDB (PDB: 4ZXB, obsoleting PDB: 3LOH, and 2DTG), with the extended model (in dimeric form) provided as Model S1.
SUPPLEMENTAL INFORMATION
Supplemental Information includes two figures, four tables, PDB model, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.str.2015.12.014.
AUTHOR CONTRIBUTIONS
T.I.C. and M.C.L. conducted crystallographic analysis; M.B.M. analyzed sequencing data; T.I.C. and B.J.S. undertook bioinformatics analysis; M.A.W. and J.W. analyzed alanine scanning data; T.I.C. developed the iMDFF approach; T.I.C., B.J.S., C.W.W., and M.C.L. wrote the paper.
REFERENCES
- Adamo M, Raizada MK, LeRoith D. Insulin and insulin-like growth factor receptors in the nervous system. Mol. Neurobiol. 1989;3:71–100. doi: 10.1007/BF02935589. [DOI] [PubMed] [Google Scholar]
- Adams TE, Epa VC, Garrett TP, Ward CW. Structure and function of the type 1 insulin-like growth factor receptor. Cell. Mol. Life Sci. 2000;57:1050–1093. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Boado RJ, Zhang Y, Pardridge WM. Humanization of anti-human insulin receptor antibody for drug targeting across the human blood-brain barrier. Biotechnol. Bioeng. 2007;96:381–391. doi: 10.1002/bit.21120. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER Version 2.10. Global Phasing Ltd; 2011. [Google Scholar]
- Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, DeLaBarre B, Davies JM, Weis WI. X-ray structure determination at low resolution. Acta Crystallogr. D Biol. Crystallogr. 2009;65:128–133. doi: 10.1107/S0907444908043795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiacchia KB. Quantitation of the class I disulfides of the insulin receptor. Biochem. Biophys. Res. Commun. 1991;176:1178–1182. doi: 10.1016/0006-291x(91)90409-z. [DOI] [PubMed] [Google Scholar]
- Craft S. Alzheimer disease: insulin resistance and AD–extending the translational path. Nat. Rev. Neurol. 2012;8:360–362. doi: 10.1038/nrneurol.2012.112. [DOI] [PubMed] [Google Scholar]
- De Meyts P. Insulin/receptor binding: the last piece of the puzzle?: what recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. Bioessays. 2015;37:389–397. doi: 10.1002/bies.201400190. [DOI] [PubMed] [Google Scholar]
- Ebina Y, Ellis L, Jarnagin K, Edery M, Graf L, Clauser E, Ou JH, Masiarz F, Kan YW, Goldfine ID, et al. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985;40:747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frank M, Lütteke T, von der Lieth CW. GlycoMapsDB: a database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res. 2007;35:287–290. doi: 10.1093/nar/gkl907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca F, Pandini G, Sciacca L, Pezzino V, Squatrito S, Belfiore A, Vigneri R. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch. Physiol. Biochem. 2008;114:23–37. doi: 10.1080/13813450801969715. [DOI] [PubMed] [Google Scholar]
- Houde D, Demarest SJ. Fine details of IGF-1R activation, inhibition, and asymmetry determined by associated hydrogen/deuterium-exchange and peptide mass mapping. Structure. 2011;19:890–900. doi: 10.1016/j.str.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jones TA. Interactive electron-density map interpretation: from INTER to O. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2115–2125. doi: 10.1107/S0907444904023509. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Bevins CL, Cama A, Ojamaa K, Marcus-Samuels B, Kadowaki H, Beitz L, McKeon C, Taylor SI. Two mutant alleles of the insulin receptor gene in a patient with extreme insulin resistance. Science. 1988;240:787–790. doi: 10.1126/science.2834824. [DOI] [PubMed] [Google Scholar]
- Kadowaki H, Kadowaki T, Cama A, Marcus-Samuels B, Rovira A, Bevins CL, Taylor SI. Mutagenesis of lysine 460 in the human insulin receptor. Effects upon receptor recycling and cooperative interactions among binding sites. J. Biol. Chem. 1990a;265:21285–21296. [PubMed] [Google Scholar]
- Kadowaki T, Kadowaki H, Rechler MM, Serrano-Rios M, Roth J, Gorden P, Taylor SI. Five mutant alleles of the insulin receptor gene in patients with genetic forms of insulin resistance. J. Clin. Invest. 1990b;86:254–264. doi: 10.1172/JCI114693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou M, Garrett TP, McKern NM, Hoyne PA, Epa VC, Bentley JD, Lovrecz GO, Cosgrove LJ, Frenkel MJ, Ward CW. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. USA. 2006;103:12429–12434. doi: 10.1073/pnas.0605395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGreevy R, Singharoy A, Li Q, Zhang J, Xu D, Perozo E, Schulten K. xMDFF: molecular dynamics flexible fitting of low-resolution X-ray structures. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2344–2355. doi: 10.1107/S1399004714013856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, Elleman TC, Richards KM, Bentley JD, Pilling PA, et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443:218–221. doi: 10.1038/nature05106. [DOI] [PubMed] [Google Scholar]
- Menting JG, Whittaker J, Margetts MB, Whittaker LJ, Kong GKW, Smith BJ, Watson CJ, Žáková L, Kletviková E, Jiráček J, et al. How insulin engages its primary binding site on the insulin receptor. Nature. 2013;493:241–245. doi: 10.1038/nature11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillutla RC, Hsiao KC, Beasley JR, Brandt J, Østergaard S, Hansen PH, Spetzler JC, Danielsen GM, Andersen AS, Brissette RE, et al. Peptides identify the critical hotspots involved in the biological activation of the insulin receptor. J. Biol. Chem. 2002;277:22590–22594. doi: 10.1074/jbc.M202119200. [DOI] [PubMed] [Google Scholar]
- Qiang G, Xue S, Yang JJ, Du G, Pang X, Li X, Goswami D, Griffin PR, Ortlund EA, Chan CB, Ye K. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes. 2014;63:1394–1409. doi: 10.2337/db13-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sawyer L, James MN. Carboxyl-carboxylate interactions in proteins. Nature. 1982;295:79–80. doi: 10.1038/295079a0. [DOI] [PubMed] [Google Scholar]
- Smith BJ, Huang K, Kong G, Chan SJ, Nakagawa S, Menting JG, Hu S-Q, Whittaker J, Steiner DF, Katsoyannis PG, et al. Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc. Natl. Acad. Sci. USA. 2010;107:6771–6776. doi: 10.1073/pnas.1001813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow LG, McKern NM, Gorman JJ, Strike PM, Robinson CP, Bentley JD, Ward CW. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 1997;272:29460–29467. doi: 10.1074/jbc.272.47.29460. [DOI] [PubMed] [Google Scholar]
- Sparrow LG, Lawrence MC, Gorman JJ, Strike PM, Robinson CP, McKern NM, Ward CW. N-linked glycans of the human insulin receptor and their distribution over the crystal structure. Proteins. 2008;71:426–439. doi: 10.1002/prot.21768. [DOI] [PubMed] [Google Scholar]
- Taylor RM, II, Hudson TC, Seeger A, Weber H, Juliano J, Helser AT. VRPN: a device-independent, network-transparent VR peripheral system. In: Shaw C, Wang W, Green M, editors. Proceedings of the ACM Symposium on Virtual Reality Software and Technology; ACM; Baniff, Alberta, Canada. 2001. pp. 55–61. [Google Scholar]
- The International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A, Bell JR, Chen EY, Herrera R, Petruzzelli LM, Dull TJ, Gray A, Coussens L, Liao YC, Tsubokawa M, et al. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature. 1985;313:756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- Waelbroeck M. The pH dependence of insulin binding. A quantitative study. J. Biol. Chem. 1982;257:8284–8291. [PubMed] [Google Scholar]
- Whittaker L, Hao C, Fu W, Whittaker J. High-affinity insulin binding: insulin interacts with two receptor ligand binding sites. Biochemistry. 2008;47:12900–12909. doi: 10.1021/bi801693h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.