Abstract
Background
New non-invasive methods are needed for sub-stratifying high-risk prostate cancer patients. Magnetic resonance spectroscopic imaging (MRSI) maps metabolites in prostate cancer, providing information on tumor aggressiveness and volume.
Purpose
To investigate the correlation between MRSI and treatment failure (TF) after radical prostatectomy (RP).
Methods
Two-hundred sixty-two patients who underwent endorectal MRI/MRSI followed by RP at our institution from 2003 to 2007 were studied. MRI stage, number of voxels in the MRSI index lesion (NILV), number of high-grade voxels (NHGV), and number of voxels containing undetectable polyamines (NUPV) were derived. Clinical outcome was followed until August, 2014. Treatment failure was defined as 1) biochemical recurrence (BCR), 2) persistently detectable PSA after RP, or 3) adjuvant therapy initiated in the absence of BCR. MRI/MRSI features and clinical parameters were compared to TF by univariate Cox Proportional Hazards Regression. After backward selection, each MRSI parameter was included in a separate regression model adjusted for NCCN-based clinical risk score (CRS), number of biopsy cores positive (NPC), and MRI stage.
Results
In univariate analysis, all clinical variables were associated with TF in addition to MRI stage, NILV, NHGV, and NUPV. In multivariate analysis, NILV, NHGV, and NUPV were also significant risk factors for TF (p=0.016, p=0.002, p=0.006, respectively). The association between the number of tumor voxels with undetectable polyamines and the probability of treatment failure has not been previously reported. The number of MRSI cancer voxels correlated with ECE (p < 0.0001).
Conclusions
MRSI was associated with post-radical prostatectomy treatment failure in models adjusted for the number of positive biopsy cores and clinical risk score. This is the first report that in radical prostatectomy patients MRSI has an association with treatment failure independent of the number of positive biopsy cores. MRSI may help the clinician determine whether patients with high risk disease who undergo RP are candidates for specialized additional treatment.
Keywords: Gleason score, MRI, spectroscopy, MRSI, prostate cancer, choline
1. Introduction
Prostate cancer (PCa) is the most common cancer in males and the second leading cause of cancer deaths in the U.S. [1]. Candidates for radical prostatectomy (RP) may have clinically low, intermediate, or high-risk disease and further prognostic data would aid the physician making treatment decisions [2,3]. Proton MR spectroscopic imaging (1H-MRSI) permits the analysis of metabolism as reflected by the levels of choline-containing compounds, polyamines (spermine and spermidine), creatine, and citrate in PCa [4]. As a result, prognosis-related features such as extracapsular extension (ECE) [5] and Gleason score [6–8] have been linked to MRSI, and several studies have suggested a correlation between MRSI and clinical outcome [9–11]. In 2010, Zakian, et. al. reported that both the degree of abnormality in the ratio of total choline plus creatine to citrate (CC/C) and the MRSI tumor volume correlated with biochemical failure after RP in 130 patients [9]. A similar result was reported in 67 patients treated with radiation therapy [10]. However, relatively coarse spatial resolution, long scan times and the widespread implementation of diffusion-weighted imaging (DW-MRI) have mitigated interest in proton MRSI of the prostate.
In this study, we sought to confirm the prognostic potential of 1H-MRSI in a large surgical population with long-term followup. To this end, we retrospectively examined MRSI data from 403 subjects who underwent endorectal MRI/MRSI followed by radical prostatectomy. The MRI/MRSI data were included with patient clinical data in predictive models of treatment failure. Because of the widespread availability of individual biopsy core information, we included the numbers of positive and negative cores in our analysis to determine the value of MRSI metabolic data in the presence of this information.
2. Material and Methods
2.1 Data Collection
Our population consisted of 403 consecutive patients who were scheduled to undergo endorectal MRI/MRSI followed by RP from July 2003 to December 2007. At that time, all patients referred for MRI were scheduled for combined MRI/MRSI. One-hundred forty-one subjects were excluded for technical or clinical reasons as follows: a) prior treatment for PCa including neoadjuvant hormonal therapy (N = 28), b) non-commercial software used for MRSI (N = 37), c) MRSI portion not performed due to the presence of a metallic hip implant or patient discomfort leading to early exam termination (N = 15), d) data not retrievable (N = 5), e) MRI reader data unavailable (N = 4), f) clinical followup or pathology data unavailable (N = 8), g) biopsy < 3 weeks prior to MRI (N = 2), h) no prior positive biopsy (N = 2), i) data acquisition error (N = 3), and j) MRSI data unusable due undetectable metabolites and/or artifact including motion and lipid contamination (N = 37). The MRSI-specific factors i ) and j) resulted in the exclusion of 40 patients (10% of the study population). A retrospective waiver of authorization was granted by our institutional review board and the study was HIPAA-compliant.
Endorectal MRI/MRSI was performed on a 1.5T scanner (Excite, G.E. Medical Systems, Waukeshau, WI). Diagnostic imaging consisted of 2D multi-slice axial T1-weighted images (TR/TE = 400–700/10–14 msec, slice thickness = 5 mm, inter-slice gap = 0 mm, field of view [FOV] = 24–26 cm, matrix 256×192), and 2D axial, coronal, and sagittal T2-weighted fast spin-echo images (TR/effective TE = 4400/102 msec, echo train length = 12, slice thickness = 3 mm, inter-slice gap = 0 mm, FOV = 14 cm, matrix = 256×192, excitations = 4). Following diagnostic MR imaging, MRSI was performed using a commercially available prostate MRSI acquisition package (PROSE, General Electric, Waukeshau, WI) which incorporated double spin-echo excitation [12] with spectral-spatial localization pulses [13]. Using 3-dimensional phase-encoding, voxel grids of 16x8x8 samples were acquired in 17 minutes with the following acquisition parameters: spectral width = 2500, time samples = 512, TR = 1000 ms, TE = 130 ms, 1 acquisition, spectroscopic volume of interest = 11.0 × 5.5 × 5.5 cm3, spatial resolution = 6.9 mm). The MRSI acquisition technique did not change over the study period.
The MRSI data were processed offline on a G.E. Advantage Workstation using the Functool software (G.E. Medical Systems, Waukeshau, WI). During post-processing, data were zero-filled to 3 mm resolution in the superior-inferior direction and 1024 points in the spectral dimension, Fourier transformed, automatically phased, and baseline corrected. Manual phasing and peak alignment were performed if necessary. Numerical integration of metabolite regions of interest was performed to determine peak areas. Voxels adjacent to the ejaculatory ducts or prostatic urethra demonstrating elevated choline were assumed to reflect glycerophosphocholine in seminal fluid and were excluded from analysis. Each patient’s MRSI data set was assigned a data quality score based on the percentage of voxels which had interpretable levels of metabolites and no lipid contamination or baseline artifact. A data quality score of 0 corresponded to fewer than 25% of voxels interpretable, while scores of 1, 2, and 3 corresponded to 25–50%, 50–75%, and 75–100% voxels interpretable, respectively. Patients having a data quality score of 0 were eliminated from the analysis.
2.2 Cancer identification on imaging and pathology
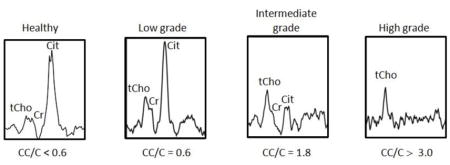
All MRI examinations were interpreted by members of our Genitourinary (GU) Radiology team who had a minimum of 3 years of experience interpreting endorectal MRI [14]. Based on the Radiologist’s report, each patient was assigned a MRI stage score indicating the extent of disease (scale 1–7) (see Appendix) [15]. MRSI spectral grids were overlaid on corresponding axial-T2 weighted images as in Figure 1a. Cancerous voxels were identified based on previously published criteria [16] by a single MR spectroscopist with greater than 10 years of experience in prostate cancer. The sextant, zone, and anterior/posterior location of each MRSI lesion were recorded. Each tumor voxel was assigned a high, intermediate, or low MRSI grade based on CC/C value (low grade: CC/C 0.5 ≤ CC/C < 0.7, intermediate grade: 0.7 ≤ CC/C < 3.0, high grade: CC/C ≥ 3.0) see Appendix [11]. We also recorded a relative polyamine (PA) score which ranged from 0 (PA peak undetectable) through 2 (PA peak ≥ tCho peak). Figure 1B includes spectra with increasing metabolic abnormality indicated by increasing total choline (tCho) and decreasing levels of polyamines and citrate. The MRSI “index lesion” [9] was defined as the largest cluster of adjoining cancer voxels.
Figure 1.


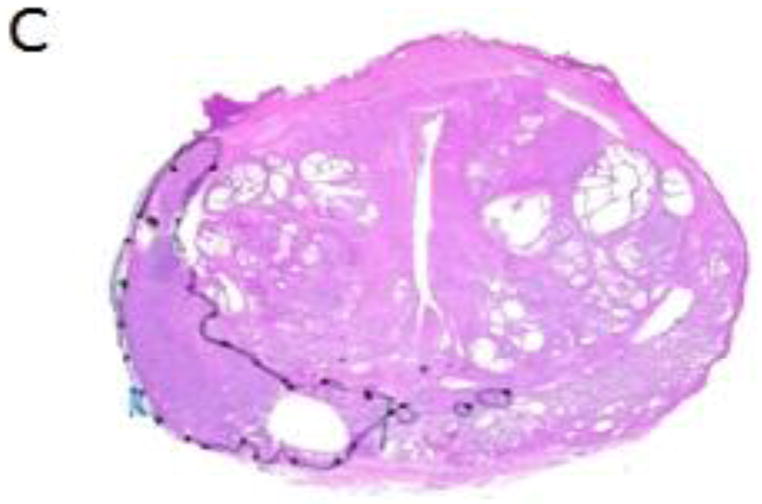
A) Example of MRSI voxel grid overlaid on a T2-weighted image of the prostate gland in a patient with prostate cancer. The voxels with metabolite levels indicating cancer are outlined in red. C) Examples of spectra from healthy peripheral zone (left) and prostate cancer (center, right). The healthy peripheral zone spectrum demonstrates a high citrate peak (Cit), as well as choline-containing compounds (tCho), polyamines (PA), and creatine/phosphocreatine (Cr). The central voxel spectrum has reduced polyamines and citrate relative to choline and was considered intermediate grade. The spectrum in the voxel on the right contains elevated choline and undetectable polyamines and was considered high grade. C) Whole-mount histopathology section of the prostate corresponding to the image in (A). The region outlined in black is the dominant lesion which has Gleason score ≥ 4+4. Within the lesion, there is artifact where fresh tissue was sampled at the time of surgery.
The prostatectomy specimen whole-mount preparation has been described previously [7]. A genitourinary pathologist with >10 years of experience outlined cancer foci in fixed, stained, microsections using green ink for predominantly Gleason score 3 morphology and black for predominantly Gleason score ≥ 4 morphology. Figure 1C contains the whole-mount prostate tumor map corresponding to the prostate section image in Figure 1A. After photography and digitization, tumor volumes were calculated using ImageJ [17].
A GU radiologist who did not participate in the original MRI readings determined the coincidence between the MRSI lesions and pathologic lesions using methodology published previously [18]. The radiologist was provided with spectral grids which indicated the location of cancer voxels. These were compared to the pathologic tumor maps to determine whether the MRSI-detected lesion matched a pathologic lesion.
2.3 Clinical Data and Treatment Failure
Clinical data including age, clinical stage, PSA value, biopsy Gleason score, and number of positive and negative biopsy cores were obtained from a hospital database. We assigned each patient a clinical risk score (CRS) (low, medium, or high) based on the NCCN guidelines [19]. The NCCN CRS is generated from a weighted combination of baseline values of PSA, clinical stage, and biopsy Gleason score to help patients and physicians make treatment decisions (See Appendix). After radical prostatectomy, PSA testing was performed every 6–12 months. Treatment failure (TF) was defined as 1) biochemical recurrence (BCR), 2) persistently detectable PSA after RP, or 3) adjuvant therapy initiated in the absence of BCR. A patient was considered to have BCR if, after achievement of a non-detectable PSA, the PSA rose to ≥ 0.1 ng/mL and remained ≥ 0.1 ng/mL on repeat assessment. The BCR date was the date of the first PSA ≥ 0.1 ng/mL. For persistently detectable PSA after RP, the date of failure was the date of the initial post-surgical PSA measurement. All patients in category (3) were treated with androgen deprivation or radiation therapy due to high-risk surgical pathology features including positive margins and/or positive lymph nodes. The failure date for these patients was the first day of adjuvant therapy. The end date for reporting was August 27, 2014.
2.4 Statistical Analysis
MRSI parameters were compared to pathologic lesion Gleason score using univariate ordinal logistic regression for clustered data, adjusting for multiple measurements per patient. Parameters examined were the number of voxels in each index lesion including all MRSI grades (NILV), number of high-grade voxels (NHGV) and number of undetectable polyamine voxels (NUPV). The pathologic Gleason scores were assigned as: 0 (negative on pathology), ≤ 3+4 and >3+4.
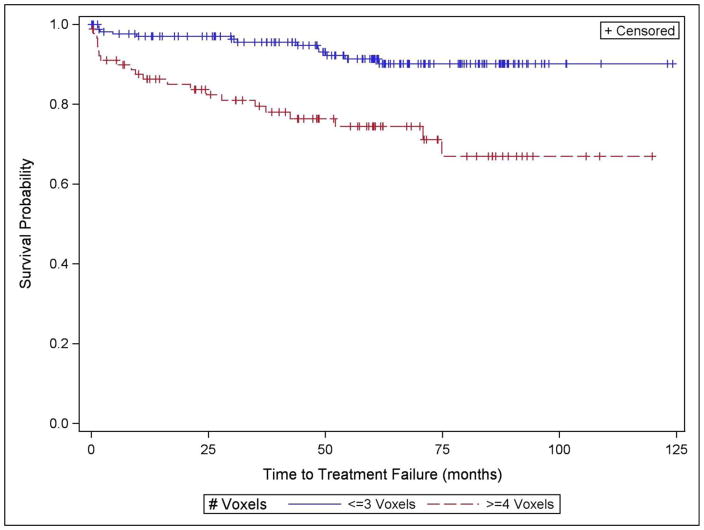
The relationship between the presence of ECE and NILV was assessed with univariate logistic regression on a per patient basis. For the purpose of comparing MR parameters to TF, biopsy Gleason score was dichotomized to ≤ 3+4 or >3+4, and clinical stage to T1–T2 or T3–T4. All other variables were treated as continuous measurements. Univariate Cox proportional hazards regression assessed the relationship between clinical, demographic and imaging variables and TF. Time to treatment failure (TTF) was defined as the interval between prostatectomy and TF date or last follow-up date. Patients without TF were censored at last follow-up. We graphically assessed the univariate relationship between NILV and TF using Kaplan-Meier curves and corresponding log-rank test, dichotomizing NILV into < 4 voxels vs. ≥ 4 voxels based on a previous finding that index lesions consisting of 4 or more voxels conferred a greater risk of biochemical recurrence [9].
Multicollinearity was examined among MRSI variables. We ran separate Cox proportional models for each MRSI parameter adjusting for clinical parameters and MRI stage. To determine appropriate clinical parameters, backward selection methods were employed on the following clinical variables with α=0.01 as the elimination criterion: age, number of negative biopsy cores, number of positive cores, percent of cores positive, and NCCN-based CRS. P-values < 0.05 were considered statistically significant. Harrell’s c-index was used to assess the discriminatory ability of the multivariable models. Analyses were performed using SAS 9.2 (SAS Institute, Cary, NC) and Stata SE 12 (StataCorp, College Station, TX USA).
3. Results
3.1 Clinical and Imaging Summary
The clinical characteristics of the 262 patients are described in Table 1. The majority of patients were clinical stage T1c (59.6%) and had biopsy Gleason score = 3+3 (67.94%). Using NCCN-based risk guidelines, 132 patients (50%) were classified as clinically low risk, 102 patients (39%) were in the intermediate risk category, and 28 (11%) were high-risk. Table 2 summarizes the clinical outcome. With a median follow up time of 57.6 months (range: 0.1–124.2 months), 35 (13.4%) patients experienced treatment failure with biochemical recurrence being the most common type of TF.
Table 1.
Clinical descriptive statistics (N=262 patients).
| Median | Range | |
|---|---|---|
| Age at Diagnosis (years) | 58.8 | 39.6–72.4 |
| Pre-Biopsy PSA (ng/mL) | 4.9 | 0.8–35.9 |
| Clinical Stage | Number | % |
| T1c | 155 | 59.16 |
| T2 unspecified | 6 | 2.29 |
| T2a | 32 | 12.21 |
| T2b | 16 | 6.11 |
| T2c | 35 | 13.36 |
| T3a | 15 | 5.73 |
| T3b | 1 | 0.38 |
| T4 | 2 | 0.76 |
| Biopsy Gleason Score | Number | % |
| 3+3 | 178 | 67.94 |
| 3+4 | 49 | 18.70 |
| 4+3 | 22 | 8.40 |
| 4+4 | 10 | 3.82 |
| 4+5 | 3 | 1.14 |
| Median | Range | |
| Number of Negative Cores (NNC) | 9.0 | 0.0–29.0 |
| Number of Positive Cores (NPC) | 2.0 | 1.0–14.0 |
| Percent of Cores Positive | 20.0 | 3.7–100.0 |
Table 2.
Clinical Outcome in 262 patients.
| Median | Range | |
|---|---|---|
| Follow-up time (months) | 57.6 | 0.1–124.2 |
| Time to failure (months) | 16.3 | 0.2–74.8 |
| Outcome | N | % |
| No Failure | 227 | 86.64 |
| Treatment Failure | 35 | 13.36 |
| Failure Type | N | % |
| BCR | 19 | 7.25 |
| Persistently Detectable PSA after RP | 10 | 3.82 |
| Adjuvant Treatment without BCR | 6 | 2.29 |
Table 3 contains the MRSI index lesion characteristics and the MRI stage for these patients. One-hundred seventy-two patients (66%) had < 4 voxels per index lesion while 90 (34%) had NILV ≥ 4. The median number of voxels in the index lesion (NILV) was 3 (range 1–79) indicating an approximate median index lesion volume of 0.428 mm3 based on an individual nominal voxel size of 0.143 cm3. The majority of MRSI lesions were located in the peripheral zone or bridged the peripheral and transition zones. Because of the very low number of TZ tumors, segregation by zone in the treatment failure predictive models was not possible and the tumors for all zones were grouped together. Two-hundred thirty-five patients (90%) had low MRI stage disease (stage 1–3) while 27 (10%) had high MRI stage disease (stage 4–7).
Table 3.
MRSI Index Lesion descriptive statistics and MRI stage in 262 patients. IQR = interquartile range.
| MRSI Index Lesions-total voxels | N | % | ||
| No lesion (0 voxels) | 79 | 30.1 | ||
| Lesions with 1–3 voxels | 93 | 35.5 | ||
| Lesions with ≥ 4 voxels | 90 | 34.4 | ||
| MRSI Index Lesion Voxel Characteristics | median | range | IQR | |
| Number of voxels (NILV) | 3 | 1 – 79 | 2.0–7.0 | |
| Number of high grade voxels (NHGV) | 0 | 0–55 | 2.0–8.0 | |
| Number of voxels with undetectable | 0 | 0–63 | 1.0–6.0 | |
| polyamines (NUPV) | ||||
| MRSI Index Lesion Zonal Location | N | % | ||
| Peripheral Zone | 143 | 54.6 | ||
| Transition Zone | 14 | 5.4 | ||
| Both PZ and TZ | 26 | 9.9 | ||
| No Lesion | 79 | 30.1 | ||
| MRSI Data Quality Score | N | % | ||
| 1 | 51 | 19.5 | ||
| 2 | 119 | 45.4 | ||
| 3 | 92 | 35.1 | ||
| MRI Stage | N | % | ||
| 1 | 29 | 11.06 | ||
| 2 | 130 | 49.62 | ||
| 3 | 76 | 29.01 | ||
| 4 | 18 | 6.87 | ||
| 5 | 1 | 0.38 | ||
| 6 | 5 | 1.91 | ||
| 7 | 3 | 1.14 |
3.2 MRI/MRSI comparison to clinical results
NILV was positively associated with the lesion Gleason score on whole-mount surgical pathology (p=0.0002); however, the lesion Gleason score was not correlated with NHGV (p=0.28), nor NUPV (p=0.055). ECE was positively associated with the total number of cancer voxels detected by MRSI (p<0.0001).
In the univariate survival analyses, as shown in Table 4, all standard clinical variables as well as the NCCN-based risk score were associated with TF (p < 0.0001 – p = 0.025). In addition the number of negative biopsy cores (NNC) (p = 0.003), number of positive cores (NPC) (p < 0.0001), and percent of cores positive (p < 0.0001) were associated with treatment failure. Patients with clinical stage 3 or 4 had a higher risk of TF than lower stage patients, and those with biopsy Gleason score >3+4 had a higher risk of TF than those with a lower score. Patients in the NCCN-based high risk category had 15 times the probability of TF compared to the low risk group [95% CI: 6.29 −37.48]. MRI stage was associated with TF (p < 0.0001) as were the MRSI measures NILV, NHGV, and NUPV (all p < 0.0001). There were 227 patients who did not have treatment failure and 35 who failed. One-hundred fifty-nine of the non-failures had NILV < 4 while 68 had NILV ≥ 4. Of the 35 patients with TF, 13 had NILV <4 voxels and 22 had NILV ≥ 4. Patients with NILV ≥ 4 had significantly shorter TTF than those with smaller MRSI index lesions (p < 0.0001) (Figure 2). There was no difference in TTF when the large index lesion group (NILV ≥ 4 voxels) was sub-stratified according to the presence or absence of at least one high-grade voxel (p = 0.500).
Table 4.
Univariate Cox Proportional Hazard analysis of clinical, biopsy and imaging variables for the prediction of treatment failure in 262 patients with 35 treatment failures. HR = Hazards ratio. REF indicates the reference category to which the other categories are compared. For the continuous variables, the Hazards ratio indicates the percentage increase in the chance of a treatment failure event for a unit increase in the variable. For example, a 1.07 HR for age at diagnosis indicates that the chance of treatment failure increases by 7% for every year of increase in age. For the categorized variables, the HR indicates the percentage increase in the chance of TF compared to the reference category. All variables were significant (p < 0.05-p < 0.0001).
| Variable type | Variable | Category | N | HR | [95% CI] |
|---|---|---|---|---|---|
| Clinical | Age at Diagnosis | 1.07 | [1.01–1.13] | ||
| Pre-treatment PSA | 1.09 | [1.03–1.15] | |||
| Clinical Stage | T1–T2 | 244 | REF | ||
| T3–T4 | 18 | 8.17 | [3.99–16.75] | ||
| Biopsy Gleason Score | ≤ 3+4 | 227 | REF | ||
| > 3+4 | 35 | 8.13 | [4.18–15.81] | ||
| NCCN-based clinical risk score (CRS) | low | 132 | REF | ||
| intermediate | 102 | 2.51 | [0.99–6.37] | ||
| high | 28 | 15.36 | [6.29–37.48] | ||
| Biopsy Core | # negative cores (NNC) | 0.86 | [0.78–0.95] | ||
| Data | # positive cores (NPC) | 1.28 | [1.16–1.41] | ||
| % of cores positive | 1.03 | [1.02–1.04] | |||
| MRI | MRI Stage | ||||
| 1–3 | 235 | REF | |||
| 4–7 | 27 | 6.57 | [3.25–13.26] | ||
| MRSI | # of index lesion voxels (NILV) | 1.08 | [1.06–1.11] | ||
| # of high grade (HG) voxels (NHGV) | 1.14 | [1.08–1.21] | |||
| # of undetectable polyamine voxels (NUPV) | 1.11 | [1.07–1.16] |
Figure 2.
Results of Kaplan-Meier analysis of the association between time to treatment-failure and the number of voxels in the MRSI index lesion (NILV). The group with greater MRSI tumor volume as represented by NILV had significantly shorter time to treatment-failure (p < 0.0001)
Based on the backward selection model, the NCCN-based clinical risk score and number of positive biopsy cores were included as predictors in the multivariate models which incorporated MRI and MRSI findings (Table 5). In model 1 (clinical model), both clinical risk score (p < 0.0001) and number of positive biopsy cores (p = 0.0004) were significantly associated with the probability of TF. MRI stage was significant (p = 0.038) when added to the clinical data (model 2). In models 3, 4, and 5, NILV (p = 0.016), NHGV (p=0.002), and NUPV (p = 0.006) were separately added to model 2 and remained significant. The c-index for model 1 was 0.79. For models 3–5 when MRI and MRSI information were added to the model, regardless of which MRSI parameters were included, the c-index was approximately 0.81 suggesting that joint information about MRI and MRSI may be predictive of treatment failure. This observation needs to be confirmed, however, in a larger, independent study.
Table 5.
Multivariate Cox Proportional Hazard Analysis of Clinical, Biopsy and Imaging Variables for the Prediction of Treatment Failure after Radical Prostatectomy. Bold type indicates statistical significance.
| Model | Variable | Category | HR | [95% CI] | p value |
|---|---|---|---|---|---|
| 1 (Clinical) | # Positive Biopsy Cores (NPC) | 1.22 | [1.09–1.37] | 0.0004 | |
| NCCN Clinical Risk Score (CRS) | <0.0001 | ||||
| Low | REF | ||||
| Intermediate | 2.02 | [0.79–5.17] | |||
| High | 11.1 | [4.46–27.63] | |||
|
| |||||
| 2 (Clinical + MRI Stage) | # Positive Biopsy Cores (NPC) | 1.19 | [1.06–1.33] | 0.0024 | |
| NCCN Clinical Risk Score (CRS) | <0.0001 | ||||
| Low | REF | ||||
| Intermediate | 2.0 | [0.78–5.12] | |||
| High | 8.75 | [3.39–22.57] | |||
| MRI Stage | 0.0378 | ||||
| 1–3 | REF | ||||
| 4–7 | 2.31 | [1.05–5.07] | |||
|
| |||||
| 3 (Clinical + MRI stage + # index lesion voxels) | # Positive Biopsy Cores (NPC) | 1.15 | [1.02–1.3] | 0.023 | |
| NCCN Clinical Risk Score (CRS) | <0.0001 | ||||
| Low | REF | ||||
| Intermediate | 1.74 | [0.66–4.57] | |||
| High | 7.41 | [2.83–19.43] | |||
| MRI Stage | 0.16 | ||||
| 1–3 | REF | ||||
| 4–7 | 1.78 | ||||
| # MRSI Voxels (NILV) | 1.04 | [1.01–1.08] | 0.0159 | ||
|
| |||||
| 4 (Clinical + MRI stage + # high grade voxels) | # Positive Biopsy Cores (NPC) | 1.16 | [1.03–1.31] | 0.0151 | |
| NCCN Clinical Risk Score (CRS) | <0.0001 | ||||
| Low | REF | ||||
| Intermediate | 1.8 | [0.69–4.7] | |||
| High | 7.86 | [3.02–20.44] | |||
| MRI Stage | 0.0458 | ||||
| 1–3 | REF | [1.02–4.94] | |||
| 4–7 | 2.24 | ||||
| # High grade MRSI voxels | 1.11 | [1.04–1.18] | 0.0016 | ||
| (NHGV) | |||||
|
| |||||
| 5 (Clinical + MRI stage + # zero polyamine voxels) | # Positive Biopsy Cores (NPC) | 1.17 | [1.04–1.32] | 0.0097 | |
| NCCN Clinical Risk Score (CRS) | <0.0001 | ||||
| Low | REF | ||||
| Intermediate | 1.78 | [0.68–4.65] | |||
| High | 7.8 | [2.98–20.43] | |||
| MRI Stage | 0.21 | ||||
| 1–3 | REF | ||||
| 4–7 | 1.71 | [0.74–3.96] | |||
| # undetectable polyamine voxels (NUPV) | 1.07 | [1.02–1.13] | 0.0056 | ||
4 Discussion
In 262 patients, our study found that the number of voxels in the pretreatment MRSI index lesion was a significant risk factor for treatment failure after radical prostatectomy in a model including NCCN-based clinical risk score and number of positive biopsy cores. This is the largest study to date relating MRSI data to long-term clinical outcome in prostate cancer. Both NUPV and NHGV were also significantly associated with TF. The association between the number of lesion voxels with undetectable polyamines (NUPV) and treatment failure has not been previously reported. Our results indicate that TTF is shorter in patients with MRSI index lesions comprised of ≥ 4 voxels, in agreement with a prior study in a separate, smaller patient population [9]. We further demonstrated that an increased number of cancer voxels was associated with ECE, supporting a previous report [5].
We and others have reported a correlation between MRSI-measured metabolite ratios containing total choline, creatine, and citrate and the pathologic Gleason score [6–8,20], and ex vivo tissue studies have related spermine, citrate, and choline-containing compounds to Gleason grade [21,22]. These data support our hypothesis that the metabolic information provided by MRSI helps predict TF by reflecting both lesion aggressiveness and volume. The new finding that the number of index lesion voxels with undetectable polyamines is associated with treatment failure tends to agree with the previously shown relationship to aggressiveness [21] which may be due to the loss of glandular tissue when tumor is present.
The finding that MRSI parameters remained significantly associated with treatment failure in models including the number of positive biopsy cores in addition to standard clinical parameters, has not been previously reported. This suggests that whole-gland coverage by MRSI provides an advantage over biopsy where the number of samples is limited. In contrast to our previous study [9], the presence of one or more high-grade voxels in large MRSI lesions did not increase the risk of TF. It is possible that the finding in the initial study with fewer patients and fewer treatment failures was fortuitous. With its relatively coarse spatial resolution, 1H-MRSI has been shown to be of limited value in low-risk, low volume prostate cancer [7]. Our finding that metabolic tumor volume is associated with treatment failure suggests that 1H-MRSI may have utility for identifying patients in the intermediate or high risk category who are most likely to experience treatment failure. Further, in patients with intermediate and high risk disease, ECE has been shown to be a strong predictor of BCR and metastases after RP [23], and we noted a correlation between the number of index lesion voxels and ECE. To enhance the chance of long-term survival in high-risk patients, new treatment options including pre-surgical systemic chemotherapy, hormonal therapy, and targeted agents are being investigated [24]. A recent study suggested that high-risk patients undergoing RP should be further stratified based on primary Gleason pattern and the number of high-risk criteria [3]. By noninvasively providing metabolic information reflecting Gleason score and tumor volume, MRSI may have the ability to aid in substratification of these patients.
A limited number of studies have assessed the ability of other MRI contrast modalities to predict treatment failure after RP [15,25]. A much larger number of studies have attempted to correlate MR imaging findings with tumor aggressiveness, and a comprehensive review is beyond the scope of this manuscript. Diffusion weighted MRI (DW-MRI) and corresponding apparent diffusion coefficient (ADC) maps have shown promise [26–31], and combined ADC and MRSI data have also been examined [20,26] although there is no consensus on whether the combination improves predictive power. The evidence that ADC correlates with PCa aggressiveness suggests that DWI data may correlate with long-term outcome. One of the major weaknesses of our current study was the lack of diffusion data. This was due to the fact that DWI was not standard-of-care when patient accrual began. As the metabolic tumor volume was found to be predictive of treatment failure, it would be extremely valuable to evaluate both MRSI tumor volume and DWI tumor volume as predictors of TF in the same patient population.
The study had several other limitations. The retrospective nature of the study introduces the potential for inherent biases. Because of the limited number of patients with high-grade tumors, we dichotomized Gleason score into ≤ 3+4 and >3+4 groups rather than segregating further. Our inclusion of 6 patients in the TF category who were given adjuvant therapy in the absence of BCR may have led to an overestimation of failure events. However, the treating physicians determined that these patients were at very high risk for failure given their surgical pathology results.
Multiple improvements in prostate 1H-MRSI have occurred since the acquisition of the data for this study. Three-Tesla scanners are widely available and the increased signal due to higher field strength could be employed to shorten scan time, improve spatial resolution, or improve spectral quality. Optimized acquisition protocols including weighted k-space sampling and compressed sensing may reduce scan times without loss of spectral quality and may permit 1H-MRSI without an endorectal coil [32–34]. These results suggest that 1H-MRSI could be integrated with other MR imaging contrast modalities to provide optimal predictive data in a clinically acceptable scan time. To overcome the need for expertise in 1H-MRSI interpretation, automated spectral interpretation methodologies are being investigated [35,36]. Alternatively, the advent of hyperpolarized 13C-MRSI has brought about the introduction of new techniques and tracers which may permit rapid measurement of prostate cancer metabolism [37]. These developments suggest that the non-invasive measurement of metabolic tumor volume could be incorporated into clinical practice.
5. Conclusions
We have shown in a population of 262 patients with up to 10 years of follow-up that the MRSI-detected metabolic tumor volume is significantly associated with treatment failure after radical prostatectomy. By noninvasively providing metabolic information reflecting both tumor aggressiveness and volume, MRSI may help to identify patients with very high risk of treatment failure who might benefit from new agents and therapeutic clinical trials.
Acknowledgments
This work was supported by: NIH R01-CA076423.
Abbreviations
- BCR
biochemical recurrence
- CC/C
ratio of total choline plus creatine to citrate
- CRS
clinical risk score
- DW-MRI
diffusion-weighted MRI
- ECE
extracapsular extension
- MRSI
magnetic resonance spectroscopic imaging
- NCCN
National Comprehensive Care Network
- NHGV
number of high-grade voxels
- NILV
number of index lesion voxels
- NPC
number of positive biopsy core
- NUPV
number of voxels with undetectable polyamines
- PA
polyamines
- PCa
prostate cancer
- PSA
prostate-specific antigen
- RP
radical prostatectomy
- tCho
total choline
- TF
treatment failure
- TTF
time to treatment failure
5. Appendix
5.1 Assignment of MRI stage score
Using the MRI exam report in the patient’s clinical record, each patient was assigned a MRI stage score based on a 7-point scale with the following values: 1) No Tumor Seen, 2) Tumor Seen; No extracapsular extension (ECE), 3) Can’t Rule Out ECE, 4) Unilateral ECE, 5) Bilateral ECE, 6) Seminal Vesicle Invasion, and 7) Lymph Node Invasion.
5.2 Assignment of MRSI grade to spectrum
Each tumor voxel was assigned a high, intermediate, or low MRSI grade based on CC/C value (low grade: CC/C 0.5 ≤ CC/C < 0.7, intermediate grade: 0.7 ≤ CC/C < 3.0, high grade: CC/C ≥ 3.0). Below are examples of spectra corresponding to each grade.
5.3 Assignment of NCCN-based Clinical Risk Score (CRS)
In patients with non-metastatic prostate cancer, the NCCN assigns 5 risk groups based on biopsy Gleason score, clinical stage, PSA, and biopsy core data when available: very low, low, intermediate, high, and very high [17]. The very low risk group is differentiated from the low risk group using biopsy core information, while the high and very high risk groups are differentiated by clinical stage (T3b or T4 indicates very high risk (locally advanced)). For the purposes of our study we used a simplified, 3-level risk assignment (low, intermediate, high) which combines the NCCN very low and low risk groups, and the high and very high risk groups. Our low risk group includes patients with clinical stage T1c or T2a disease, biopsy Gleason score ≤ 6 and PSA < 10 ng/mL. The intermediate group has clinical stage T2b or T2c, or PSA 10–20 ng/mL or biopsy Gleason score = 7. Our high risk group is characterized by clinical stage ≥ T3a, PSA ≥ 20 ng/mL, or biopsy Gleason score ≥ 8.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kristen L. Zakian, Email: zakiank@mskcc.org.
William Hatfield, Email: wnhat1@student.monash.edu.
Omer Aras, Email: araso@mskcc.org.
Kun Cao, Email: kun-cao@hotmail.com.
Derya Yakar, Email: derya.yakar@radboudumc.nl.
Debra A. Goldman, Email: goldmand@mskcc.org.
Chaya S. Moskowitz, Email: moskowc1@mskcc.org.
Amita Shukla-Dave, Email: davea@mskcc.org.
Yousef Mazaheri Tehrani, Email: mazahery@mskcc.org.
James Eastham, Email: easthamj@mskcc.org.
Hedvig Hricak, Email: hricakh@mskcc.org.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Hong SK, Vertosick E, Sjoberg DD, Scardino PT, Eastham JA. Insignificant disease among men with intermediate-risk prostate cancer. World Journal of Urology. 2014;32(6):1417–1421. doi: 10.1007/s00345-014-1413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jo JK, Kook HR, Byun SS, Lee SE, Hong SK. Stratification of Contemporary Patients Undergoing Radical Prostatectomy for High-risk Prostate Cancer. Ann Surg Oncol. 2014 doi: 10.1245/s10434-014-4183-7. [DOI] [PubMed] [Google Scholar]
- 4.Kurhanewicz J, Vigneron DB, Hricak H, et al. Three-dimensional H-1 MR Spectroscopic Imaging of the in Situ Human Prostate with High (0.24–0. 7-cm3) Spatial Resolution Radiology. 1996;198(3):795–805. doi: 10.1148/radiology.198.3.8628874. [DOI] [PubMed] [Google Scholar]
- 5.Yu KK, Scheidler J, Hricak H, et al. Prostate cancer: prediction of extracapsular extension with endorectal MR imaging and three-dimensional proton MR spectroscopic imaging. Radiology. 1999;213(2):481–488. doi: 10.1148/radiology.213.2.r99nv26481. [DOI] [PubMed] [Google Scholar]
- 6.Kurhanewicz J, Vigneron DB, Nelson SJ. Three-dimensional magnetic resonance spectroscopic imaging of brain and prostate cancer. Neoplasia. 2000;2(1–2):166–189. doi: 10.1038/sj.neo.7900081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakian KL, Sircar K, Hricak H, et al. Correlation of proton MR spectroscopic imaging with gleason score based on step-section pathologic analysis after radical prostatectomy. Radiology. 2005;234(3):804–814. doi: 10.1148/radiol.2343040363. [DOI] [PubMed] [Google Scholar]
- 8.Kobus T, Hambrock T, Hulsbergen-van de Kaa CA, et al. In vivo assessment of prostate cancer aggressiveness using magnetic resonance spectroscopic imaging at 3 T with an endorectal coil. Eur Urol. 2011;60(5):1074–1080. doi: 10.1016/j.eururo.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Zakian KL, Hricak H, Ishill N, et al. An exploratory study of endorectal magnetic resonance imaging and spectroscopy of the prostate as preoperative predictive biomarkers of biochemical relapse after radical prostatectomy. The Journal of Urology. 2010;184(6):2320–2327. doi: 10.1016/j.juro.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph T, McKenna DA, Westphalen AC, et al. Pretreatment endorectal magnetic resonance imaging and magnetic resonance spectroscopic imaging features of prostate cancer as predictors of response to external beam radiotherapy. Int J Radiat Oncol Biol Phys. 2009;73(3):665–671. doi: 10.1016/j.ijrobp.2008.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pucar D, Koutcher JA, Shah A, et al. Preliminary assessment of magnetic resonance spectroscopic imaging in predicting treatment outcome in patients with prostate cancer at high risk for relapse. Clin Prostate Cancer. 2004;3(3):174–181. doi: 10.3816/cgc.2004.n.028. [DOI] [PubMed] [Google Scholar]
- 12.Bottomley P. Selective volume method for performing localized NMR spectroscopy. 4,480,228. USA patent. 1984
- 13.Schricker AA, Pauly JM, Kurhanewicz J, Swanson MG, Vigneron DB. Dualband spectral-spatial RF pulses for prostate MR spectroscopic imaging. Magn Reson Med. 2001;46(6):1079–1087. doi: 10.1002/mrm.1302. [DOI] [PubMed] [Google Scholar]
- 14.Mullerad M, Hricak H, Wang L, et al. Prostate cancer: detection of extracapsular extension by genitourinary and general body radiologists at MR imaging. Radiology. 2004;232(1):140–146. doi: 10.1148/radiol.2321031254. [DOI] [PubMed] [Google Scholar]
- 15.Fuchsjager MH, Shukla-Dave A, Hricak H, et al. Magnetic resonance imaging in the prediction of biochemical recurrence of prostate cancer after radical prostatectomy. BJU Int. 2009;104(3):315–320. doi: 10.1111/j.1464-410X.2009.08406.x. [DOI] [PubMed] [Google Scholar]
- 16.Shukla-Dave A, Hricak H, Moskowitz C, et al. Detection of prostate cancer with MR spectroscopic imaging: an expanded paradigm incorporating polyamines. Radiology. 2007;245(2):499–506. doi: 10.1148/radiol.2452062201. [DOI] [PubMed] [Google Scholar]
- 17.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vargas HA, Akin O, Shukla-Dave A, et al. Performance Characteristics of MR Imaging in the Evaluation of Clinically Low-Risk Prostate Cancer: A Prospective Study. Radiology. 2012 doi: 10.1148/radiol.12120041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohler JL, Kantoff PW, Armstrong AJ, et al. Prostate Cancer, version 2. 2014. Journal of the National Comprehensive Cancer Network : JNCCN. 2014;12(5):686–718. doi: 10.6004/jnccn.2014.0072. [DOI] [PubMed] [Google Scholar]
- 20.Thormer G, Otto J, Horn LC, et al. Non-invasive estimation of prostate cancer aggressiveness using diffusion-weighted MRI and 3D proton MR spectroscopy at 3.0 T. Acta Radiol. 2014 doi: 10.1177/0284185113520311. [DOI] [PubMed] [Google Scholar]
- 21.Giskeodegard GF, Bertilsson H, Selnaes KM, et al. Spermine and citrate as metabolic biomarkers for assessing prostate cancer aggressiveness. PLoS One. 2013;8(4):e62375. doi: 10.1371/journal.pone.0062375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson MG, Vigneron DB, Tabatabai ZL, et al. Proton HR-MAS spectroscopy and quantitative pathologic analysis of MRI/3D-MRSI-targeted postsurgical prostate tissues. Magn Reson Med. 2003;50(5):944–954. doi: 10.1002/mrm.10614. [DOI] [PubMed] [Google Scholar]
- 23.Bill-Axelson A, Holmberg L, Filen F, et al. Radical prostatectomy versus watchful waiting in localized prostate cancer: the Scandinavian prostate cancer group-4 randomized trial. J Natl Cancer Inst. 2008;100(16):1144–1154. doi: 10.1093/jnci/djn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha EK, Eastham JA. Chemotherapy and novel therapeutics before radical prostatectomy for high-risk clinically localized prostate cancer. Urol Oncol. 2015 doi: 10.1016/j.urolonc.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Park JJ, Kim CK, Park SY, et al. Prostate cancer: role of pretreatment multiparametric 3-T MRI in predicting biochemical recurrence after radical prostatectomy. AJR Am J Roentgenol. 2014;202(5):W459–465. doi: 10.2214/AJR.13.11381. [DOI] [PubMed] [Google Scholar]
- 26.Kobus T, Vos PC, Hambrock T, et al. Prostate cancer aggressiveness: in vivo assessment of MR spectroscopy and diffusion-weighted imaging at 3 T. Radiology. 2012;265(2):457–467. doi: 10.1148/radiol.12111744. [DOI] [PubMed] [Google Scholar]
- 27.Donati OF, Afaq A, Vargas HA, et al. Prostate MRI: evaluating tumor volume and apparent diffusion coefficient as surrogate biomarkers for predicting tumor Gleason score. Clin Cancer Res. 2014;20(14):3705–3711. doi: 10.1158/1078-0432.CCR-14-0044. [DOI] [PubMed] [Google Scholar]
- 28.Jung SI, Donati OF, Vargas HA, et al. Transition zone prostate cancer: incremental value of diffusion-weighted endorectal MR imaging in tumor detection and assessment of aggressiveness. Radiology. 2013;269(2):493–503. doi: 10.1148/radiology.13130029. [DOI] [PubMed] [Google Scholar]
- 29.Tamada T, Kanomata N, Sone T, et al. High b value (2,000 s/mm2) diffusion-weighted magnetic resonance imaging in prostate cancer at 3 Tesla: comparison with 1,000 s/mm2 for tumor conspicuity and discrimination of aggressiveness. PLoS One. 2014;9(5):e96619. doi: 10.1371/journal.pone.0096619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, Jiang Y, Antic T, et al. Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology. 2014;271(2):461–471. doi: 10.1148/radiol.14131320. [DOI] [PubMed] [Google Scholar]
- 31.Razzak M. Prostate cancer: aggressiveness--MRI can tell. Nature reviews Urology. 2013;10(8):433. doi: 10.1038/nrurol.2013.140. [DOI] [PubMed] [Google Scholar]
- 32.Scheenen TWJ, Heijmink SWTPJ, Roell SA, et al. Three-dimensional Proton MR Spectroscopy of Human Prostate at 3 T without Endorectal Coil: Feasibility1. Radiology. 2007;245(2):507–516. doi: 10.1148/radiol.2451061444. [DOI] [PubMed] [Google Scholar]
- 33.Geethanath S, Baek HM, Ganji SK, et al. Compressive sensing could accelerate 1H MR metabolic imaging in the clinic. Radiology. 2012;262(3):985–994. doi: 10.1148/radiol.11111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tayari N, Steinseifer IK, Fu CX, et al. Robust 3D 1H MRSI of the Prostate without Endorectal Coil at 3T. Toronto: 2015. p. 1051. [Google Scholar]
- 35.Matulewicz L, Jansen JF, Bokacheva L, et al. Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of (1) H magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2014;40(6):1414–1421. doi: 10.1002/jmri.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zechmann CM, Menze BH, Kelm BM, et al. Automated vs manual pattern recognition of 3D (1)H MRSI data of patients with prostate cancer. Acad Radiol. 2012;19(6):675–684. doi: 10.1016/j.acra.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Science translational medicine. 2013;5(198):198ra108. doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]