Abstract
A major virulence attribute of Candida albicans is its ability to form biofilms, densely packed communities of cells adhered to a surface. These biofilms are intrinsically resistant to conventional antifungal therapeutics, the host immune system, and other environmental factors, making biofilm-associated infections a significant clinical challenge. Here, we review current knowledge on the development, regulation, and molecular mechanisms of C. albicans biofilms.
Keywords: Candida albicans, biofilm, fungi, pathogen, microbiota, microbial community, infection, transcriptional regulation
1. Introduction
Over the last two decades, research in the field of microbial biofilms has gained increasing momentum and emerging scientific evidence has initiated a change in the way we think about microbial life [1,2]. Although microorganisms have traditionally been studied in free-floating (planktonic) cultures or as colonies grown on the surfaces of nutrient agar plates, it is now accepted that biofilms are the preferred and probably the “natural” state of growth for most microorganisms [1–3]. A biofilm is a community of microbial cells that are adhered to a surface (or found at air-liquid interface), are surrounded by an extracellular matrix, and have properties that are distinct from their free-floating counterparts [1,3]. The first documented scientific report about a biofilm was in 1683 by Antonie van Leeuwenhoek in an article written for the Royal Society of London, where he made the following microscopic observation about dental plaque: “The number of these animalcules in the scurf of a man’s teeth are so many that I believe they exceed the number of men in a kingdom” [4]. Today, microbial biofilms have been observed in a diverse set of environments that include both biotic surroundings (e.g. aquatic environments, plant tissues and mammalian tissues), as well as abiotic surroundings (e.g. catheters, prosthetic devices and biomaterials). Biofilm structure, development, and unique properties are characteristic of the microbial species that forms (or are members of) the biofilm and while most species, including Candida sp., Staphylococcus sp., Streptococcus sp., and Escherichia coli, form biofilms on solid surfaces, some species, for example Bacillus sp. and Mycobacterium sp., form biofilms at air-liquid interfaces [5]. However, a near-universal feature of biofilms is their increased resistance to chemical and physical injury, making them very difficult to combat in clinical settings and a burden to overcome from the standpoint of human health [5,6].
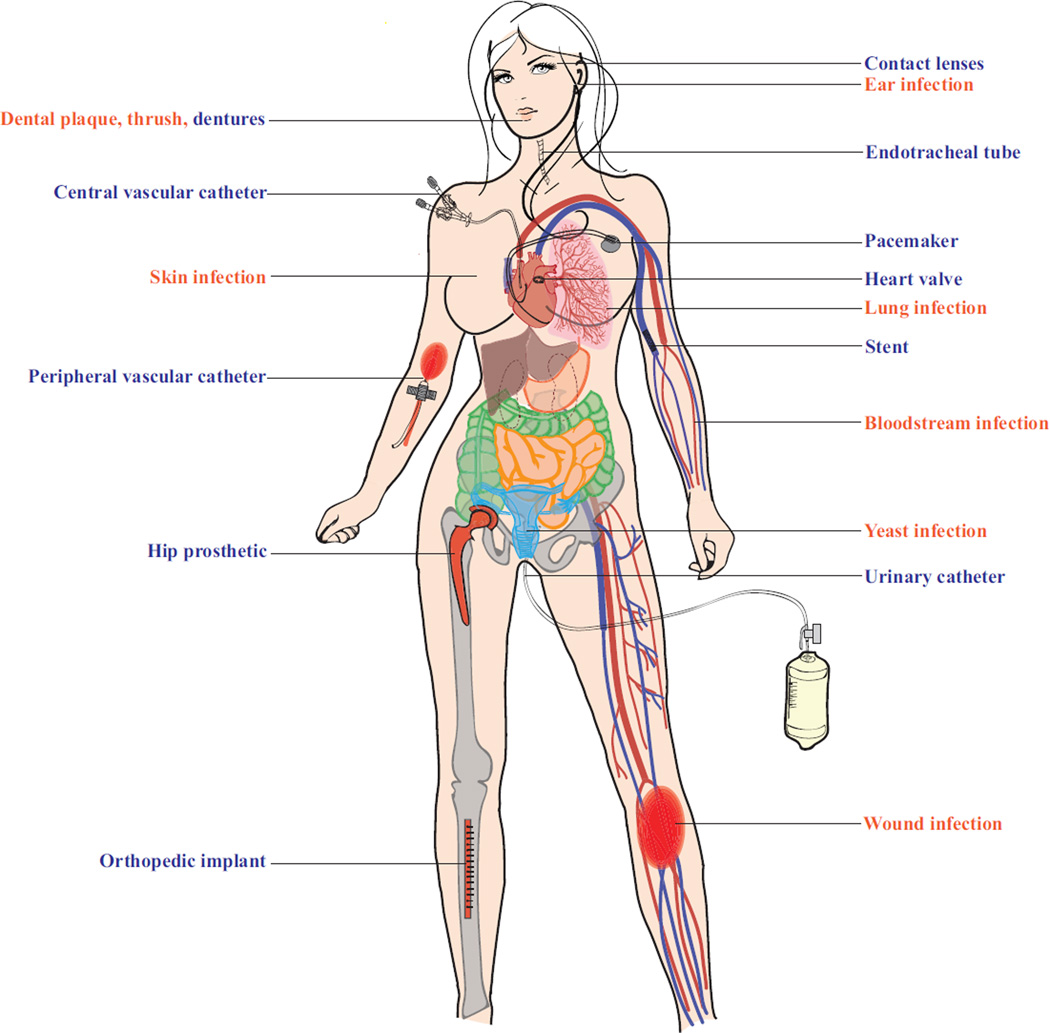
Recent estimates by the National Institutes of Health indicate that pathogenic biofilms are responsible, directly or indirectly, for over 80% of all microbial infections [1,7]. Further, the recalcitrant nature of microorganisms within biofilms adds to the trouble of eradicating these infections. In this review, we focus on Candida albicans biofilms, which can colonize mucosal surfaces, such as those coating the oral and vaginal epithelia, and implanted medical devices, such as prosthetics, heart valves, and catheters, and can seed systemic infections in humans (see Fig 1 for common C. albicans biofilm-associated infections occurring through the use of a medical device directly colonized by a biofilm or from a localized or disseminated infection originating from a biofilm).
Figure 1.
Common C. albicans biofilm-associated infections. Schematic highlighting areas of the female body susceptible to C. albicans infection occurring through the use of a medical device directly colonized by a biofilm (labeled in blue) or from a localized or disseminated infection originating from a biofilm (labeled in orange).
C. albicans is a member of the healthy human microbiota, asymptomatically colonizing several niches in the body, including but not limited to, the gastrointestinal (GI) tract, female reproductive tract, oral cavity, and skin [6,8]. In most individuals with a healthy immune system, C. albicans is a harmless commensal that exists in harmony with other members of the microbiota. However, disturbances to this delicate balance, resulting, for example, from variations in the local environment (pH shifts or nutritional changes), use of antibiotics, or alterations in the immune system (caused by an infection or immunosuppressant therapy), can enable C. albicans to rapidly proliferate and cause infection [6,9]. These infections range from superficial mucosal and dermal infections, such as thrush, diaper rash, and vaginal yeast infections (75% of women will have a yeast infection at least once in their lifetime), to more serious hematogenously disseminated infections with sizable mortality rates (approaching 47% in some cases) [6]. C. albicans is a leading cause of hospital-acquired infections, it accounts for 15% of all sepsis cases, and is the cause of 40% of bloodstream infections in clinical settings [1]. While C. albicans can infect both immunocompetent and immunocompromised individuals, these infections are especially serious in the latter group, which includes AIDS patients, patients undergoing cancer chemotherapy treatments or immunosuppression therapies, and individuals with implanted medical devices [6,10].
C. albicans forms highly structured biofilms composed of multiple cell types (round budding yeast-form cells, oval pseudohyphal cells, and elongated hyphal cells) encased in an extracellular matrix [11]. C. albicans is the predominant fungal species isolated from medical device infections, including urinary and central venous catheters, pacemakers, mechanical heart valves, joint prostheses, contact lenses, and dentures [10] (Fig 1). Once a C. albicans biofilm is formed on an implanted medical device, it acts as a reservoir for pathogenic cells, is highly resistant to drugs and the host immune system, and has the potential to seed disseminated bloodstream infections (candidemia) that can lead to invasive systemic infections of tissues and organs. Each year in the United states, over five million central venous catheters are placed and currently – even with recent improved clinical approaches – biofilm infection occurs in over 50% of these catheters [6]. With an estimated 100,000 deaths and $6.5 billion in excess expenditure annually in the United States alone, these infections have serious health and economic consequences [1]. Additionally, as these fungal biofilms are largely resistant to known antifungal drugs, the current standard of care to treat these infections involves the removal of the colonized medical device, oftentimes through surgery, combined with administration of high doses of antifungal agents [12,13]. Removal of some of these devices (e.g. artificial heart valves and joints) can be costly and, in some cases, dangerous to the patient, and the administration of high doses of antifungal agents (typically given intravascularly), can result in further complications, including kidney and liver damage [10,13]. Oftentimes, these treatments are not even possible, as many critically-ill patients are unable to tolerate them, leaving these patients with few available options and underscoring the need to find better therapeutic and diagnostic therapies to combat these biofilms.
2. C. albicans biofilm development
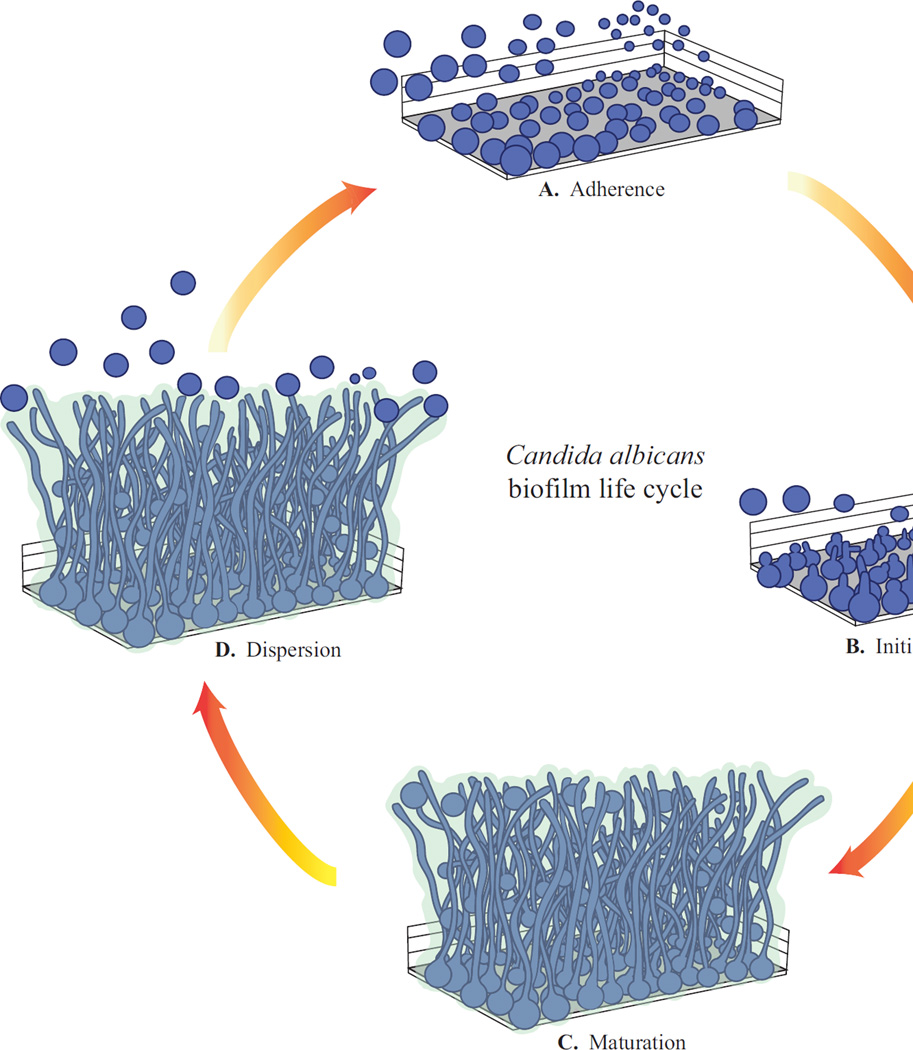
Most of our knowledge of C. albicans biofilm formation originates from the study of monospecies biofilms, which have been characterized in both in vitro and in vivo systems and consist of four distinct phases of development [9,11] (Fig 2). C. albicans biofilm formation begins with the adherence of round yeast cells to a solid surface (in the laboratory, a small silicone disc, the material of common intravascular catheters, or a polystyrene microtiter plate, are often used). Typically, a culture of C. albicans is added to the solid surface to initiate the adherence phase (60–90 minutes) and non-adhered or loosely adhered cells are then washed away, resulting in the formation of a basal layer of anchoring yeast cells (Fig 2A). This stage is often referred to as the “seeding” step and is essential for normal biofilm development. The next stage in biofilm development consists of cell proliferation and early-stage filamentation of the adhered cells (Fig 2B). This is followed by biofilm maturation, resulting in a complex network of several layers of polymorphic cells, including hyphal cells (chains of cylindrical cells), pseudohyphal cells (ellipsoidal cells joined end to end), and round yeast cells, encased in an extracellular matrix, giving the biofilm a thick and structured appearance as well as providing protection from chemical and physical injury (Fig 2C). A mature biofilm typically forms by 24 hours, and can be visualized by eye as a cloudy surface structure on top of the solid surface, and under a microscope, as an organized collection of different cell types. Throughout these stages of biofilm development, the growth media is kept constantly shaking, to prevent free-floating cells from settling on the surface, or is continuously flowing over the biofilm, to mimic flow conditions commonly present in catheters. The final step of biofilm development is termed the dispersal stage, where some round yeast cells disperse from the biofilm to seed new sites (Fig 2D); this is the least studied phase of C. albicans biofilm development. Several models of in vitro C. albicans biofilm formation have been reported, and studies have focused on analyzing the impact of different types of substrates, nutritional media, and the presence of flow or static conditions, on biofilm development [14]. In the laboratory, C. albicans biofilms can develop on several different substrates and in many different types of media, indicating an inherent robustness of biofilm development to a wide range of environmental conditions.
Figure 2.
C. albicans biofilm life cycle. A. Adherence of round yeast cells to a surface. B. Initiation of biofilm formation, where cells proliferate to form a basal layer of adhered cells. C. Maturation of the biofilm, where complex layers of polymorphic cells develop and become encased in an extracellular matrix. D. Dispersion, where round yeast cells leave the mature biofilm to seed new sites.
In general, C. albicans in vitro biofilm formation has correlated well with in vivo and ex vivo biofilm models; they follow a similar time course in phases of development and also appear architecturally similar to biofilms retrieved from patients with infections. For example, Candida biofilms obtained from denture stomatitis patients and from patients with infected intravascular catheters confirm the presence of yeast, hyphae and extracellular matrix [1]. One of the advantages of in vivo models is the opportunity to study C. albicans biofilm formation in the presence of the host immune system, which can provide additional mechanistic insights into host-pathogen interactions. Biofilm architecture in rat and rabbit central venous catheter models, indwelling urinary catheter models, and rat denture stomatitis models are also similar to the in vitro biofilm structure, including numerous yeast cells in the basal region, and hyphae and extracellular matrix extending throughout the biofilm [15–17]. Vaginal mucosal in vivo mouse models (vaginal mucosa inoculated with C. albicans in live mice) and ex vivo models (vaginas excised from euthanized mice that are inoculated with C. albicans in tissue culture plates) also show similar biofilm architectures with yeast cells, hyphae, and extracellular matrix evident throughout the biofilms, formed on top of mucosal layers [18]. Other animal models for monitoring biofilm formation include rodent oral mucosal, oropharyngeal, subcutaneous, and burn wound models [19,20]. Development of newer model systems is underway, and will aid us in visualizing the temporal and spatial progression of biofilm infections in live animals using bioluminescence imaging. For example, recently, a codon optimized C. albicans luciferase bioreporter was used in a vulvovaginal candidiasis model to observe biofilm formation in real-time in the vaginal lumen [21]. Other C. albicans bioluminescent biofilm models include oropharyngeal, cutaneous, subcutaneous, and implanted catheter models [22,23].
3. Genetic regulation of C. albicans biofilm formation
Saccharomyces cerevisiae diverged from C. albicans over 200 million years ago, and while the ease of genetic manipulation of S. cerevisiae is well established, C. albicans is not genetically tractable in the conventional sense (for example, its parasexual cycle is cumbersome to use in the lab). However, some tools have been developed to genetically manipulate C. albicans using recombinant DNA technologies and, to date, nearly 1,000 gene knockout mutants of this organism have been constructed (out of a total gene count of ~6,000), and many of these deletion mutants have also been screened for biofilm formation [1,7]. Other approaches to identify biofilm-specific genes and proteins in C. albicans include genome-wide transcriptional profiling and proteomics techniques, respectively [24,25]. These studies have revealed that many hundreds of mRNAs and proteins are differentially expressed between biofilms and planktonic cells. In this section we review these genome-wide studies with particular emphasis on the “master” regulators that orchestrate biofilm formation, as well as some of the key non-regulatory genes that have been genetically validated to play important roles in biofilm formation. Genes whose deletions cause broad phenotypes (such as slow growth), have been excluded from this discussion, as we hypothesize that their effects on biofilm formation are likely to be indirect. As the available deletion libraries are enriched for knockouts of transcriptional regulators, it follows that most of our current knowledge is on the transcriptional control of biofilm formation, and we do not know as much about the process itself. Based on the literature to date, we can identify 50 transcriptional regulators and 101 non-regulatory genes that have functionally validated roles in biofilm formation (See [1] for these gene lists).
3.1 “Master” regulators of C. albicans biofilm development
In 2012, a comprehensive study of the large and complex transcriptional network controlling the development of C. albicans biofilms was described [26]. This network is comprised of six “master” transcriptional regulators (Efg1, Tec1, Bcr1, Ndt80, Brg1, and Rob1), each of which is required for normal biofilm development, both in vitro, under standard laboratory conditions, and in vivo, in rat catheter and rat denture models [26]. Further analysis of these master regulators was carried out by genome-wide chromatin immunoprecipitation and gene expression profiling experiments, revealing a complex transcriptional circuit where these regulators control each other as well as a large number of direct and indirect downstream target genes. Taken together, these six master regulators directly bind to the promoters of, and likely regulate the expression of, approximately 1,000 target genes, some of which are additional transcriptional regulators, providing a framework of the vast, complex and intertwined network of genes involved in biofilm formation. Work carried out in both planktonic and biofilm conditions have indicated that various target genes play distinct roles in different processes of biofilm development, such as adhesion, hyphal formation, drug resistance, and the production of extracellular matrix (see [1,7] for a summary), all of which are important characteristics of biofilms. However, the majority of newly identified target genes in the biofilm network have not yet been studied; many have no overt sequence similarity to any previously characterized genes from any organism, and may be genes unique to C. albicans or the Candida/CTG clade. Furthermore, based on orthology mapping, the entire set of target genes is significantly enriched for “young” genes, suggesting that the ability of C. albicans to form biofilms evolved relatively recently with respect to evolutionary timescales. This inference provides an explanation as to why C. albicans and its closely related species are only a few of the many fungal species that have the ability to form biofilms within a mammalian host. With this, undoubtedly incomplete outline of the biofilm network, it is now possible to mechanistically dissect how biofilm formation is orchestrated and to systematically study the roles of the non-regulatory target genes in biofilm development.
In addition to the six master transcriptional regulators discussed above, 44 additional transcriptional regulators have been identified, whose deletion has been shown to affect at least some aspect of C. albicans biofilm formation [1,7,27]. Interestingly, the majority of these regulators are directly bound by at least one of the six master biofilm network regulators, suggesting that they may be directly regulated by the core biofilm circuit.
Although the complexity of the transcriptional network controlling biofilm development in C. albicans may seem inordinately complex, it is quite typical of several other eukaryotic transcriptional networks. To name a few, mammalian stem cell maintenance, Drosophila eye development, and Arabidopsis circadian clock rhythms are all controlled by multiple transcriptional regulators that regulate each other and several additional target genes [1]. Complex transcriptional circuits are also known to govern pseudohyphal growth and the response to osmotic stress in baker’s yeast [28] as well as the white-opaque cell-type switch in C. albicans [29]. While the significance of the complexity and interwoven nature of these transcriptional circuits is not fully understood, this level of complexity seems to be a common feature of most networks that coordinate morphological changes. In the sections below, we briefly review the known proteins playing roles in different stages of biofilm formation in C. albicans.
3.2 Adherence of C. albicans cells to different surfaces
The ability of C. albicans to form biofilms on both biotic and abiotic surfaces is a significant contributing factor to the robustness of C. albicans biofilms. The initial process of adherence begins when cells attach to each other and to surfaces (Fig 2A), which can be hard (such as the biomaterials that are part of a prosthetic device or denture surface), or soft (such as a mucosal epithelial layer in the oral cavity or vagina). This step is the first phase in the formation of a C. albicans biofilm and is crucial for all later stages of biofilm development. The master regulator Bcr1, as well as some of its downstream targets, including the cell wall proteins Als1, Als3, and Hwp1, are all required for adherence during biofilm formation [30–33]. Several additional transcriptional regulators have also been recently implicated in adherence [34]. The majority of these adherence regulators were identified from a recent study that screened a library of transcription regulator deletion mutants using an in vitro quantitative flow cell assay [34]. In this study, 30 transcriptional regulators were identified to be important for adherence to a silicone substrate under these flow conditions. Of these 30, four (Bcr1, Ace2, Snf5, and Arg81) were also required for biofilm formation under commonly used conditions for in vitro biofilm formation (on polystyrene microtiter plates with shaking). These and other studies clearly indicate that biofilm formation by C. albicans occurs over a broad range of conditions and that the genetic requirements may vary from one condition to the next.
3.3 Hyphal cells of C. albicans and their roles in biofilm development
After the initial adherence of the round yeast cells to a surface to form a basal layer, the next phase of biofilm development is the growth and proliferation of hyphal cells (Fig 2B). C. albicans is a polymorphic fungus and is distinguished from many other fungal species by its ability to form both yeast and hyphal cells under many different environmental conditions. (This was the basis of the early classification of C. albicans as dimorphic.) Although readily formed in planktonic cultures, in the presence of specific nutritional and/or environmental cues, hyphae are a characteristic feature and an important structural component of C. albicans biofilms. Thus, it is not surprising that proteins involved in hyphal growth in suspension cultures are also required for proper biofilm formation. These include the transcriptional regulators Efg1, Tec1, Ndt80, and Rob1 [26,35]. The hyphae in biofilms contribute to the overall architectural stability of the biofilm, and act as a support scaffold for yeast cells, pseudohyphae, other hyphae, and other microbial cells in the context of polymicrobial biofilms (see below) [11,36]. Thus, the ability to form hyphae as well as the ability of these hyphae to adhere to one another and other cell morphologies is critical for normal biofilm development and maintenance [1]. Indeed, the master regulator Bcr1 is not required for hyphal formation per se, but it is needed for the hyphae to adhere to one another in the context of a biofilm [30].
3.4 Extracellular matrix of C. albicans biofilms
An important feature of C. albicans biofilms is the presence of an extracellular matrix that is formed during the maturation phase of biofilm development and encompasses the complex network of yeast, pseudohyphal and hyphal cells, providing protection from host immune defenses and antifungal drugs, and contributing to the three dimensional architectural stability of the biofilm (Fig 2C). Although the extracellular matrix is partly self-produced and secreted by C. albicans cells within the biofilm, it may also contain environmental aggregates, such as structural components from lysed C. albicans and host cells, specific host cells that are associated with or recruited to the area and incorporated into the biofilm, such as erythrocytes, epithelial cells, urothelial cells, and neutrophils [37], and thus can vary widely depending on the location of the biofilm within the host [38]. One recent study identified biofilm-matrix-associated proteins by liquid-chromatography mass spectrometry analysis of the proteins collected from the matrix of C. albicans biofilms grown in three different in vivo models (rat venous catheter, denture, and urinary catheter) [37]. Fourteen host proteins were found to be abundant in all three in vivo settings, including heme-related proteins and inflammatory and leukocyte-associated proteins, such as hemoglobin, myeloperoxidase, C-reactive protein, and alarmin S100-A9. A few studies have focused on the composition of the C. albicans biofilm matrix under in vitro settings, where it was found that the matrix is largely comprised of glycoproteins (55%), carbohydrates (25%), lipids (15%), and nucleic acids (5%) [1,25]. Polysaccharides make up the second major fraction of the matrix, which include glucose, mannose, rhamnose and N-acetylglucosamine, however the largest fraction consists mainly of mannan-glucan complexes made predominantly of α-1,6-linked mannan and α-1,2-linked side chains complexed to β-1,6-glucan [25]. While studies have historically focused on the chemical breakdown of the matrix, several recent studies have investigated the genetic regulation of matrix production in C. albicans biofilms. There are currently two known regulators of biofilm matrix production in C. albicans: Rlm1 and Zap1. Deletion of RLM1 causes a reduction in matrix levels [39], while deletion of ZAP1, leads to an increase in the accumulation of extracellular matrix material, probably by upregulating two glucoamylase enzymes, Gca1 and Gca2 [40]. Over 500 proteins have been identified in the matrix [25], most of which are predicted to be enzymes, including hydrolyzing enzymes, suggesting that the matrix may play an active role in breaking down biopolymers. It is certainly intriguing to consider the biofilm matrix as an extracellular, enzymatically-active, element of a C. albicans biofilm – one that can breakdown molecules as both a protective response and a nutrient source [1].
3.5 Dispersal of cells from a C. albicans biofilm
The ability of certain cells within a C. albicans biofilm to disperse into the environment is perhaps the least understood phase in the biofilm life cycle (Fig 2D). Observations of in vitro biofilm development indicate that cells are probably dispersed continuously throughout biofilm formation, and they are thought to be primarily (if not exclusively) in the round yeast-form [41]. Although these dispersed cells morphologically resemble the round yeast cells seen in the planktonic mode of growth, they have distinct characteristics. For example, the dispersed cells have increased adherence properties, and have a higher capacity to form biofilms, relative to planktonic cells. Further, these cells also display elevated virulence levels in mouse models of infection [41]. Three transcriptional regulators of C. albicans biofilm dispersal have been identified, Nrg1, Pes1 and Ume6; transcriptional overexpression of UME6 reduced the number of dispersed cells, whereas overexpression of PES1 and NRG1 increased the number of dispersed cells actively released from the biofilm [41,42]. Nrg1 likely acts through the conserved Set3 chromatin-modifying complex [42]. The mutant strains of individual Set3 complex members formed extra-strong, “rubbery” biofilms that were unable to undergo normal biofilm dispersal and were especially recalcitrant to mechanical perturbation [43]. Additionally, Nrg1 and Set3 complex deletion mutants are hyperfilamentous, consistent with their inability to generate yeast-form cells [42–44].
The molecular chaperone Hsp90 has also been implicated in C. albicans biofilm dispersal, as depletion of Hsp90 markedly reduces the number of dispersed cells from a biofilm [45]. Depletion of Hsp90 also induces filamentation by relieving Hsp90-mediated repression of the cAMP-PKA signaling pathway [46]. Another protein identified as playing a role in biofilm dispersion is the cell wall protein Ywp1, where deletion of YWP1 leads to decreased biofilm dispersal and increased biofilm adhesiveness [47]. Based on the evidence in the literature to date, one may predict that a mutation that favors filamentous cells over yeast-form cells may reduce biofilm dispersal. Thus, both the yeast and hyphal cell types and the ability to transition between them are critical, not only for biofilm formation and maintenance, but also for biofilm dispersal and spread.
4. Multispecies biofilms formed between C. albicans and other species
Microbial infections are often thought of, and treated, as if a single microbial species were acting alone, yet most infections occur in the presence of the members of the human microbiota – the collection of microorganisms inhabiting the body. Humans are host to a large and diverse population of microbial species; in fact, microbial cells are estimated to outnumber human cells by at least a factor of ten in an average human body [48]. The human microbiota includes members from all three kingdoms of life: bacteria, archaea, and fungi, and in healthy individuals these microbes exist in a delicate balance, forming a complex ecosystem. Perturbations to this balance, such as those caused by changes in diet, genetic background of the host, alterations in host immunity, and transient environmental perturbations, such as changes in pH, viscosity of mucosal layers, and the use of broad-spectrum antimicrobial agents, can alter this balance and result in infection due to the overgrowth of certain microorganisms over others. The vast diversity of microorganisms occupying specialized niches in the human body provides numerous opportunities for physical and chemical interactions to occur between microbial species in these polymicrobial environments that have developed over the course of millions of years of coevolution with humans. Many infections, including those of the oral cavity (e.g. periodontitis) and ear (e.g. otitis media), wound infections, chronic infections in the lung (e.g. cystic fibrosis), urinary tract infections, and catheter infections, are all polymicrobial in nature [49], typically resulting (or originating) from polymicrobial biofilms in which multiple species of microbes form a complex and recalcitrant community of interacting cells.
Although, C. albicans is the most frequently isolated fungal pathogen from mixed bacterial-fungal infections, there has been only incremental progress in understanding dual-species biofilms formed between C. albicans and common bacterial species that are likely to interact with C. albicans in humans and cause infection. Most of the analyses of such polymicrobial infections are limited to observational studies, occurring largely in immunocompromised patients. Given that immunocompromised individuals are highly susceptible to infections and are faced with many additional medical risk factors, it is difficult to assess the extent of these polymicrobial interactions on the clinical prognosis of patients [49], and even more difficult to deduce molecular mechanisms from these types of studies. Other studies have looked at the in vitro interactions of C. albicans with bacterial species that have been isolated from the oral cavity, abdomen, vagina, and skin. For example, several studies have investigated dual-species biofilms formed between C. albicans and bacteria commonly isolated from denture stomatitis, peritonitis, periodontitis, and dental caries, such as Streptococcus mutans, Streptococcus gordonii, Staphylococcus aureus, Actinomyces viscosus, and Fusobacterium species [50–53]. C. albicans and P. aeruginosa are both commonly isolated together in infections arising from catheters, chronic lung infections (e.g. cystic fibrosis), and skin infections (e.g. burn wounds) [54]. Studies using animal models indicate that co-infection with C. albicans and other bacterial species may increase virulence. For example, one study found that mice infected with a sub-lethal inoculum of P. aeruginosa and C. albicans together had a higher mortality rate compared with mice that were infected with only one of either species alone [55]. Another study using an in vitro mucosal model recently demonstrated that the synergistic interactions between C. albicans and one of the oral bacteria Streptococcus oralis, Streptococcus sanguinis, or Streptococcus gordonii grown in dual-species biofilms, mutually benefitted both species in their abilities to invade and form pathogenic biofilms on mucosal surfaces [36]. Additionally, C. albicans has also been found to interact with several bacterial species found largely in the gut, such as Enterococcus and Escherichia species, and bacterial species prevalent in the vagina, such as Lactobacillus species [56–58]. It is evident that C. albicans and bacteria can interact with each other in several ways that impact their survival and virulence. Common modes of interactions include, for example, the secretion of signaling molecules that influence the behavior of one species towards the other, direct physical contact between microbial cells (e.g. hyphae provide a site of attachment for bacterial cells within polymicrobial biofilms), and chemical alterations of the local environment that influence the other species (e.g. alterations in pH and oxygen content) [36,49,59,60]. Examples of these types have been observed in studies of the vaginal microbiota, especially in the interactions between C. albicans and Lactobacillus species, as growth of the bacteria produces lactic acid that lowers the local pH, inhibiting the growth of C. albicans on the vaginal mucosal surface [61]. In another example, P. aeruginosa is known to secrete a 12-carbon acyl homoserine lactone that modulates hyphal growth of C. albicans [54]. Evidence of these interactions, taken in context with human health, challenges us to reexamine our thinking and approaches towards microbial infection. This is more evident in light of a recent study that examined dual-species biofilms formed between C. albicans and each of five prevalent human gut microbiota members: Escherichia coli, Klebsiella pneumonia, Enterococcus faecalis, Bacteroides fragilis, and Clostridium perfringens [62]. This study showed that biofilms formed by C. albicans provided a protective hypoxic microenvironment that supported the growth of the two strictly anaerobic bacteria, B. fragilis and C. perfringens – even though the biofilms were grown under oxygen-rich conditions and the oxygen-rich conditions do not support the survival of either bacterial species grown on its own. Thus, the presence of a C. albicans biofilm can act as a sanctuary for anaerobic pathogens in oxygen-rich environments. Moreover, even when cultured together in suspension cultures, and under ambient oxygen conditions, these anaerobic bacteria could induce C. albicans to form “mini-biofilms” structures which, in turn, could protect the bacteria inside the mini-biofilms, allowing them to proliferate under otherwise toxic conditions. Investigating interactions occurring between different species in polymicrobial biofilms and their relevance to human health is clearly a research area of great interest and further studies are much needed to develop new strategies to target these complex infections.
5. C. albicans interactions with the host immune system and host environment
As an opportunistic pathogen, C. albicans can cause a wide range of infections, from superficial mucosal and skin infections, to hematogenously disseminated candidiasis [1]. However, C. albicans is typically a commensal microorganism, existing as a normal member of the human microbiota, and usually causing no infection in people with healthy immune systems and balanced microbiota. The extent to which C. albicans is able to cause infections in humans is likely closely linked with the ability of the immune system to discriminate between the commensal versus pathogenic forms of C. albicans. Although it is not yet clear exactly how this discrimination occurs, there are multiple immune mechanisms that are known to function to prevent the overgrowth and invasion of C. albicans in host tissue, events which lead to infection. For the discussion to follow, we will refer to this overgrowing and invasive form of C. albicans as the “pathogenic” form.
5.1 Innate immune responses to C. albicans infection
Cells of the innate immune system are able to recognize pathogen- associated molecular patterns (PAMPs) from the pathogenic form of C. albicans, resulting in a cascade of host signaling events that ultimately leads to destruction and clearance of the C. albicans cells. The host cells involved in this process include epithelial cells, neutrophils, macrophages, and dendritic cells. Currently we know of ten surface receptors, including two Toll-like receptors (TLR2 and TLR4), six C-type lectin receptors (Dectin-1, Dectin-2, MR, DC-SIGN, Mincle, and MBL), and two internal receptors (TLR9 and NLRP3) that are implicated in the recognition of C. albicans [6]. These receptors typically bind to sugars found on the C. albicans cell surface, such as mannose derivatives and β-1,3-glucans. These binding events initiate several downstream signaling cascades that lead to increased cytokine and chemokine production, and ultimately, phagocytosis of the fungal cell. Internalization of C. albicans cells by neutrophils, macrophages, or dendritic cells, in turn, leads to the activation of internal receptors, resulting in TLR9 or NLRP3 inflammasome activation. While the innate immune response is a key player in combatting C. albicans infection, the adaptive immune response also plays an important role, where antibodies to extracellular proteins, such as mannan and C. albicans-specific proteins, are also critical in eliminating C. albicans and preventing its overgrowth [6].
5.2 C. albicans biofilms subvert the innate immune response
Although the molecular mechanisms of how the growth of C. albicans in the biofilm state affects interactions with the host immune system are currently unknown, there are clues that indicate that aspects of biofilm formation may provide protection against host defenses. For example, despite the fact that mature C. albicans biofilms are recognized and often surrounded by neutrophils in vivo, the surrounding neutrophils tend to be inactive and fail to trigger a reactive oxygen species response [63]. This resistance of C. albicans biofilms to neutrophil killing is attributed to the presence of glucans in the extracellular matrix inhibiting neutrophil activation. Hyphal cells are another important component of biofilms that contribute to subversion of the innate immune response. Not only can hyphae penetrate epithelial cell layers during invasive growth, they are also able to mediate escape of the C. albicans cell from within phagocytic cells by physically piercing the phagocytic cell [64]. The proteins Pra1, Gpd2, and members of the secreted aspartyl protease (Sap) family are highly expressed during biofilm formation, and are individually capable of blocking complement activation [65]. Msb2 is another protein highly expressed in biofilms, and is involved in binding and blocking host-secreted antimicrobial peptides [6]. It is also known that deletion of one of the major transcriptional regulators of biofilm development, Bcr1, causes C. albicans to be more susceptible to damage by leukocytes [66]. Although it is yet to be demonstrated at the molecular level that C. albicans biofilms interact differently with the immune system than planktonic cells, there is evidence to support the idea that biofilms are geared to be more resistant to the host immune system.
5.3 C. albicans adaptations to the host environment
As C. albicans shares its environment with other commensals that are part of the human microbiota, it has evolved nutrient acquisition mechanisms to sequester nutrients from neighboring microbes. One such mechanism involves the release of Saps by C. albicans that can cause injury to host tissues and, in turn, free nutrients, such as amino acids and carbon sources [67]. Unlike many other commensal microbes, which are much more specialized to grow in particular niches of the human body and require specific carbon sources (e.g. glucose) to proliferate, C. albicans is able to utilize several different carbon sources, and is thus able to proliferate in many distinct and dynamic environmental regions of the body. There is also evidence indicating that growth of C. albicans on physiologically relevant alternative carbon sources (e.g. lactic acid), can cause significant changes in the properties of the fungal cell wall, rendering it much more resistance to stresses like osmotic pressure, antifungal drug exposure, and other cell wall stresses, compared to cells grown on more traditional carbon sources (e.g. glucose) [68]. C. albicans can also readily adapt to varying pH conditions that it encounters in different body niches. For example, the cell wall proteins Phr1 and Phr2 are required for adaptation to changing pH levels in systemic blood stream and vaginal infections, respectively, providing an advantage to C. albicans over other commensals lacking such adaptations. In this section, we have provided a mere snapshot of the various mechanisms through which C. albicans can interact with the host immune system and the local host environment to promote its proliferation and cause infection. We have only just begun to explore how the commensal versus pathogenic forms of C. albicans interact with the host, and this is an area that will certainly expand in future studies.
5.4 Mucus and C. albicans biofilms
Epithelial tissues covered with mucus are the first contact for most microorganisms that enter mammalian hosts, and have consequently evolved multiple mechanisms to defend against invading microorganisms. By acting as a barrier to the deeper interiors of the host, mucosal epithelial tissues are critical for protecting the host against disseminated infection. Although the exact protective mechanisms of these host tissues specifically against penetration by C. albicans (as well as other microorganisms) are largely unknown, we do know that the viscoelastic properties of mucus, predominantly attributed to large, heavily O-glycosylated membrane-bound and secreted glycoproteins called mucins, play critical roles in providing this protection. There are five major secreted gel-forming mucins in the human body: MUC2, MUC5AC, MUC5B, MUC6, and MUC19 [69]. A recent report indicated that the mucins MUC2, MUC5AC, and MUC5B can suppress certain C. albicans virulence traits, such as hyphal growth and biofilm development [70]. These mucins were found to impede the development of biofilm formation in C. albicans in two major ways: by suppressing filamentation and by inhibiting adhesion of C. albicans cells to surfaces [70]. Further characterization of the mechanisms through which microbes interact with mucin is an exciting avenue for future studies.
6. C. albicans biofilm resistance to antifungal drugs
Unlike the diversity seen in antibiotic drugs that is the result of the numerous known antibiotic classes and several distinct modes of action against different bacterial targets, current antifungal drugs are extremely limited in spectrum. Only four major classes of antifungal drugs are used to treat the majority of fungal infections: azoles, polyenes, echinocandins, and nucleoside analogs [12]. Azoles (e.g. fluconazole), which are fungistatic, are the most commonly prescribed class of antifungals used to treat both systemic and topical infections. Azoles inhibit ergosterol biosynthesis by targeting the demethylase Erg11, leading to a buildup of toxic sterol pathway intermediates. Polyenes (e.g. amphotericin B), which are fungicidal, are the oldest class of antifungals used to treat severe infections. Polyenes work by intercalating into the ergosterol of the cell membrane, forming pores that abolish the proton gradient of the cell, resulting in ion leakage and destabilization of the cell membrane. Echinocandins (e.g. caspofungin), which are fungicidal against most Candida species, are the newest class of antifungals used to treat persistent infections through intravascular administration. Echinocandins work by targeting the synthesis of β-1,3-glucans, which are critical polysaccharide components of fungal cell walls. Nucleoside analogs (e.g. 5-flucytosine (5-FC)) are antimetabolites that mimic nucleosides during nucleic acid synthesis. 5-FC specifically acts as a pyrimidine analogue, disrupting fungal RNA, DNA, and protein synthesis, ultimately causing cell cycle arrest. 5-FC does not have antifungal activity itself, but becomes an active antifungal agent only when it is converted into 5-fluorouracil (5-FU); this conversion occurs in the presence of a cytosine deaminase, which is inside fungal (but not host) cells.
C. albicans biofilms are inherently resistant to the majority of known antifungal drugs, making these infections particularly difficult to combat. The azoles and the classic formulation of the polyenes, for example, are not effective against C. albicans biofilms, which further limits the drugs that can be used to treat these infections and highlights the need for the development of new antifungal therapies with efficacy against the biofilm mode of growth. Resistance of C. albicans biofilms to classic antifungal drugs is multifactorial and mechanistically complex, but is largely due to three major factors: the upregulation of efflux pumps, the presence of the extracellular matrix, and the existence of recalcitrant, metabolically inactive cells referred to as “persister” cells. Below, we briefly review each of these resistance factors in the context of C. albicans biofilms.
6.1 Efflux pumps
There are two major classes of efflux pumps that modulate drug exportation in C. albicans: the ATP binding cassette (ABC) transporter superfamily (including Cdr1 and Cdr2) and the major facilitator (MF) transporter superfamily (including Mdr1) [71–74]. In planktonic cells, these efflux pumps are typically upregulated in response to antifungal drugs; however, in biofilms, they are upregulated within the first few hours of adhesion and remain upregulated throughout biofilm development, even in the absence of an antifungal drug [1,26]. This immediate upregulation of efflux pumps that occurs in the early stages of biofilm development is a key contributor to the early recalcitrance of biofilms to antifungal agents. One possibility is that this rapid transcriptional response may have evolved in response to inhibitory molecules produced by other microbial species that are competing to occupy the same environmental niche within the host. A better structural understanding of how these efflux pumps export known antifungal agents during biofilm formation may make it feasible to design inhibitors of this transport, and thereby render C. albicans biofilms vulnerable to existing antifungal drugs.
6.2 Extracellular matrix
The biofilm extracellular matrix is another major contributor to antifungal drug resistance in C. albicans biofilms. The matrix acts as both a physical barrier to drug penetration and as a stabilizer of the overall architecture of the biofilm [1,75]. One known constituent of the biofilm matrix that contributes to its drug resistant properties is the polysaccharide β-1,3-glucan [75]. Treatment of biofilms with β-1,3-glucanase increases the susceptibility of biofilms to fluconazole, and addition of exogenous β-1,3-glucans increases the tolerance of planktonic cells to fluconazole [75]. There is also evidence to suggest that β-1,3-glucans of the matrix can bind specifically to amphotericin B, thereby preventing this antifungal drug from exerting its effect on the matrix-encased fungal cells within a biofilm [76]. Specifically targeting or enzymatically degrading the biofilm matrix may prove to be a useful approach in the development of novel therapeutics against C. albicans biofilms.
6.3 Persister cells
Another factor contributing to the drug resistant properties of C. albicans biofilms is the presence of persister cells. These cells are a minor subset of metabolically dormant yeast cells that stochastically arise as phenotypical variants within biofilms, and are extremely resistant to antifungal drugs [1,77]. Persister cells were discovered to exist in C. albicans biofilms, when upon treatment with Amphotericin B, a biphasic killing of cells was observed [77,78]. Although little is known about the formation and roles of persister cells during C. albicans biofilm development, we do know that the drug resistance of persister cells is independent of cell membrane composition and efflux pump expression, but is rather the result of the metabolically dormant state of the cells [1]. Understanding how persister cells are mechanistically regulated and controlled is an intriguing area for future study, which could lead to the development of novel therapeutic approaches to blocking their formation or survival.
7. Developing novel therapeutics for C. albicans biofilm infections
No biofilm-specific drugs exist today for the treatment of any biofilm-based microbial infection, making treatment of these infections particularly problematic. The resistance of biofilms to standard antifungal drugs is multifactorial; as described above, not only do biofilms provide physical protection from the host immune defenses and antifungal drugs, cells in biofilms are intrinsically resistant to drugs due to their constitutive up-regulation of drug efflux pumps and their altered metabolic states. It is these biofilm-specific properties that make developing effective therapeutics for biofilm infections particularly challenging. A better understanding of the molecular mechanisms underlying biofilm formation and maintenance is the key to the development of new therapeutic agents that specifically target the biofilm state. For example, a better understanding of the molecular mechanisms of adherence, both between C. albicans cells and a surface and between cells within the biofilm, could lead to strategies for preventing biofilms from forming or for disrupting existing biofilms. Identification of the molecular mechanisms behind biofilm dispersal could lead to a drug-based strategy to prevent cells from leaving a biofilm or enhance cell dispersal to the extent that a robust biofilm cannot develop. Also, a better understanding of the molecular basis of metabolic dormancy of subpopulations of cells might enable strategies to reverse the cell physiology or prevent the survival of these recalcitrant cells. Finally, studies into how microbes interact with host protective factors, such as how biofilms form on mucosal epithelial tissues, and identification of the factors that hamper C. albicans virulence in the presence of these protective factors, may lead to new therapeutic strategies to target these interactions. These are only a handful of the many possible mechanism-based strategies that could be exploited to develop new biofilm-specific therapeutics in the future.
In addition to these futuristic approaches, current work is ongoing to identify compounds that disrupt biofilm formation and/or maintenance through screens of chemical libraries. Although initial results from this approach are encouraging, it remains to be seen whether it will lead to the development of useful therapeutics. It is worth noting that strategies that weaken C. albicans biofilm formation or maintenance could render biofilms susceptible to conventional antifungal drugs, a prospect that could lead to combination therapies. Currently, there are only two specific targets of antifungal drugs (ergosterol biosynthesis and β-1,3-glucan synthesis), thus the development of antifungal drugs with new mechanisms of actions is clearly needed. Ultimately, future treatments will take advantage of our developing mechanistic knowledge of the biofilm and planktonic states and their interactions with the host to effectively clear biofilm-based infections and reduce relapse infection rates.
Acknowledgments
This work was supported by NIH grant R00AI100896.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
C.J.N. is a co-founder of BioSynesis, Inc., a company developing inhibitors and diagnostics of C. albicans biofilm formation.
References
- 1.Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 4.Jass J, Surman S, Walker J. Medical biofilms detection, prevention, and control. Chichester, UK: John Wiley & Sons, Ltd; 2003. [Google Scholar]
- 5.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 6.Fox EP, Nobile CJ. The role of Candida albicans biofilms in human disease. In: Dietrich LA, Friedmann TS, editors. Candida albicans symptoms, causes and treatment options. Nova Science Publishers; 2013. pp. 1–24. [Google Scholar]
- 7.Fox EP, Nobile CJ. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription. 2012;3:315–322. doi: 10.4161/trns.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14:380–385. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 10.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox EP, Singh-babak SD, Hartooni N, Nobile CJ. Biofilms and antifungal resistance. In: Coste AT, Vandeputte P, editors. Antifungals from genomics to resistance and the development of novel agents. Caister Academic Press; 2015. pp. 71–90. [Google Scholar]
- 13.Andes DR, Safdar N, Baddley JW, Playford G, Reboli AC, Rex JH, et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis. 2012;54:1110–1122. doi: 10.1093/cid/cis021. [DOI] [PubMed] [Google Scholar]
- 14.Tournu H, Van Dijck P. Candida biofilms and the host: models and new concepts for eradication. Int J Microbiol. 2012;2012:845352. doi: 10.1155/2012/845352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 2004;72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CC, Yu A, Lee H, Fidel PL, Jr, Noverr MC. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun. 2012;80:1736–1743. doi: 10.1128/IAI.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nett JE, Brooks EG, Cabezas-Olcoz J, Sanchez H, Zarnowski R, Marchillo K, et al. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect Immun. 2014;82:4931–4940. doi: 10.1128/IAI.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Noverr MC. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS One. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricicova M, Kucharikova S, Tournu H, Hendrix J, Bujdakova H, Van Eldere J, et al. Candida albicans biofilm formation in a new in vivo rat model. Microbiology. 2010;156:909–919. doi: 10.1099/mic.0.033530-0. [DOI] [PubMed] [Google Scholar]
- 21.Doyle TC, Nawotka KA, Kawahara CB, Francis KP, Contag PR. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb Pathog. 2006;40:82–90. doi: 10.1016/j.micpath.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Vande Velde G, Kucharikova S, Schrevens S, Himmelreich U, Van Dijck P. Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cell Microbiol. 2014;16:115–130. doi: 10.1111/cmi.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vande Velde G, Kucharikova S, Van Dijck P, Himmelreich U. Bioluminescence imaging of fungal biofilm development in live animals. Methods Mol Biol. 2014;1098:153–167. doi: 10.1007/978-1-62703-718-1_13. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Gomariz M, Perumal P, Mekala S, Nombela C, Chaffin WL, Gil C. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics. 2009;9:2230–2252. doi: 10.1002/pmic.200700594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, et al. Novel entries in a fungal biofilm matrix encyclopedia. MBio. 2014;5:e01333–e01314. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonhomme J, d’Enfert C. Candida albicans biofilms: building a heterogeneous, drug-tolerant environment. Curr Opin Microbiol. 2013;16:398–403. doi: 10.1016/j.mib.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Ni L, Bruce C, Hart C, Leigh-Bell J, Gelperin D, Umansky L, et al. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 2009;23:1351–1363. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol. 2013;90:22–35. doi: 10.1111/mmi.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 31.Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5:1604–1610. doi: 10.1128/EC.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29:99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nett JE, Zarnowski R, Cabezas-Olcoz J, Brooks EG, Berhardt J, Marchillo K, et al. Host contributions to construction of three device-associated Candida biofilms. Infect Immun. 2015;83:4630–4638. doi: 10.1128/IAI.00931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobile CJ, Mitchell AP. Microbial biofilms: e pluribus unum. Curr Biol. 2007;17:R349–R353. doi: 10.1016/j.cub.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell. 2011;10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Kohler JR, et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 2010;9:1531–1537. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nobile CJ, Fox EP, Hartooni N, Mitchell KF, Hnisz D, Andes DR, et al. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio. 2014;5:e01201–e01214. doi: 10.1128/mBio.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hnisz D, Bardet AF, Nobile CJ, Petryshyn A, Glaser W, Schock U, et al. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012;8:e1003118. doi: 10.1371/journal.pgen.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, et al. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011;7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granger BL. Insight into the antiadhesive effect of yeast wall protein 1 of Candida albicans. Eukaryot Cell. 2012;11:795–805. doi: 10.1128/EC.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 49.Peleg AY, Hogan Da, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 50.Peters BM, Noverr MC. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun. 2013;81:2178–2189. doi: 10.1128/IAI.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jack AA, Daniels DE, Jepson MA, Vickerman MM, Lamont RJ, Jenkinson HF, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindsay AK, Hogan DA. Candida albicans: Molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol Rev. 2014;28:85–96. [Google Scholar]
- 55.Neely AN, Law EJ, Holder IA. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun. 1986;52:200–204. doi: 10.1128/iai.52.1.200-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bandara HM, Yau JY, Watt RM, Jin LJ, Samaranayake LP. Escherichia coli and its lipopolysaccharide modulate in vitro Candida biofilm formation. J Med Microbiol. 2009;58:1623–1631. doi: 10.1099/jmm.0.012989-0. [DOI] [PubMed] [Google Scholar]
- 57.Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun. 2013;81:189–200. doi: 10.1128/IAI.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falagas ME, Betsi GI, Athanasiou S. Probiotics for prevention of recurrent vulvovaginal candidiasis: a review. J Antimicrob Chemother. 2006;58:266–272. doi: 10.1093/jac/dkl246. [DOI] [PubMed] [Google Scholar]
- 59.Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. 2015;17:484–492. doi: 10.1016/j.micinf.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strus M, Kucharska A, Kukla G, Brzychczy-Włoch M, Maresz K, Heczko PB. The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida. Infect Dis Obstet Gynecol. 2005;13:69–75. doi: 10.1080/10647440400028136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. Anaerobic Bacteria Grow within Candida albicans Biofilms and Induce Biofilm Formation in Suspension Cultures. Curr Biol. 2014;24:2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie Z, Thompson A, Sobue T, Kashleva H, Xu H, Vasilakos J, et al. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis. 2012;206:1936–1945. doi: 10.1093/infdis/jis607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh S, Navarathna DHMLP, Roberts DD, Cooper JT, Atkin AL, Petro TM, et al. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun. 2009;77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol. 2009;47:465–475. doi: 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 66.Dwivedi P, Thompson A, Xie Z, Kashleva H, Ganguly S, Mitchell AP, et al. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One. 2011;6:e16218. doi: 10.1371/journal.pone.0016218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NAR, et al. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012;14:1319–1335. doi: 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD, Ribbeck K. Mucins suppress virulence traits of Candida albicans. MBio. 2014;5:e01911. doi: 10.1128/mBio.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramage G, Bachmann S, Patterson TF, Wickes BL, Lopez-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49:973–980. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- 73.Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;3:547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- 74.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 75.Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, et al. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007;51:510–520. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vediyappan G, Rossignol T, D’Enfert C. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother. 2010;54:2096–2111. doi: 10.1128/AAC.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathe L, Van Dijck P, Mathé L, Van Dijck P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 2013;59:251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.LaFleur MD, Kumamoto CA, Lewis K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother. 2006;50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]