Abstract
Bacteroides fragilis is the leading cause of anaerobic bacteremia and sepsis 1. Enterotoxigenic strains producing B. fragilis toxin (BFT, fragilysin) contribute to colitis 2 and intestinal malignancy 3, yet are also isolated in bloodstream infection 4,5. It is not known whether these strains harbor unique genetic determinants that confer virulence in extra-intestinal disease. We demonstrate that BFT contributes to sepsis and identify a B. fragilis protease, fragipain (Fpn), which is required for endogenous activation of BFT through removal of its auto-inhibitory prodomain. Structural analysis of Fpn reveals a His-Cys catalytic dyad characteristic of C11 family cysteine proteases that are conserved in multiple pathogenic Bacteroides spp and Clostridium spp. Fpn-deficient enterotoxigenic B. fragilis is attenuated in its ability to induce sepsis, however Fpn is dispensable in B. fragilis colitis wherein host proteases mediate BFT activation. Our findings define a role for B. fragilis enterotoxin and its activating protease in the pathogenesis of bloodstream infection, indicating a greater complexity of cellular targeting and action of BFT than previously appreciated. The expression of fpn by both toxigenic and non-toxigenic strains suggests this protease may contribute to anaerobic sepsis beyond its role in toxin activation, potentially serving as a target for disease modification.
Anaerobic bacteremia caused by Bacteroides fragilis group organisms afflicts patients with acute intestinal injury and underlying disease including malignancy, an immunocompromised state, inflammatory bowel disease, congestive heart failure, and diabetes 6,7. Disruption of intestinal barrier function in these predisposing conditions precipitates extra-intestinal infection including bacteremia and sepsis 8,9, resulting in a ∼3-5-fold greater mortality risk, increased hospital length of stay, and the need for more aggressive therapeutic and surgical interventions especially in the setting of diverticulitis and overt colonic injuries 6,10-12. B. fragilis is the leading cause of anaerobic sepsis6,7,13. While experimental immunization targeting B. fragilis capsular polysaccharide protects against intra-abdominal sepsis-associated bacteremia 14, the virulence factors that contribute to severe bloodstream infection are not well-defined. Rising rates of antimicrobial resistance prompt the need to further understand the mechanisms that underlie B. fragilis sepsis 13,15,16.
Nearly 20% of B. fragilis bloodstream isolates harbor a 6 kb pathogenicity island encoding B. fragilis enterotoxin (BFT, fragilysin) that is absent from non-toxigenic B. fragilis (NTBF) 4,5,17. Toxin-producing isolates are enriched in the bloodstream relative to other sites of extra-intestinal infection 4, suggesting that these strains exhibit enhanced bloodstream survival and virulence. BFT is a zinc-dependent metalloprotease with structural homology to eukaryotic ADAM metallopeptidases 2,3,18-21. BFT is the only recognized virulence factor in enterotoxigenic B. fragilis (ETBF), promoting colonic injury and inflammation through E-cadherin cleavage 3,20,22. Proteolytic cleavage of cadherins by ADAM10 contributes to the pathogenesis of Staphylococcus aureus infection through tissue barrier disruption 23-25. Given the similarity of BFT to ADAM metalloproteases21, we examined whether BFT impacts lethal sepsis. Mice were infected intravenously with 1 × 109 colony-forming units (CFU) wild-type ETBF (WT), an isogenic bft mutant (Δbft), or the Δbft strain complemented with bft (cbft). WT and cbft infection led to lethal sepsis, whereas mice infected with Δbft survived (Fig. 1a). BFT was sufficient for lethality as mice succumbed to intoxication with purifiedBFT (Fig. 1b), but not a catalytically inactive mutant (iBFT) 26.
Fig. 1. BFT contributes to lethal sepsis. (a).
Kaplan-Meier curves demonstrating survival following intravenous infection of 4 week-old female C57Bl/6 mice with 1 × 109 wild-type (WT) ETBF strain 43859 (n = 9), an isogenic Δbft mutant (n = 10), or the Δbft mutant with genomic complementation of bft (cbft, n = 8). Results are representative of three independent experiments. (b) Kaplan-Meier curves of survival following intravenous injection of 4 week-old female C57Bl/6 mice (n = 5 per group) with 2 μg purified, active BFT or an enzymatically inactive variant (iBFT). **, P < 0.01, ****, P < 0.0001, Mantel-Cox log rank test. Results are representative of two independent experiments.
BFT is produced as a 45 kDa zymogen with an N-terminal prodomain that facilitates folding and enzyme latency 21. During toxin maturation, an unidentified protease mediates prodomain removal C-terminal to R211 27. To define factors required for active BFT production, we examined supernatants from transposon mutant ETBF clones for an inability to disrupt intracellular junctions in colonic epithelial HT-29 cells (Supplementary Fig. 1a and ref. 20). Of 84 defective mutants, secondary screening identified two lacking the cleaved 20 kDa active BFT moiety (BFT*) (Fig. 2a).
Fig. 2. BFT is activated by a novel B. fragilis cysteine protease. (a).
Anti-BFT western blot analysis of 8G7 and 57G6 mutants to evaluate active BFT in supernatants (BFT*). (b) Transposon insertion sites mapped to the intergenic region upstream of a clostripain-related protein in strain NCTC 9343. (c) ClustalX alignment of clostripain-related protein sequence from ETBF (ATCC 43858), NTBF strains (NCTC 9343, YCH46, 638R), other Bacteroides spp., and C. histolyticum clostripain noting the catalytic dyad (asterisks) and the C11 cysteine protease H-G-(Xaa)n-A-C motif (blue boxes). (d) Fpn expression in wild-type (WT) ETBF strain 43859, an isogenic fpn deletion mutant (Δfpn), and the Δfpn mutant with genomic complementation of fpn (cfpn). (e) Western blot analysis of full-length BFT (FL-BFT) expression in cell pellets of WT, Δfpn, and cfpn ETBF strains, and NCTC 9343 (NTBF)(left panel), and analysis of culture supernatants for active BFT (BFT*) (right panel). (f) E-cadherin staining in HT-29 cell monolayers treated with supernatants from WT, Δbft, Δfpn, and cfpn cultures. Green, E-cadherin; blue, nuclei (DAPI). Scale bars = 10 μm. (g) Anti-BFT western blot analysis (upper panel) of reaction products generated following incubation of purified, recombinant forms of Fpn and BFT, a BFT cleavage site mutant (BFTR211A) or predicted Fpn active site dyad mutants (FpnH135A and FpnC180A), noting non-specific cleavage fragments (bracket). N-terminal sequencing of BFT cleavage products (encircled) by Edman degradation and documentation of the Fpn cleavage site in BFT and BFTR211A. Western blot analysis of cleaved E-cadherin in culture supernatants following incubation of HT-29 cells with reaction products from above (lower panel, lanes 3-4).
Transposon insertion mapping revealed two sites upstream of a gene annotated in B. fragilis NTBF strain BF9343 as a peptidase similar to the Bacteroides thetaiotaomicron clostripain-related protein (Fig. 2b). RTQ-PCR confirmed a reduction in peptidase expression (Supplementary Fig. 1b). The clostripain-related peptidase is a 393 amino acid protein of predicted molecular weight 44,160 Da (Supplementary Fig. 2, GenBank KT867680). Sequence alignment of this peptidase with homologous proteins in NTBF, other Bacteroides species, and Clostridia histolyticum clostripain revealed conservation of the His-Cys catalytic dyad 28 (Fig. 2c and Supplementary Fig. 2a, asterisks) in the context of a -HisGly-(Xaa)n-AlaCys- sequence motif (blue boxes) common to clan CD cysteine proteases 29, and also revealed an N-terminal lipobox motif conserved in B. fragilis (orange box). B. fragilis 638R is predicted to encode a clostripain-like polypeptide lacking 15 N-terminal residues. Analysis of the 638R genome compared to NCTC 9343 revealed an insertional mutation (Supplementary Fig. 2b), resulting in loss of protein production (Supplementary Fig. 2c). The peptidase appears as two immunoreactive species, consistent with the clostripain heavy and light chain heterodimer formed upon autocatalytic cleavage 30. C. histolyticum clostripain is a C11 cysteine protease that exhibits specificity for an Arg-Xaa peptidyl bond 29. We hypothesized that the clostripain-like protein activates BFT and designated it fragipain (Fpn).
To evaluate necessity of Fpn for BFT activation, a fpn deletion was generated in ETBF (Δfpn) and complemented by genomic insertion of fpn (cfpn). These strains displayed in vitro growth properties indistinguishable from the parental strain (Supplementary Fig. 3a). Fpn transcript and Fpn protein production were verified through RTQ-PCR (Supplementary Fig. 3b) and immunoblot analysis (Fig. 2d). Fpn deletion resulted in the absence of active BFT in culture supernatants (Fig. 2e, BFT*) and loss of E-cadherin cleavage in HT-29 cells (Fig. 2f), both of which were restored by complementation.
Using recombinant proteins, Fpn was sufficient for native BFT cleavage. Edman degradation of the BFT reaction product demonstrated cleavage at R211 as observed in culture supernatants (Fig. 2g) 27. Mutation of BFT R211 (BFTR211A) resulted in the generation of a species with higher molecular weight than BFT* through R162 cleavage on a surface-exposed loop 21. Fpn variants harboring mutations of predicted active site residues (FpnH135A, FpnC180A, and FpnH135AC180A) were unable to cleave BFT. Fpn-activated BFT facilitated E-cadherin cleavage on HT-29 cells (Fig. 2g, lower panel), which was reduced in BFTR211A upon Fpn exposure.
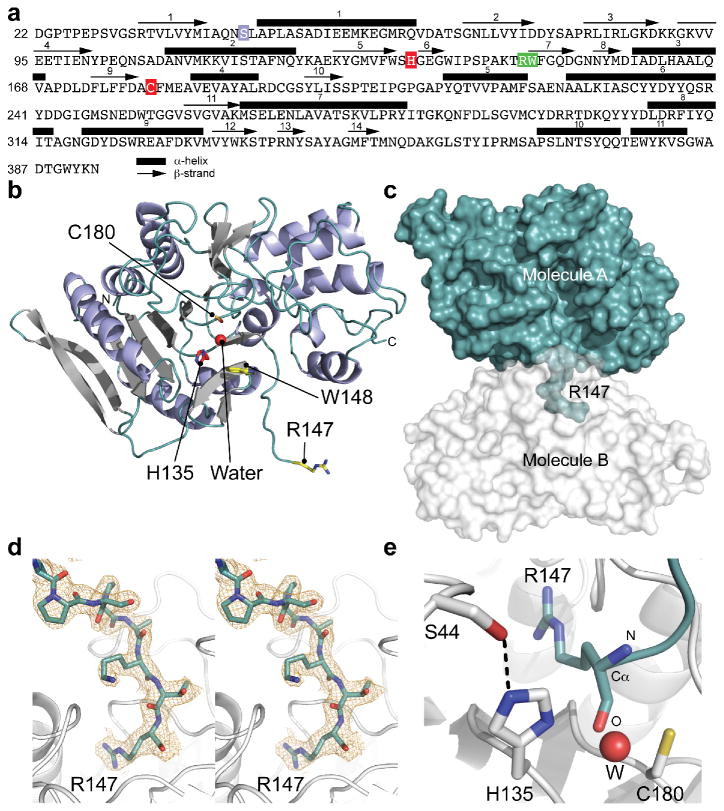
Recombinant Fpn was crystallized for X-ray diffraction and structure determination. The structure was solved by molecular replacement using a homology model based on a Parabacteroides merdae putative protease (37% sequence identity, PDB code: 3UWS) and refined to 2.48 Å resolution (Fig. 3 and Supplementary Table 1, PDB code: 5DYN). Fpn is composed of 11 α-helices surrounding a β-scaffold consisting of 9 β-strands (β1-β5, β9-β12); 5 additional β-strands (scaffold β6-β8, hairpin β13-β14) are present at the surface (Fig. 3a,b). Fpn is cleaved at the Arg-Trp peptidyl bond (Fig 3a, green) in the loop connecting β-strands 6 and 7. The cleaved loop of Fpn projects into the catalytic site of an adjacent Fpn molecule (Fig. 3c), demonstrated in a composite omit map of the electron density of the active site (Fig. 3d) that reveals a cysteine protease cleavage product bound in trans. Residues H135 and C180 (Fig. 3a, red) that form the catalytic dyad are positioned 2.9 Å and 3.1 Å from the main chain oxygen of substrate residue R147 (Fig. 3e). The structure suggests autocatalytic cleavage of Fpn and a classical cysteine protease mechanism whereby H135 deprotonates C180, promoting C180 attack on the substrate carbonyl. S44 (Fig. 3a, purple) makes a close interaction with H135 (Fig. 3e), which may position the imidazole ring of H135 for C180 deprotonation. The structure reveals water contact to H135 (Fig. 3c) and the oxygen of the arginine main chain. As in papain, this water molecule may deacylate the acyl-enzyme intermediate and release the cleaved carboxy peptide 31-33. Consistent with this mechanism of action, Fpn exhibits optimal activity at neutral pH (Supplementary Fig. 3c).
Fig. 3. Sequence and molecular structure of B. fragilis fragipain (Fpn). (a).
Fpn amino acid sequence where α-helices are numbered and delimited by a black rectangle and β-strands are numbered and delimited by a black arrow. The active site residues H135 and C180 are boxed in red. Residue S44, which makes contact with the imidazole ring of H135 is boxed in purple. The cleavage site (R147-W148) is boxed in green. (b) X-ray crystal structure of Fpn demonstrating α-helices in purple; β-strands in grey. The active site residues H135 and C180 are in red. The water molecule (W) present in the active site is represented as a red sphere. Residues at the cleavage site (R147-W148) are yellow. (c) Surface representation of Fpn molecules (A and B) in adjacent unit cells; R147 of molecule A is bound in the active site of molecule B. (d) Stereoview of the simulated annealing composite omit map (contoured at 2σ) of the R147 region. (e) Magnified view of the Fpn active site. Residues H135 and C180 are shown as sticks; contact between S44 and H135 is shown as a dotted line. R147 from the molecule in the next unit cell is shown in cyan. The water molecule (W) present in the active site is represented as a red sphere.
To examine the role of Fpn in BFT-mediated disease, antibiotic-pretreated mice received 1 × 109 CFU of WT, Δbft, Δfpn, or cfpn B. fragilis via oral gavage and were examined for colitis. While fecal CFU and weight gain were similar across groups (Supplementary Fig. 4a,b), mice infected with WT, Δfpn, and cfpn exhibited crypt hypertrophy and epithelial shedding (Fig. 4a) and an increase in the colon weight to length ratio (Fig. 4b). Δbft-infected mice did not demonstrate signs of colitis. Consistent with the lack of requirement for Fpn in BFT-mediated intestinal injury, colonic mucus harvested from specific pathogen-free (SPF) and germ-free (GF) mice mediated BFT cleavage (Fig. 4c, BFT*). In contrast, Fpn was required for lethal sepsis progression. B. fragilis Δfpn was avirulent upon intravenous infection (Fig. 4d), mirroring the phenotype observed upon BFT deletion (Fig. 1a), and demonstrating a tissue-specific requirement for Fpn in toxin activation.
Fig. 4. Selective requirement for Fpn in lethal sepsis. (a).
Colonic tissue sections stained with hematoxylin and eosin 7 days following orogastric inoculation of 4 week-old female C57Bl/6 mice with 1 × 109 wild-type (WT), an isogenic ETBF Δfpn mutant, or the Δfpn with genomic complementation of fpn (cfpn). Scale bar = 100 μm. (b) Colonic weight to length ratio 7 days post-inoculation in mice infected with WT (n = 9), Δbft ETBF (n = 10), Δfpn mutant (n = 9), or the cfpn (n = 10) strains as in (a). ****, P < 0.0001, Student's unpaired two-sided t test. Results are representative of three independent experiments. Error bars, mean ± s.e.m. (c) Analysis of active BFT production (BFT*) following incubation of purified full-length BFT (FL BFT) with colonic mucous prepared in PBS following harvest from specific pathogen-free (SPF) or germ-free (GF) female C57Bl/6 mice. A small amount of BFT cleavage is observed upon incubation with PBS alone (lane 1). Results are representative of two independent experiments. (d) Kaplan-Meier curves demonstrating survival following intravenous infection of 4 week-old female C57Bl/6 mice with 1 × 109 wild-type (WT, n = 7), an Δfpn (n = 8), or cfpn (n = 8) ETBF. **, P < 0.01, ****, P < 0.0001, Mantel-Cox log rank test. Results are representative of three independent experiments. (e) VE-cadherin staining in human pulmonary artery endothelial cell monolayers treated with BFT compared to monolayers treated with media alone or iBFT. Green, VE-cadherin; blue, nuclei (DAPI). Scale bars = 10 μm. Results are representative of two independent experiments.
While the colonic epithelial barrier is recognized as an important target of BFT 20, the role of BFT and Fpn in sepsis suggested a broader cellular tropism. Loss of endothelial barrier integrity is a primary pathophysiologic alteration in sepsis 34. We therefore examined whether the vascular endothelium is directly injured by BFT. In contrast to media or inactive toxin, treatment of human pulmonary artery endothelial cells (HPAECs) with BFT resulted in loss of junctional VE-cadherin (Fig. 4e), confirmed by generation of a C-terminal cadherin fragment consistent with metalloprotease-dependent cleavage (Supplementary Fig. 5 and ref. 23).
These studies define a role for BFT in sepsis, revealing the requirement of a novel B. fragilis protease for extra-intestinal toxin activation to confer virulence. While the molecular pathogenesis of life-threatening systemic disease associated with staphylococcal enterotoxins and Shiga toxins is well-characterized 35,36, little insight is available on systemic complications from enterotoxins produced by anaerobic intestinal commensals. The structural similarity of BFT with ADAM metalloproteinases 21 and our demonstration that BFT targets VE-cadherin indicates that activated BFT functions to disrupt barrier integrity akin to the role of ADAM10 in S. aureus sepsis 23,24. These studies highlight the role of ADAM-family metalloproteases in sepsis-associated endovascular injury, caused directly by BFT or following catalytic activation of host ADAM10 induced by S. aureus α-toxin 23-25,37. As expression-enhancing human ADAM10 polymorphisms are associated with increased susceptibility to severe sepsis 38, differential levels of active BFT expression may modulate B. fragilis sepsis severity. The identification of a role for BFT in sepsis therefore provides an opportunity to understand the molecular pathogenesis of anaerobic infection and examine clinical outcomes in individuals infected with toxin-producing strains, including in the context of polymicrobial infections. As B. fragilis remains the leading cause of anaerobic sepsis 6,11, our observations are anticipated to stimulate investigation of novel preventive and therapeutic interventions applicable to an ever-growing population receiving advanced medical care. Of keen interest is the potential development of targeted probiotic therapy to eradicate toxigenic B. fragilis in individuals at high risk for sepsis.
Further studies are needed to develop a mechanistic understanding of BFT and Fpn bloodstream activation, identify cellular targets of toxin action, and examine whether fpn expression by non-toxigenic B. fragilis promotes disease. To our knowledge, Fpn is the first C11 peptidase implicated in sepsis. C11 family cysteine proteases are widely present in pathogenic Bacteroides spp and Clostridium spp (http://merops.sanger.ac.uk/cgi-bin/famsum?family=c11) 39, together responsible for over 50% of anaerobic bacteremia 1,10. The unique relevance of Fpn in BFT activation within the bloodstream may therefore also prompt a broader investigation of C11 proteases in extra-intestinal infection.
Online Methods
Bacterial and eukaryotic cell culture
The ETBF strains used in these studies were ATCC 43858 and ATCC 43859. The NTBF strains used were NCTC 9343 (ATCC 25285) and 638R (TM4000). B. fragilis strains were propagated anaerobically at 37 °C in Brain Heart Infusion (BHI) medium supplemented with 0.0005% hemin and 0.5 μg/mL vitamin K1 (BHIS). Media was supplemented with 200 mg/L gentamicin and 5 mg/L erythromycin or 5 mg/L clindamycin for selection of strains harboring drug resistance loci. E. coli S17 was used for propagation and conjugation of plasmids that were introduced into B. fragilis, while E. coli Origami-2 (Novagen) was used for protein purification. E. coli were maintained in LB supplemented with 50 mg/L kanamycin for strains containing the pET-28b vector or 100 mg/L ampicillin for all other vectors. For conjugations, equal volumes of late log phase E. coli and B. fragilis cultures were washed once with BHI, mixed, plated as a puddle on BHIS or LB agar, and incubated aerobically at 37 °C overnight, modified from a previously published protocol 40. Bacteria were then resuspended in BHI, plated onto BHIS agar containing gentamicin and erythromycin or clindamycin, and grown anaerobically. HT-29 cells were obtained from the American Type Culture Collection (ATCC) and propagated in DMEM supplemented with 10% FBS and 2 mM L-glutamine. HT-29 cells were authenticated by ATCC through short tandem repeat profiling and cell morphology, and confirmed to be contaminant-free prior to distribution. Primary human pulmonary artery endothelial cells (HPAECs; Lonza) were cultured in Endothelial Basal Medium-2 (EBM-2) BulletKit media (Lonza).
Animal Modeling
All animal studies were conducted in accord with all ethical regulations under protocols approved by the University of Chicago Animal Care and Use Committee and Institutional Biosafety Committee. Female C57BL/6 mice were bred in-house from mice originally purchased from Jackson Laboratory or purchased from Jackson Laboratory and maintained under specific pathogen-free conditions for use in experimentation at 4 weeks of age. Sample size estimates for animal experiments were based on prior animal modeling studies utilized within the laboratory for investigation of lethal infectious disease and colitis. At the time of weaning, animals were randomly distributed for use in experimentation. There was no investigator blinding in animal experimentation, and no animals were excluded from analysis.
Intravenous infection
Overnight cultures of B. fragilis were subcultured at a 1:100 ratio into fresh BHIS and grown for 14-15 hours. Cells from 25 mL culture were sedimented and resuspended in 15 mL PBS. These suspensions were adjusted by the addition of PBS until a 1:5 dilution yielded an OD600 = 0.55. Bacteria from 9 mL of suspensions were sedimented, resuspended in a volume of PBS empirically determined to yield a cell density of 1 × 1010 CFU/mL. Prior to the retro-orbital delivery of 0.1 mL of the bacterial inoculum, mice were anesthetized with ketamine (100 mg/kg) delivered via intraperitoneal route. The number of CFU in each inoculum was confirmed by serial dilution plating on BHIS agar. Mice were euthanized at time points indicated in the results or when moribund in accord with criteria established in the animal use protocol.
Colitis modeling
Mice were pre-treated with 100 mg/L clindamycin in drinking water for one week prior to and throughout the course of infection. To prepare inocula, overnight cultures of B. fragilis were subcultured at a 1:50 ratio into fresh BHIS and grown until they reached OD600 ≈ 0.5. Bacteria from 50 mL culture were sedimented and processed as for intravenous infection. After the final centrifugation, bacteria were resuspended in 1.8 mL 0.1 N sodium bicarbonate to yield a concentration of 1010 CFU/mL. Infection was accomplished by oral gavage of 0.1 mL of each inoculum. The number of CFU in each inoculum was confirmed by serial dilution plating on BHIS agar. To analyze fecal CFU following oral inoculation, fecal pellets were collected from individual mice, weighed, and vortexed in 1 mL PBS to achieve complete resuspension. Serial 10-fold dilutions were plated on BHIS agar containing gentamicin and clindamycin to determine CFU/g feces. For histopathologic analysis of infected colonic tissue, mice were euthanized 1 week following infection and colons were dissected from the mice and immediately injected with 10% formalin. Following the removal of fecal pellets, the colon was opened longitudinally and Swiss-rolled around a toothpick starting from the distal colon with the mucosal surface facing outward. Tissue rolls were fixed in 10% formalin overnight. Paraffin embedding, tissue sectioning, and H&E staining were performed by Nationwide Histology. Pathologic analysis was performed by Dr. Jerrold Turner. Imaging was performed at the University of Chicago Integrated Light Microscopy Facility. Digital image files were created with a 3D Histech Pannoramic Scan whole slide scanner (Perkin Elmer, Waltham, MA) with a Stingray F146C color camera (Allied Vision Technologies, Stadtroda, Germany). Individual images were created with the 3D Histech Pannoramic Viewer software (Perkin Elmer, Waltham, MA).
Generation of polyclonal antisera
Purified cleaved BFT and Fpn were administered to New Zealand White rabbits. A total of 2.25 mg protein was administered in 3 injections given in 3-week intervals. Complete Freund's adjuvant was used for the initial immunization and incomplete Freund's adjuvant for the remaining boosts. Serum was collected starting 3 weeks after the last boost, and reactivity to the proteins of interest determined by immunoblot analysis of purified protein.
Preparation of intestinal mucus
Germ-free mice and specific-pathogen free mice were fasted for 8 hours prior to dissection. The colon was harvested from each mouse and opened longitudinally with removal of residual fecal matter to permit preparation of intestinal mucus as previously described 41.
Bacterial studies
Transposon mutagenesis screen
To generate transposon mutants, the pSAM-Bt vector 42 was introduced to ATCC 43858 via conjugation with a donor E. coli S17 strain. Clones with transposon insertions were selected on BHIS agar containing gentamicin and erythromycin, used to inoculate BHIS liquid media containing antibiotics, and grown overnight in deep 96-well plates. 1:100 dilutions of culture supernatants in serum-free DMEM were used to treat semi-confluent HT-29 cell monolayers for 3 hours. Cells were fixed in 10% formalin, stained with 0.05% Crystal Violet for 5 minutes, washed with water, and screened visually. Screening PCR of transposon mutants was performed using the GoTaq system (Promega). Cloning and inverse PCR was performed using the Phusion system (ThermoFisher). For inverse PCR, genomic DNA was prepared using the Wizard Genomic DNA Extraction kit (Promega), digested with BstNI (NEB), and then treated with T4 ligase (NEB). PCR was performed on this DNA using primers that annealed in the transposon. PCR products were gel extracted and sequenced. Supplementary Table 2 describes all primers utilized for cloning and PCR-based analysis.
Reverse Transcription Quantitative PCR
Bacteria were harvested at the indicated times using RNA Protect (Qiagen). RNA was extracted using the RNeasy kit (Qiagen) and reverse transcribed with random priming using the iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed on cDNA, RNA, and water controls using the SYBR Green kit (Bio-Rad) and the Bio-Rad CFX96 real-time system with a C1000 thermal cycler. Standard curves for PCR efficiency determination were derived from 10-fold serial dilutions of pooled cDNA. Expression levels were compared by normalizing fpn transcript to the amount of 16S transcript present in each sample.
Bacterial plasmids and generation of bacterial strains
To generate isogenic Δbft and Δfpn strains, 1 kb genomic regions flanking the target gene were cloned from ETBF ATCC 43859, fused together, and ligated into the pKNOCK suicide vector 43 using KpnI and BamHI sites. The vector was conjugated into ETBF ATCC 43859 that were then plated on BHIS agar containing gentamicin and clindamycin. Serial passaging of the resulting clones was performed, and clones were screened for loss of pKNOCK by clindamycin sensitivity and PCR with primers to document deletion of the targeted locus. Loss of protein expression was confirmed by western blot analysis.
To restore bft and fpn gene expression in isogenic deletion mutants, we generated the pAH1 vector that enables stable insertion into the genome. We identified a targetable insertion site in an intergenic region present in the genomes of multiple published strains of B. fragilis. This site is downstream of genes on the 5′ and 3′ ends to prevent the disruption of native loci. The selected intergenic region was cloned into the suicide vector pKNOCK to generate the vector pAH1. To generate cbft and cfpn B. fragilis strains, bft or fpn loci with the corresponding upstream regions were cloned and fused downstream of the rpoD promoter, then ligated into pAH1. These constructs were introduced to Δbft or Δfpn strains via conjugation, and integration of the construct into the correct site and protein expression were confirmed by PCR and western blot, respectively. To generate clindamycin resistant Δbft and Δfpn strains for mouse infection, pAH1 was introduced via conjugation.
Generation of recombinant bacterial proteins
E. coli Origami-2 carrying pET-28b expression vectors were used for protein purification. For BFT, a cDNA was generated by PCR to encode residue S20 through the native stop, appended with an N-terminal His6-tag. The cDNA was cloned into pET-28b using the NcoI and XhoI sites. To clone fpn, a cDNA was generated by PCR to encode residue Q20 through the last coding residue, appended with a C-terminal His6-tag followed by a stop codon. The cDNA was cloned into pET-28b using the NcoI and NotI sites. For both BFT and Fpn, the N-terminal 19 amino acids of the native proteins were omitted from the tagged recombinant proteins as these constitute the predicted lipoprotein signal sequence. Mutant alleles of bft and fpn were generated using overlap PCR and cloned into pET-28b using the same sites as their WT counterpart. To purify recombinant proteins, overnight cultures were subcultured at a ratio of 1:20, grown until they reached OD600 = 0.4-0.6, then induced with 0.5 mM IPTG at room temperature overnight. Protein was purified as previously described, except in the absence of protease inhibitors 21. To generate purified, cleaved BFT, immobilized TPCK trypsin (ThermoFisher) and purified BFT were co-incubated at a ratio of 100 μl immobilized trypsin bead slurry:1 mg protein in PBS for 3 hours at room temperature with mixing. The trypsin beads were removed and the remaining solution was co-incubated with Ni-NTA for 1 hour at 4 °C in a column. The flow-through containing cleaved BFT was collected. Proteins were dialyzed into PBS, concentration was determined using a BCA assay (Pierce), and purity was assessed by SDS-PAGE analysis followed by Coomassie blue staining.
Protein and cellular assays
In vitro cleavage of BFT
For Fpn cleavage of BFT, equimolar amounts of WT or mutant variants of BFT and Fpn were reacted in PBS at 37 °C for 30 minutes. For mouse mucus cleavage of BFT, 2 μg of BFT was mixed with 20 μl mucus and incubated for 20 minutes at 37 °C. 500–1,000 ng protein was applied per lane of SDS-PAGE. To prepare samples for Edman degradation, samples were incubated with Ni-NTA beads for 1 hour at 4 °C after reaction in order to remove Fpn and uncleaved BFT. Proteins that remained in solution were separated by SDS-PAGE and transferred to a PVDF membrane that was then stained with Coomassie Blue. The indicated bands were cut out of the membrane, and Edman degradation was performed at Tufts University Core Facilities.
Fragipain enzymatic analysis
200 nM Fpn was reacted with 250, 500, 750, 1,000, 1,250, 1,500, and 1,750 μM Nα-benzoyl-L-arginine ethyl ester (BAEE) substrate (Sigma-Aldrich) at 37 °C in 67 mM NaH2PO4 across a pH gradient. Absorbance (OD253) was read at 5-minute intervals using a Tecan Infinite 2000 Pro plate reader.
Preparation of B. fragilis cell lysates and supernatants
Cell lysates were prepared by centrifuging 0.7 mL culture at 5,000 × g for 5 minutes and lysing the cell pellet in 80 μL of 2X Laemmli's sample buffer at 95 °C for 10 minutes. Supernatants were precipitated with 58% saturated ammonium sulfate at –20°C overnight. 10μL of cell lysate or the entire amount of precipitated supernatant were analyzed per lane.
Cadherin cleavage assays
For E-cadherin cleavage analysis, HT-29 cells (3 × 106/well) were plated in 6-well plates 2 days prior to treatment. Cells were treated with 3 μg of BFT in 1 mL PBS for 20 minutes at room temperature. 0.9 mL supernatants were precipitated with 30% saturated ammonium sulfate and protein was resuspended in 30 μl 2X Laemmli's sample buffer. The N-terminal fragment of E-cadherin was detected by Western blot (ECCD-2, ThermoFisher). For VE-cadherin cleavage analysis, HPAEC cells (2.5 × 106/well) were plated in 6-well plates 2 days prior to treatment. Cells were treated with 3 μg BFT or iBFT in 1 mL unsupplemented EBM-2 for 4 hours at 37 °C. For biochemical studies, cell lysates were immunoprecipitated with anti-VE-cadherin antibody (Santa Cruz Biotechnology) and protein G sepharose (Pierce) at 4 °C overnight, then analyzed by immunoblotting according to published protocols 25. Two independent biological experiments were conducted for cadherin cleavage analyses.
Immunofluorescence staining
For E-cadherin staining of HT-29 cell monolayers, cells were plated onto cover slips in 24-well plates 2 days prior to treatment with bacterial supernatants. Cells were washed twice with PBS then treated with a 1:10 dilution of overnight bacterial supernatants in PBS for 40 minutes at room temperature. After treatment, cells were washed once with PBS then fixed in 4% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were blocked using a solution of 1% BSA and 0.2% Tween-20 in PBS for 1 hour at room temperature. A 1:50 dilution of mouse monoclonal E-cadherin antibody (BD Transduction Laboratories, clone 36/E-cadherin) in blocking buffer was bound to cells for 1 hour at room temperature. Cells were washed twice with PBS. Anti-mouse Alexa Fluor 588-conjugated antibody (Life Technologies) was bound at a 1:1,000 dilution in blocking buffer for 2 hours at room temperature. Cells were washed twice with PBS then mounted onto slides using ProLong Gold Anti-Fade with DAPI (ThermoFisher). For HPAEC cell analysis, 1×105 cells/well were plated in 24-well plates on sterile glass coverslips 2 days prior to treatment. Monolayers were treated as above, rinsed, methanol-fixed for 10 minutes, stained with an anti-VE-cadherin antibody and visualized according to published protocols 23,25. Images were captured using an Olympus DSU spinning disk confocal and analyzed with ImageJ (http://imagej.nih.gov/ij/). Two independent biological experiments were conducted for immunofluorescence studies.
Western blot analysis
For all Western blot analysis, proteins were separated by SDS-PAGE, transferred onto PVDF membrane (ThermoFisher), and blocked with 10% non-fat skim milk in Tris-buffered saline with 0.1% Tween-20 (TBST). Primary antibodies used were as follows:α-Fpn rabbit polyclonal antisera (1:2,500),α-BFT rabbit polyclonal antisera (1:2,000), ratα-E-cadherin monoclonal antibody (1:100, ECCD-2, ThermoFisher), and α-RpoA (1:20,000, gift of Dr. Dominique Missiakas). Secondary antibodies (1:10,000) were as follows: α-rabbit Alexa Fluor 680 (Life Technologies) or α-rat Alexa Fluor 680 (Life Technologies). Membranes were visualized using the Licor Odyssey imaging system. Two independent biological experiments were conducted for all western blot studies.
Crystallographic studies
Protein purification
Recombinant His6-tagged Fpn was expressed in E. coli BL21(DE3)pLysS (Novagen). A 50 ml overnight culture in Luria-Bertani (LB) medium supplemented with 50 μg/ml kanamycin (LabScientific) (LB-Kan50) was used to inoculate 2 liters of LB-Kan50; this culture was incubated at 37 °C in a rotary shaker at 220 rpm. Transcription of recombinant genes was induced at a culture density of 0.8 (monitored spectrophotometrically at OD600 nm) by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, GoldBio Technology). After 4 h of induction, the cells were harvested by centrifugation at 12,000 × g for 20 min at 4 °C. Cell pellets were resuspended in 30 ml of lysing/binding buffer (10 mM Tris pH 7.4, 150 mM NaCl, 10 mM imidazole supplemented with DNaseI (Sigma-Aldrich) and Phenyl-Methyl-Sulfonyl-Fluoride (PMSF; Sigma-Aldrich)). Cells were disrupted by three passages in a French pressure cell, and the cell debris was removed by centrifugation for 20 min at 25,000 × g. The supernatant was loaded onto a Ni2+ Sepharose affinity column (GE Life Sciences) pre-equilibrated with the binding buffer. Two washing steps were performed using 10 mM and 75 mM of imidazole (Fisher Scientific) followed by two elution steps with 200 mM and 500 mM imidazole in the binding buffer. Protein purity was estimated at 95% as assessed by 14% SDS-PAGE stained with Coomassie brilliant blue. The protein solution was then dialyzed against 2 liters of 10 mM Tris pH 7.4, 150 mM NaCl buffer to remove imidazole. All purification steps were carried out at 4 °C.
Fpn crystallization
Purified Fpn was concentrated in a centrifugal filter (3 kDa MWCO, Amicon-Millipore) and used for crystallization. Initial crystallization screening was carried out using the sitting-drop, vapor-diffusion technique in 96-well microplates (Nunc). Trays were set up using a Mosquito robot (TTP LabTech) and commercial crystallization kits (Nextal-Qiagen and Microlytic). Hanging drops were set by mixing equal volumes (0.1 μl) of the protein and the precipitant solutions equilibrated against 75 μl of the precipitant solution. In all trials, the protein concentration was 53 mg/ml. In approximately 2–3 weeks at 19°C, small crystals appeared in condition 63 of the MCSG1 crystallization kit (Microlytic). All manual crystallization attempts were carried out using the hanging-drop, vapor-diffusion technique in 24-well plates (Hampton). The drops were set by mixing different ratios (1:1, 1:2 and 2:1 μl) of the protein (at 10 mg/ml) and the crystallization solutions equilibrated against 500 μl of the precipitant solution. After manual refinement, the best crystals were obtained at 19°C by mixing 1 μl of the protein solution (at 35 mg/ml) with 2 μl of the following crystallization solution: 200 mM ammonium citrate dibasic (pH 5) (Fisher Scientific), 25% (w/v) PEG 3350 (Acros Organics). Crystals grew to their final size in two to three weeks. Before flash freezing with liquid nitrogen, crystals were cryo-protected by soaking in the crystallization solution containing 30% glycerol (Fisher Scientific).
Crystallographic data collection and data processing
Diffraction from the majority of crystals was poor. The best crystal diffracted to dmin = 2.48 Å; a single dataset (180 images) was collected from this crystal at a temperature of 100 K using a 1 degree oscillation range on beamline 21-ID-F (LS-CAT, Advanced Photon Source, Argonne, Illinois). Images were collected on a MAR Mosaic 225 detector. Diffraction images were processed using the xia2 data reduction suite 44 applying the -3da XDS 45 and Aimless 46 options, which were accessed through the SBGrid consortium 47. Geometric refinement and examination of the scaled amplitudes revealed that Fpn crystals belong to the orthorhombic space group P21212, with cell dimensions a = 124.32 Å, b = 48.90 Å, c = 60.88 Å (α=β=γ= 90°) (Supplementary Table 1). The structure was solved by molecular replacement in Phenix 48 using a model of Fpn that was based on the structure of a hypothetical protein from Parabacteroides merdae (PDB code: 3UWS). For purposes of the molecular replacement search, the number of Fpn molecules in the asymmetric unit was estimated using the Matthews Probability Calculator web server (http://www.ruppweb.org/mattprob/). The Fpn search model was generated using the Expasy Swiss-model Workspace server 49. Molecular replacement revealed one Fpn molecule in the crystallographic asymmetric unit. The initial structural model was manually examined and corrected; ions, and water molecules were added, and refinement of the structure was conducted iteratively using Coot 50 and phenix.refiine 48. The final structural model was refined to an Rwork of 17.5 % and Rfree of 22.3%. Crystallographic data and refined model statistics are presented in Supplementary Table 1.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Pairwise comparisons were performed using the Student's unpaired two-sided t test, or the Mantel-Cox log rank test for comparison of survival following Kaplan-Meier estimation. Comparison of variances was performed in GraphPad utilizing the F test, providing documentation of similar variance between groups.
Supplementary Material
Footnotes
Author Contributions: V.M.C. developed the experimental concepts and approach, generated all bacterial strains, performed transposon mutagenesis screening and in vivo and vitro studies on colonic epithelial injury and sepsis, and wrote the manuscript; J.H. generated and analyzed the Fpn crystal structure and wrote the manuscript; A.L.H. contributed essential bacterial genetic reagents and expertise; W.P.T. performed studies on the endothelium; J.R.T. performed pathology analysis of colonic tissues; S.C. analyzed the Fpn crystal structure and wrote the manuscript; J.B.W. developed the experimental concepts and approach wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Robert R, Deraignac A, Le Moal G, Ragot S, Grollier G. Prognostic factors and impact of antibiotherapy in 117 cases of anaerobic bacteraemia. Eur J Clin Microbiol Infect Dis. 2008;27:671–678. doi: 10.1007/s10096-008-0487-5. [DOI] [PubMed] [Google Scholar]
- 2.Rhee KJ, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Medicine. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato N, Kato H, Watanabe K, Ueno K. Association of enterotoxigenic Bacteroides fragilis with bacteremia. Clin Infect Dis. 1996;23(Suppl 1):S83–86. doi: 10.1093/clinids/23.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 5.Claros MC, et al. Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates. Anaerobe. 2006;12:17–22. doi: 10.1016/j.anaerobe.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Redondo MC, Arbo MD, Grindlinger J, Snydman DR. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin Infect Dis. 1995;20:1492–1496. doi: 10.1093/clinids/20.6.1492. [DOI] [PubMed] [Google Scholar]
- 7.Ngo JT, et al. Population-based assessment of the incidence, risk factors, and outcomes of anaerobic bloodstream infections. Infection. 2013;41:41–48. doi: 10.1007/s15010-012-0389-4. [DOI] [PubMed] [Google Scholar]
- 8.Rocha ER, Smith CJ. Ferritin-like family proteins in the anaerobe Bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2013;26:577–591. doi: 10.1007/s10534-013-9650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brook I, Frazier EH. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J Med Microbiol. 2000;49:827–830. doi: 10.1099/0022-1317-49-9-827. [DOI] [PubMed] [Google Scholar]
- 10.Vena A, et al. Are incidence and epidemiology of anaerobic bacteremia really changing? Eur J Clin Microbiol Infect Dis. 2015;34:1621–1629. doi: 10.1007/s10096-015-2397-7. [DOI] [PubMed] [Google Scholar]
- 11.Ani C, Farshidpanah S, Bellinghausen Stewart A, Nguyen HB. Variations in organism-specific severe sepsis mortality in the United States: 1999-2008. Crit Care Med. 2015;43:65–77. doi: 10.1097/CCM.0000000000000555. [DOI] [PubMed] [Google Scholar]
- 12.Moghadamyeghaneh Z, et al. A comparison of outcomes of emergent, urgent, and elective surgical treatment of diverticulitis. American journal of surgery. 2015;210:838–845. doi: 10.1016/j.amjsurg.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Snydman DR, et al. Lessons learned from the anaerobe survey: historical perspective and review of the most recent data (2005-2007) Clin Infect Dis. 2010;50(Suppl 1):S26–33. doi: 10.1086/647940. [DOI] [PubMed] [Google Scholar]
- 14.Kasper DL, Onderdonk AB, Crabb J, Bartlett JG. Protective efficacy of immunization with capsular antigen against experimental infection with Bacteroides fragilis. J Infect Dis. 1979;140:724–731. doi: 10.1093/infdis/140.5.724. [DOI] [PubMed] [Google Scholar]
- 15.Hartmeyer GN, Soki J, Nagy E, Justesen US. Multidrug-resistant Bacteroides fragilis group on the rise in Europe? J Med Microbiol. 2012;61:1784–1788. doi: 10.1099/jmm.0.049825-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang FD, Liao CH, Lin YT, Sheng WH, Hsueh PR. Trends in the susceptibility of commonly encountered clinically significant anaerobes and susceptibilities of blood isolates of anaerobes to 16 antimicrobial agents, including fidaxomicin and rifaximin, 2008-2012, northern Taiwan. Eur J Clin Microbiol Infect Dis. 2014;33:2041–2052. doi: 10.1007/s10096-014-2175-y. [DOI] [PubMed] [Google Scholar]
- 17.Franco AA, et al. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J Bacteriol. 1999;181:6623–6633. doi: 10.1128/jb.181.21.6623-6633.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moncrief JS, et al. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect Immun. 1995;63:175–181. doi: 10.1128/iai.63.1.175-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco AA, et al. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect Immun. 1997;65:1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci U S A. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulas T, Arolas JL, Gomis-Ruth FX. Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog. Proc Natl Acad Sci U S A. 2011;108:1856–1861. doi: 10.1073/pnas.1012173108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JM, et al. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130:59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers ME, Kim HK, Wang Y, Bubeck Wardenburg J. ADAM10 mediates vascular injury induced by Staphylococcus aureus alpha-hemolysin. J Infect Dis. 2012;206:352–356. doi: 10.1093/infdis/jis192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoshima I, et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med. 2011;17:1310–1314. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers ME, Becker RE, Sailer A, Turner JR, Bubeck Wardenburg J. Synergistic Action of Staphylococcus aureus alpha-Toxin on Platelets and Myeloid Lineage Cells Contributes to Lethal Sepsis. Cell Host Microbe. 2015;17:775–787. doi: 10.1016/j.chom.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco AA, Buckwold SL, Shin JW, Ascon M, Sears CL. Mutation of the zinc-binding metalloprotease motif affects Bacteroides fragilis toxin activity but does not affect propeptide processing. Infect Immun. 2005;73:5273–5277. doi: 10.1128/IAI.73.8.5273-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Tassell RL, Lyerly DM, Wilkins TD. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect Immun. 1992;60:1343–1350. doi: 10.1128/iai.60.4.1343-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labrou NE, Rigden DJ. The structure-function relationship in the clostripain family of peptidases. Eur J Biochem. 2004;271:983–992. doi: 10.1111/j.1432-1033.2004.04000.x. [DOI] [PubMed] [Google Scholar]
- 29.McLuskey K, Mottram JC. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem J. 2015;466:219–232. doi: 10.1042/BJ20141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dargatz H, Diefenthal T, Witte V, Reipen G, von Wettstein D. The heterodimeric protease clostripain from Clostridium histolyticum is encoded by a single gene. Mol Gen Genet. 1993;240:140–145. doi: 10.1007/BF00276893. [DOI] [PubMed] [Google Scholar]
- 31.Shokhen M, Khazanov N, Albeck A. Challenging a paradigm: theoretical calculations of the protonation state of the Cys25-His159 catalytic diad in free papain. Proteins. 2009;77:916–926. doi: 10.1002/prot.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivares D, Arnoux P, Pignol D. A papain-like enzyme at work: native and acyl-enzyme intermediate structures in phytochelatin synthesis. Proc Natl Acad Sci U S A. 2005;102:18848–18853. doi: 10.1073/pnas.0505833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei D, Huang X, Liu J, Tang M, Zhan CG. Reaction pathway and free energy profile for papain-catalyzed hydrolysis of N-acetyl-Phe-Gly 4-nitroanilide. Biochemistry. 2013;52:5145–5154. doi: 10.1021/bi400629r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 35.Ortega E, Abriouel H, Lucas R, Galvez A. Multiple roles of Staphylococcus aureus enterotoxins: pathogenicity, superantigenic activity, and correlation to antibiotic resistance. Toxins. 2010;2:2117–2131. doi: 10.3390/toxins2082117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer CL, Leibowitz CS, Kurosawa S, Stearns-Kurosawa DJ. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins. 2012;4:1261–1287. doi: 10.3390/toxins4111261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoshima N, Wang Y, Wardenburg JB. Genetic requirement for ADAM10 in severe Staphylococcus aureus skin infection. J Invest Dermatol. 2012;132:1513–1516. doi: 10.1038/jid.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui L, et al. An ADAM10 promoter polymorphism is a functional variant in severe sepsis patients and confers susceptibility to the development of sepsis. Crit Care. 2015;19:73. doi: 10.1186/s13054-015-0796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comstock LE, et al. Analysis of a capsular polysaccharide biosynthesis locus of Bacteroides fragilis. Infect Immun. 1999;67:3525–3532. doi: 10.1128/iai.67.7.3525-3532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melton-Celsa AR, Darnell SC, O'Brien AD. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. [Google Scholar]
- 45.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.