Abstract
METTL3 is a RNA methyltransferase implicated in mRNA biogenesis, decay, and translation control through N6-methyladenosine (m6A) modification. Here we find that METTL3 promotes translation of certain mRNAs including epidermal growth factor receptor (EGFR) and the Hippo pathway effector TAZ in human cancer cells. In contrast to current models that invoke m6A reader proteins downstream of nuclear METTL3, we find METTL3 associates with ribosomes and promotes translation in the cytoplasm. METTL3 depletion inhibits translation, and both wild-type and catalytically inactive METTL3 promote translation when tethered to a reporter mRNA. Mechanistically, METTL3 enhances mRNA translation through an interaction with the translation initiation machinery. METTL3 expression is elevated in lung adenocarcinoma and using both loss- and gain-of-function studies we find that METTL3 promotes growth, survival, and invasion of human lung cancer cells. Our results uncover an important role of METTL3 in promoting translation of oncogenes in human lung cancer.
Keywords: METTL3, N6-methyladenosine, m6A, Translation, EGFR, Ribosome, Cancer
Graphical abstract
INTRODUCTION
Biochemical evidence from forty years ago identified that mammalian messenger RNAs (mRNAs) contain N6-methyladenosine (m6A) that predominantly occurs close to the 3′ end of the mRNA (Desrosiers et al., 1974; Horowitz et al., 1984; Perry et al., 1975). The recent mapping of m6A sites throughout the transcriptome of mammalian cells revealed that thousands of mRNAs are modified at a consensus sequence motif and that m6A peaks are located across the mRNA gene body and typically found close to the stop codon (Dominissini et al., 2012; Ke et al., 2015; Meyer et al., 2015; Meyer et al., 2012; Schwartz et al., 2014; Wang et al., 2014b; Zhou et al., 2015). These methylation marks can be dynamically regulated and m6A patterns can vary between cell types (Geula et al., 2015; Meyer et al., 2012). METTL3 (Methyltransferase like 3, also known as MTA70) was originally identified as a methyltransferase responsible for m6A modification (Bokar et al., 1997). Although METTL3 alone is catalytically active and methylates the adenosine within a ‘GGACU’ consensus sequence-containing substrate RNA, a complex containing two catalytic components METTL3, METTL14, and the accessory WTAP (Wilms’ tumor 1 associated protein) has been recently characterized (Bokar et al., 1997; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014; Wang et al., 2014c). METTL3 (as well as the other complex components) are observed predominantly in nuclear speckles by immunofluorescence analyses, although m6A methyltransferase activity has been detected in both nuclear and cytoplasmic extracts (Bokar et al., 1997; Harper et al., 1990; Ping et al., 2014). Consistent with the dynamic control of m6A RNA modification two specific demethylases, which also localize to nuclear speckles, FTO (Fat mass and obesity-associated protein) and ALKBH5 (alkB homolog 5, RNA demethylase) have been identified (Jia et al., 2011; Zheng et al., 2013).
It is emerging that m6A plays important and diverse biological functions in plants, yeast, flies, and mammals. For example, m6A manipulation via knockdown or deletion of the methyltransferase or demethylase impacts plant development and growth, yeast meiosis, Drosophila oogenesis, mouse spermatogenesis, body mass and metabolism, synaptic signaling, circadian clock regulation, early embryonic development, and stem cell self-renewal and differentiation (Batista et al., 2014; Chen et al., 2015; Fustin et al., 2013; Geula et al., 2015; Iles et al., 2013; Lin and Gregory, 2014; Wang et al., 2014c; Zhao et al., 2014; Zheng et al., 2013), (reviewed in (Meyer and Jaffrey, 2014; Yue et al., 2015)). Despite this growing appreciation of the biological significance of this pathway the mechanisms for how m6A might regulate gene expression remain poorly understood. Emerging models implicate the so-called ‘reader’ proteins, the best characterized of which are the YTH domain-containing proteins, YTHDF1, -2, and -3 (YTH domain-containing family 1, -2, and -3), that selectively bind m6A-modified RNAs (Dominissini et al., 2012; Li et al., 2014; Theler et al., 2014; Wang et al., 2015; Xu et al., 2014; Zhu et al., 2014). In the case of YTHDF2, binding to m6A-modified RNAs leads to their transport to processing-bodies (P-bodies) in the cell cytoplasm and subsequent degradation (Wang et al., 2014b). In contrast, YTHDF1 has been implicated in the control of translation through an interaction with the eIF3 initiation factor (Wang et al., 2015). Other putative methyl reader proteins have been identified and m6A can alter RNA conformation to influence interaction with RNA-binding proteins (Alarcon et al., 2015a; Dominissini et al., 2012; Liu et al., 2015).
In this study, we report an important role of METTL3 in promoting translation of a subset of target mRNAs independent of its catalytic activity and downstream m6A reader proteins. In addition, we uncovered a role of METTL3 in promoting oncogene expression, cancer cell growth, survival, and invasion, suggesting that METTL3 could be a potential target to suppress oncoprotein expression as a possible cancer therapy.
RESULTS
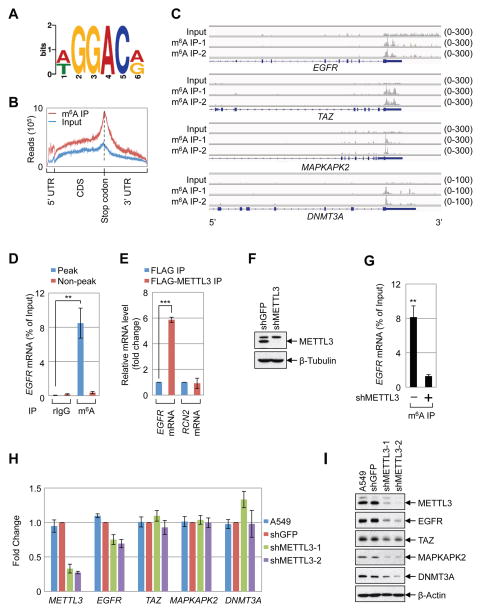
METTL3 regulates protein expression
Here we investigated the molecular and cellular role of METTL3 in human cancer cells. Global profiling of m6A target genes using meRIP-Seq identified 9298 m6A peaks (in 5099 genes) in A549 human lung cancer cells (Supplemental Table 1). Our m6A profiles correlate well between replicates and with a published dataset (Supplementary Figure 1A–B) (Ke et al., 2015). Comparison of the m6A profiles between A549 and H1299 cells revealed a substantial overlap of m6A-modified mRNAs in both cell types (Supplementary Figure 1C, Supplementary Table 1), suggesting that there are many common but also some distinct m6A genes in those two cell lines. Consistent with published studies a ‘GGAC’ sequence motif was enriched in the immunopurified RNA, and a metagene analysis revealed that m6A peaks were predominantly localized near to stop codons, with a subset of m6A peaks located in the 5′ UTR and internal exons (Figure 1A, B) (Dominissini et al., 2012; Meyer et al., 2015; Meyer et al., 2012; Schwartz et al., 2014; Wang et al., 2014c; Zhou et al., 2015). The mRNAs of several oncogenes including EGFR, TAZ, MAPKAPK2 (MK2), and DNMT3A were found to have one or more m6A peaks near the stop codon, whereas other mRNAs including HRAS have a m6A peak further downstream in the 3′ UTR, or in the case of c-MYC have m6A more broadly distributed m6A across multiple exons (Figure 1C, Supplemental Figure 1D–E, Supplemental Table 1). Quantitative reverse transcriptase PCR (q.RT-PCR) using primers to amplify either the m6A peak region or a control (non-peak) region of EGFR and other m6A containing mRNAs was used to validate the meRIP-Seq data (Figure 1D, Supplementary Figure 1F). We next investigated the role of METTL3 in the regulation of gene expression. We found EGFR mRNA (but not a non-methylated control mRNA (RCN2) specifically associated with FLAG-METTL3-containing ribonucleoprotein complexes (Figure 1E). In addition, analysis of the published METTL3 PAR-CLIP and m6A CLIP data revealed binding of METTL3 within the m6A peak region near the EGFR stop codon (Supplementary Figure 1G)(Ke et al., 2015; Liu et al., 2014). Moreover, depletion of METTL3 using shRNA resulted in substantially reduced levels of m6A-modified EGFR mRNA, supporting that METTL3 binds to EGFR mRNA and is responsible for its m6A modification (Figure 1F, G). Since m6A reportedly modulates mRNA stability we next measured the global impact of METTL3-depletion on steady state RNA levels by performing RNA-Seq on RNA collected from control- or METTL3 knockdown cells. This revealed that METTL3 knockdown had minimal effect on mRNA levels throughout the transcriptome in both A549 as well as H1299 cells (Supplemental Figure 2A–C). q.RT-PCR analysis confirmed that METTL3-depletion had little effect on the abundance of individual m6A target mRNAs (Figure 1H). Interestingly however, when we examined the expression of the corresponding proteins by Western blot, we found that METTL3 depletion results in decreased protein levels of EGFR, TAZ, MAPKAPK2, and DNMT3A (Figure 1I). Metabolic labeling experiments in control- and METTL3-depleted cells revealed a widespread requirement for METTL3 in protein synthesis (Supplemental Figure 2D). However, not all proteins were dependent on METTL3 since the level of MYC and RAS protein expression was unaffected in METTL3 knockdown cells (Supplemental Figure 2E–F). These results indicate that METTL3 might somehow regulate protein expression of a subset of m6A-modified mRNAs. We also found EGFR and TAZ protein expression to be similarly regulated in HeLa and H1299 cells (Figure 2A, B, and data not shown). In summary, we found that METTL3 knockdown had minimal impact on mRNA levels and instead found a substantial decrease in protein abundance for several m6A-containing mRNAs. Considering this, together with the observed m6A peaks at the translation stop codon, we postulated that METTL3 might regulate mRNA translation.
Figure 1. METTL3 regulates protein expression.
(A–B) Global profiling of m6A in A549 cells. (A) Sequence motif identified from the top 1000 m6A peaks. (B) Distribution of m6A peaks across all mRNAs. (C) Integrative Genomics Viewer (IGV) plots of m6A peaks at individual mRNAs. The y-axis shows sequence read number, blue boxes represent exons, and blue lines represents introns. (D) q.RT-PCR analysis of α-m6A IP in A549 cells using indicated PCR primers. (E) q.RT-PCR analysis of FLAG-METTL3 RNA IP in A549 cells with indicated primers. (F) Western blot with indicated antibodies. (G) q.RT-PCR analysis of EGFR m6A levels upon depletion of METTL3 in A549 cells. (H, I) Expression of METTL3 and m6A targets in the METTL3 knockdown cells. (H) q.RT-PCR analysis of indicated mRNA levels. mRNA was first normalized to β-Actin mRNA. Relative ratio (fold-change) obtained in the presence of shGFP was set to 1. (I) Western blotting with indicated antibodies. All q.RT-PCR data are presented as ±SEM. n=3. **p<0.01, ***p<0.001. See also Figures S1 and S2, and Supplementary Table 1.
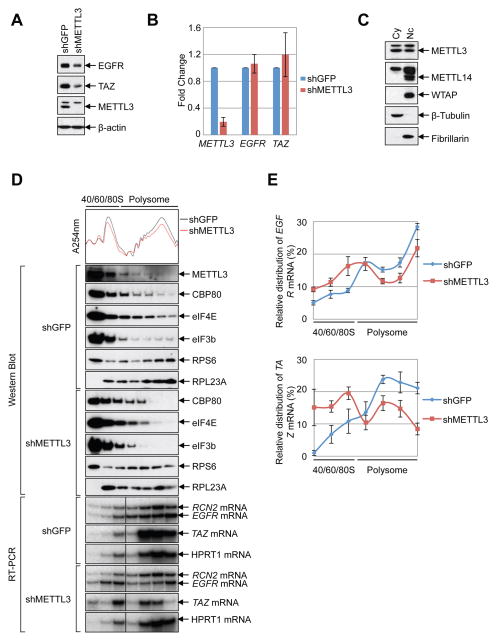
Figure 2. METTL3 enhances translation.
(A–B) METTL3 knockdown in HeLa cells (A) Western blot using indicated antibodies. (B) q.RT-PCR of indicated mRNAs normalized to β-Actin mRNA. Relative ratio (fold-change) obtained in the presence of shGFP was set to 1. Data are presented as ±SEM. n=3. (C) Western blot analysis of METTL3 in nuclear and cytoplasmic fractions using β-Tubulin (cytoplasmic, Cy) and Fibrillarin (nuclear, Nc) as controls. (D–E) Cytoplasmic extracts from control or METTL3-depleted cells were subjected to sucrose grandient centrifugation. (D) Polysome-fractionated samples analyzed by Western blot using the indicated antibodies and RT-PCR performed with α-[32P]-dCTP. (E) Relative levels of EGFR or TAZ mRNAs in each ribosome fraction were quantified and normalized to RCN2 mRNA and plotted as a percentage of the total. Data are from three independent polysome-profiling experiments. Error bars = mean ±SEM, n=3.
METTL3 enhances translation
To test whether METTL3 might be directly involved in translational control we first examined whether any METTL3 localizes to the cell cytoplasm where translation occurs. Lysates were prepared from nuclear and cytoplasmic fractions and examined by Western blot. This revealed that in addition to its nuclear localization, METTL3 protein is readily detectable in cytoplasmic fractions (Figure 2C), similar to recent reports in mouse ESCs and human cancer cells (Alarcon et al., 2015b; Chen et al., 2015). In contrast however, the localization of other known METTL3-interacting proteins including the alternative m6A methyltransferase METTL14 and the WTAP cofactor was found to be restricted to the nuclear fraction (Figure 2C). Next, we used sucrose gradient centrifugation to resolve polysome fractions from cytoplasmic extracts prepared from control- or METTL3-depleted cells. Analysis by Western blot revealed that METTL3 was detected in most of the fractions (Figure 2D). Moreover, METTL3 depletion considerably altered the polysome profile with an increase in the 60S peak and corresponding decrease in 80S and polysome peaks (Figure. 2D). Western blot analysis of different ribosome fractions revealed that the nuclear m7G cap-binding protein CBP80, cytoplasmic m7G cap-binding protein eIF4E, and the translation initiation factor eIF3 core subunit eIF3b were detected in most fractions of the control (shGFP) cells. The distribution of these factors shifted from heavy polysome fractions to lighter sub-polysome fractions upon METTL3 knockdown (Figure 2D). Next we used RT-PCR to examine the distribution of endogenous METTL3 target or non-target mRNAs in the ribosome fractions. We found that the relative distribution of EGFR and TAZ mRNAs was shifted from polysome to sub-polysome fractions in the METTL3-depleted cells compared to the RCN2 control mRNA (Figure 2D, E). Additionally, the polysome distribution of HPRT1, another control mRNA that is not m6A-modified (our data and (Wang et al., 2014b)), was unaffected by METTL3 knockdown (Figure 2D). Altogether these data support that METTL3 plays a role in promoting translation and raise the possibility that the METTL3 protein itself, rather than possible downstream m6A reader proteins including YTHDF1 or YTHDF2, might participate in translational control of a subset of m6A containing mRNAs.
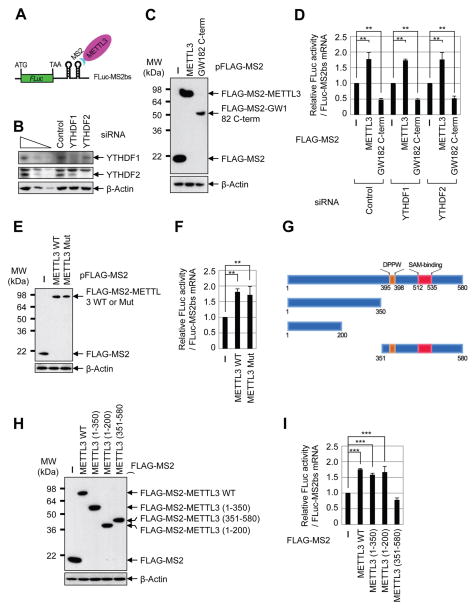
METTL3 directly promotes translation independently of methyltransferase activity or downstream m6A reader proteins
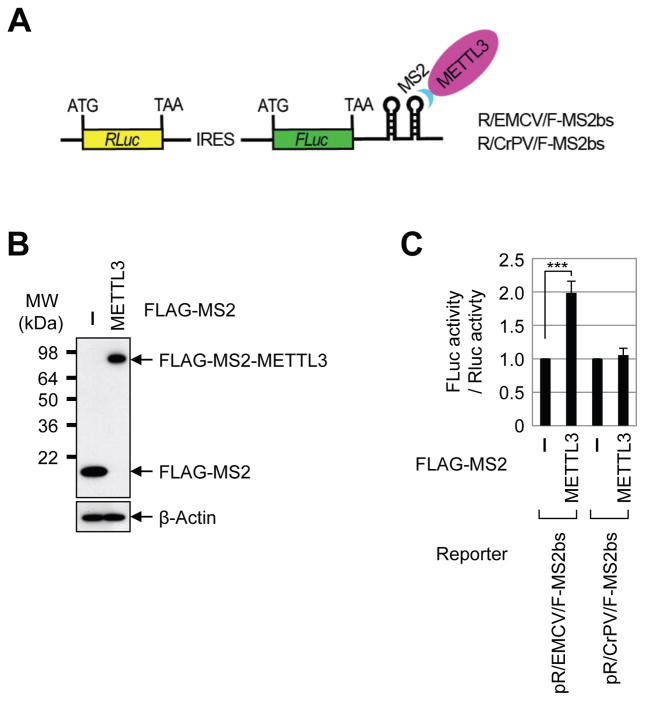
To directly test the possible role of METTL3 in promoting translation and to examine the requirement of YTHDF1 or YTHDF2 we performed tethering experiments using a luciferase reporter mRNA containing two MS2-binding sites located just downstream of the stop codon (Figure 3A) in control and YTHDF1- and YTHDF2-depleted cells (Figure 3B). The relative expression level of effector proteins including FLAG-MS2 (negative control), FLAG-MS2-METTL3, and FLAG-MS2-GW182 C-term (positive control that suppresses translation) were measured by Western blot (Figure 3C). Intriguingly, we found that directly tethering FLAG-MS2-METTL3 to the 3′ UTR robustly enhanced translation efficiency by around 1.8 fold without changing the mRNA level (Figure 3D, Supplemental Figure 3A). FLAG-MS2-GW182 inhibited translation as expected (Chang et al., 2012). Knockdown of YTHDF1 or YTHDF2 had no effect on METTL3-mediated enhanced translation, thereby uncoupling the role of METTL3 as an m6A ‘writer’ and its role in translation (Figure 3D). We next directly tested whether METTL3 might promote translation independently of its m6A catalytic activity. To test this we performed additional tethering experiments using plasmids to express either wild-type or catalytic mutant (aa395–398, DPPW → APPA) METTL3. Interestingly, this analysis revealed that the m6A catalytic activity of METTL3 is dispensable for its role in promoting translation in these tethering assays (Figures 3E–F, Supplemental Figure 3B). We then generated a series of domain deletion mutant versions of METTL3 and tested their role in promoting translation (Figure 3G). We found that the N-terminal of METTL3 was sufficient to promote translation, while the catalytic domain containing C-terminal of METTL3 had no effect in promoting translation, further confirming that METTL3 promotes translation independent of its catalytic activity (Figures 3H–I, Supplemental Figure 3C). Moreover, METTL14 and WTAP were also found to be dispensable for the enhanced translation that is mediated by METTL3 in tethering assays (Supplementary Figures 3D–G). Taken together, these results provide strong evidence that METTL3 associates with the translational machinery and enhances translation of target mRNAs independent of its methyltransferase activity, binding-partners METTL14 and WTAP, or m6A reader proteins including YTHDF1 and YTHDF2.
Figure 3. METTL3 directly promotes translation independently of catalytic activity or downstream reader proteins.
(A) Schematic diagram of tethering reporter assay. (B) Western blot analysis of YTHDF1 and YTHDF2 expression in control and siRNA knockdown cells. (C) αFLAG western blot of indicated proteins. (D) Tethering assay to measure translation efficiency of reporter mRNAs. Firefly luciferase (FLuc) activity was measured and normalized to the Renilla luciferase (RLuc) activity. Relative FLuc activity was normalized to the relative FLuc-MS2bs mRNAs. The normalized FLuc activity (translation efficiency) in the presence of MS2 and control siRNA for each set was set to 1. **P<0.01. Error bars = mean ±s.d., n=3. (E) αFLAG western blot of indicated proteins. (F) Tethering assay to measure translation efficiency of reporter mRNAs. (G) Schematic diagram of METTL3 deletion mutants. (H) α-FLAG western blot of indicated proteins. (I) Tethering assay to measure translation efficiency of reporter mRNAs. See also Figure S3.
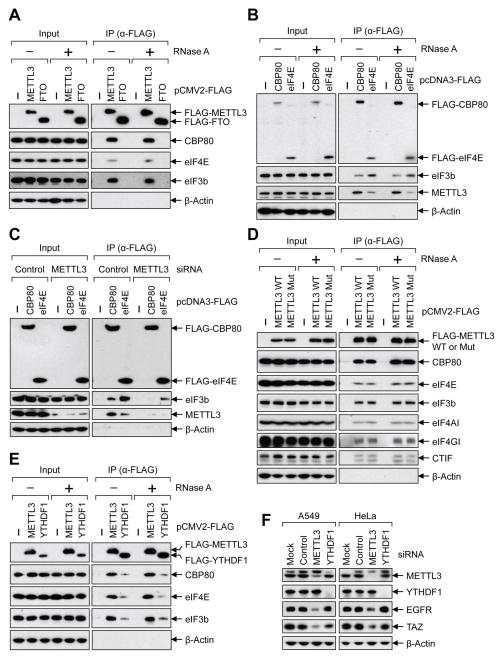
METTL3 enhances translation of target mRNAs by recruiting eIF3 to the translation initiation complex
We next sought to understand the mechanism by which METTL3 promotes translation. Considering that initiation is typically the rate-limiting step of translation, as well as our data that METTL3-depletion disrupted the global translation efficiency and effected both CBP80/20-dependent translation as well as eIF4E-dependent translation, we tested whether METTL3 might interact with translation initiation factors. To this end, we performed co-IP experiments with FLAG-affinity-purified complexes and tested for the association of METTL3 with translation initiation proteins. This revealed that CBP80, eIF4E, and the eIF3 subunit eIF3b were co-immunopurified with FLAG-METTL3 in an RNA-independent manner (Figure 4A and Supplemental Figure 4A). However, none of these factors were found associated with FLAG-FTO (Figure 4A). Reciprocal co-IPs performed using FLAG-CBP80, and FLAG-eIF4E confirmed these specific interactions. This also revealed that though METTL3 associates with both m7G cap-binding complexes there was more METTL3 associated with CBP80 than eIF4E (Figure 4B). eIF3b (positive control) was detected in both FLAG-CBP80- and FLAG-eIF4E-containing complexes as expected. Since METTL3 impacts both the CBP80/20-dependent translation as well as eIF4E-dependent translation and that eIF3 is a common factor in both steps of translation initiation we hypothesized that METTL3 might be involved in the recruitment of eIF3 to translation initiation complexes. To test this possibility, we examined the association of eIF3 with FLAG-CBP80 or FLAG-eIF4E by co-IP from control- or METTL3-knockdown cells. We found considerably less eIF3b associated with FLAG-CBP80 or FLAG-eIF4E in the METTL3 knockdown samples thereby supporting our model that METTL3 helps recruit eIF3 to promote translation initiation (Figure 4C). Interestingly, and consistent with the tethering assays, both wild-type and catalytically inactive METTL3 displayed comparable association with these translation initiation factors (Figure 4D).
Figure 4. METTL3 recruits eIF3 to the translation initiation complex.
(A–E) Co-IPs of indicated FLAG-tagged proteins analyzed by Western blot using the indicated antibodies. Where indicted lysates were treated with RNase A. (E) Co-IPs performed using lysates collected from either control- or METTL3 siRNA transfected cells. (F) Western blot performed on cell lysates collected from indicated cells transfected with control-, METTL3-, or YTHDF1 siRNA. See also Figure S4.
It was recently reported that YTHDF1 binds to the m6A sites and can enhance translation by interacting with eIF3 (Wang et al., 2015). We therefore carried out additional co-IP experiments to examine the association of METTL3 and YTHDF1 with the translation initiation complex. Consistent with published work we found that FLAG-YTHDF1 interacts with eIF3b as well as both CBP80 and eIF4E in an RNA-independent manner (Figure 4E). However, a substantially higher level of these translation initiation factors was detected in METTL3-containing complexes (Figure 4E). Importantly, no interaction between YTHDF1 and METTL3 was detected by co-IP suggesting that these proteins might function in distinct translation initiation complexes (Supplemental Figure 4B). Despite these co-IP results and data from tethering assays showing that METTL3 can enhance translation independent of YTHDF1 or YTHDF2 (Figure 3E), it remained possible that the effects of METTL3 depletion on mRNA translation might be mediated (at least in part) by YTHDF1. To address this, we individually depleted METTL3 and YTHDF1 using siRNAs and examined expression of METTL3 target genes by Western blot. Strikingly, this revealed that while transient METTL3 knockdown suppressed expression of EGFR and TAZ proteins in both A549 and HeLa cells as expected, the depletion of YTHDF1 had no effect on the levels of EGFR and TAZ protein levels (Figure 4F).
To test whether METTL3 might promote translation by recruiting translation initiation factors we performed METTL3 tethering assay using bicistronic reporter mRNAs containing either Encephalomyocarditis Virus (EMCV) internal ribosome entry site (IRES) or Cricket ParalysisVirus (CrPV) IRES in between Renilla luciferase (RLuc) and Firefly luciferase (FLuc) (Figure 5A). Since EMCV IRES requires all translation initiation factors except for eIF4E whereas CrPV IRES-dependent translation does not need any translation initiation factors, these different IRES reporters allowed us to interrogate the specific role of METTL3 in promoting translation. We found that direct tethering of METTL3 to the 3′ UTR of the different reporters specifically enhanced the translation of EMCV IRES-dependent translation of Fluc compared to cap-dependent RLuc translation. In contrast, METTL3 tethering did not enhance CrPV IRES-dependent FLuc translation. This specific functional effect of METTL3 tethering on EMCV- but not CrPV-IRES reporters is consistent with a role of METTL3 in recruiting translation initiation factors. Of note, direct tethering of METTL3 enhanced only the proximal open reading frame (i.e. not the upstream RLuc) (Figure 5B–C), which is consistent with the enrichment of m6A sites near the stop codon of endogenous target mRNAs. In summary, our results illuminate an important role of METTL3 in directly enhancing translation - a pathway that is distinct from the downstream m6A-binding protein YTHDF1.
Figure 5. METTL3 promotes translation by recruiting translation initiation factors.
(A) Schematic diagram of tethering reporter assay. (B) α-FLAG western blot of indicated proteins. (C) Tethering assay to measure translation efficiency of reporter mRNAs. FLuc activity was measured and normalized to the RLuc activity. ***P<0.001. Error bars = mean ±s.d., n=3.
METTL3 promotes cancer cell growth, survival, and invasion
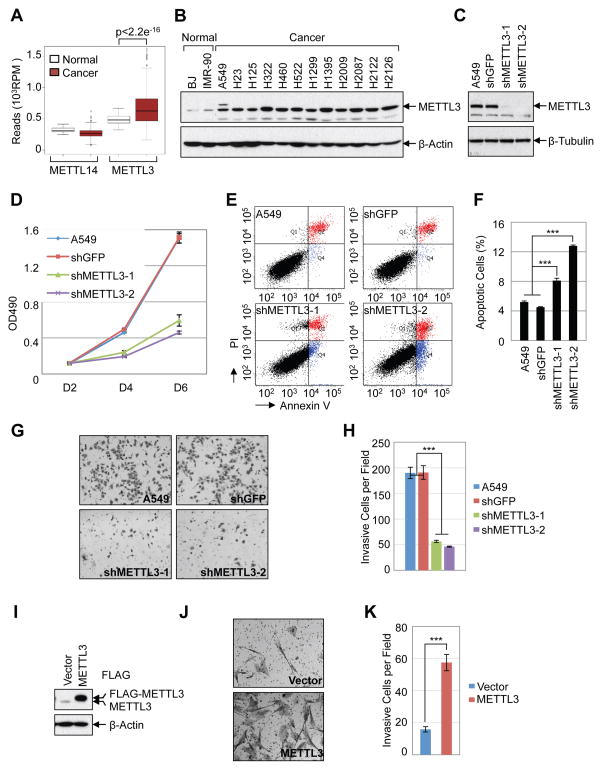
Considering our findings that METTL3 promotes translation and expression of several important oncogenes we were motivated to explore the role of METTL3 in cancer. We first examined the expression of METTL3 in the Cancer Genome Atlas (TCGA) datasets http://cancergenome.nih.gov. We found that METTL3 mRNA expression is significantly elevated in lung adenocarcinoma (LUAD) and colon adenocarcinoma (COAD) compared with the normal tissues (Figure 6A, Supplemental Figure 5A). In contrast, expression of the alternative methyltransferase METTL14 was not elevated in these patient samples. We further determined the relative levels of METTL3 protein in human lung cancer cell lines by Western blot. We found that the expression of METTL3 is elevated in lung adenocarcinoma cancer cell lines compared to the non-transformed human fibroblasts cell lines BJ and IMR-90 (Figure 6B). Next we studied the function of METTL3 in regulation of cancer cell survival using shRNA-mediated stable knockdown in lung cancer cells (A549, H1299, H1792) and HeLa cells (Figure 6C, Supplemental Figure 5B). METTL3 depletion results in strong inhibition of cancer cell growth (Figure 6D and Supplemental Figure 5C), increases cell apoptosis (Figure 6E–F, and significantly decreases the invasive ability of cancer cells (Figure 6G–H, Supplemental Figure 5D–E). Conversely, METTL3 overexpression in human fibroblasts promotes cell invasion (Figure 6I–K, Supplemental Figure 5F–H). Overall, our data support an important role of METTL3 in controlling cancer cell growth, survival and invasion.
Figure 6. METTL3 promotes cancer cell growth, survival, and invasion.
(A) Expression of METTL3 and METTL14 in normal tissue (n=58) and lung andenocarcinoma (n=513) from TCGA-LUAD dataset. (B) Western blot analysis of METTL3 expression in normal human fibroblasts and lung adenocarcinoma cell lines. (C–H) Knockdown of METTL3 regulates cellular proliferation, survival and invasion of A549 cells. (C) Westen blot shows stable knockdown of METTL3 by shRNAs. (D) MTS assay of cellular proliferation in A549 cells. (E) Annexin V/PI staining of METTL3 knockdown and control A549 cells by FACS. (F) Quantification of apoptotic cells, numbers represent the sum of early and late apoptotic cells. (G) In vitro cell invasion assay. (H) Quantification of invasive cells. (I–K) METTL3 overexpression regulates cell invasion in IMR-90 cells. (I) Western blot. (J) In vitro cell invasion assay. (K) Quantification of invasive IMR-90 cells. Data are presented as ±SEM. n=3. **p<0.01, ***p<0.001. See also Figure S5.
DISCUSSION
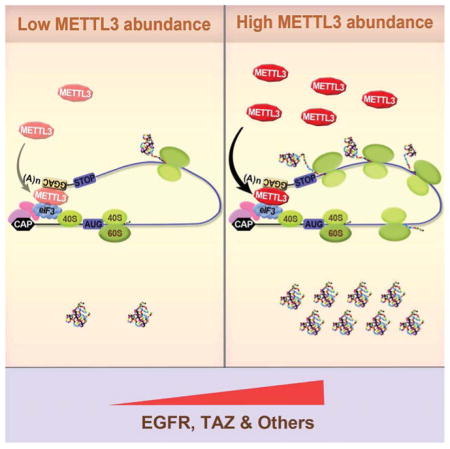
Our results uncover a direct role for METTL3 in promoting translation. This deduction is based on the following. First, we observe that METTL3 depletion leads to decreased protein expression without substantial changes in mRNA abundance. Second, we find that METTL3 associates with ribosomes and its depletion impacts polysome profiles and leads to decreased translation of target mRNAs. Third, tethering METTL3 to a luciferase mRNA enhances translation independently of methyltransferase activity, known co-factors, or downstream m6A reader proteins. Fourth, METTL3 interacts with translation initiation factors in an RNA-independent manner. Fifth, METTL3 knockdown inhibits the recruitment of eIF3 to both the CBP80- and eIF4E-cap binding proteins. Sixth, METTL3 specifically promotes translation of initiation factor-dependent reporter mRNAs. We also reveal that cancer cells exploit this pathway for elevated expression of known oncoproteins for increased cell proliferation, survival, and invasion. Indeed, an earlier study reported an ~8-fold increased m6A methyltransferase activity in nuclear extracts prepared from transformed cells compared to non-transformed cells (Tuck et al., 1996). Interestingly, variants in the FTO demethylase have been identified by human genome wide association studies (GWAS) to be linked with breast cancer and melanoma progression, thereby implicating altered m6A homeostasis with cancer risk (Garcia-Closas et al., 2013; Iles et al., 2013).
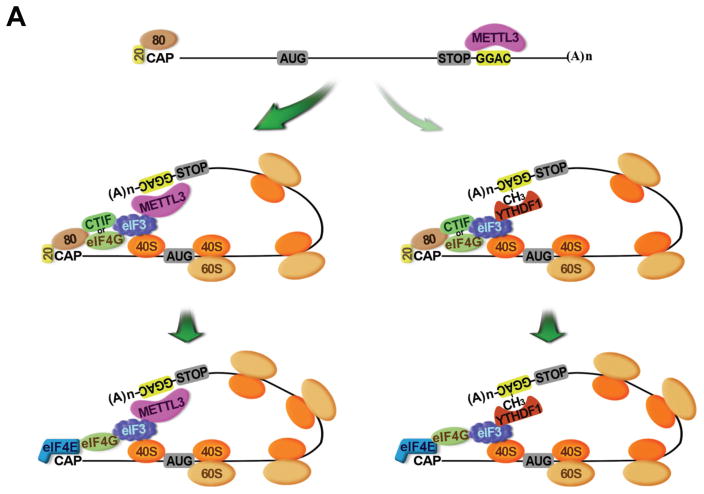
Our demonstration that METTL3 localizes to polysomes is consistent with earlier work that detected methyltransferase activity in crude ribosome extract but not in ribosome-depleted cytoplasmic extract (Tuck, 1992). Our results are further supported by biochemical experiments performed using a rabbit reticulocyte translation system where it was demonstrated that translation of the in vitro transcribed Dihydrofolate reductase (DHFR) mRNA was enhanced by incubation with partially purified METTL3 from HeLa cells (Heilman et al., 1996). Although m6A has been recently implicated in regulating translation through the binding of YTHDF1, we find that METTL3 promotes EGFR and TAZ protein expression in a YTHDF1-independent manner (Wang et al., 2015). We propose a model in which METTL3 enhances both CBP80/20- and eIF4E-dependent translation by binding to a specific subset of mRNAs and helping to recruit eIF3 to translation initiation complexes. A separate pathway involving the remodeling of mRNA-ribonucleoprotein complexes, replacement of METTL3, and binding of YTHDF1 to the m6A modified sites might promote translation of a distinct subset of mRNA targets (Figure 7). Most recently, two papers reported that dynamic m6A modification in the 5′ UTR of certain mRNAs promotes stress-induced translation (Meyer et al., 2015; Zhou et al., 2015). m6A in the 5′ UTR of Hsp70 mRNA was found to be induced by stresses including heat shock. This stress-induced m6A helps recruit eIF3 to promote Hsp70 translation in a cap-independent manner (Meyer et al., 2015; Zhou et al., 2015). Our results are clearly distinct from these since we found that: METTL3 regulates translation of specific mRNAs containing m6A peaks near the stop codon (and not in the 5′ UTR, Supplementary Figure 1G), METTL3 promotes translation of these mRNAs under normal cell culture conditions, METTL3 physically and functionally associates with translation initiation factors, and METTL3 m6A methyltransferase activity can be experimentally uncoupled from its role in promoting translation. Our results provide therefore an alternative model and independent pathway for the role of METTL3 in translational control.
Figure 7. Model of the role of METTL3 in promoting translation initiation of target mRNAs.
AUG, translation initiation codon; CAP, Cap; 20, CBP20; 80, CBP80; STOP, translation stop codon; CH3, m6A; (A)n, poly(A) tail; 40S, 40S ribosomal subunit; 60S, 60S ribosomal subunit.
Since we found that METTL3 does not regulate the translation of all the mRNAs containing m6A peaks that we tested it will be interesting to explore the global effects of METTL3 on translation and to determine the molecular determinants conferring mRNA specificity. Genome-wide profiling of METTL3 binding sites using PAR-CLIP revealed that METTL3 is only bound to about 22% of the methylated GGAC sites detected by meRIP-Seq (Liu et al., 2014), suggesting that METTL3 might display selectively towards binding and translational regulation of a specific subset of mRNAs. In addition, It’s worth mentioning that though in this study we reported that METTL3 promotes translation of a subset of mRNAs that have m6A peaks near the stop codon (Figure 7), METTL3 might also bind to other regions (5′ UTR, CDS) of other mRNAs to promote translation, given that there are also significant amount of METTL3 binding sites located in 5′UTR and CDS regions of mRNA (Liu et al., 2014), Furthermore, while our tethering results experimentally uncouple the m6A catalytic activity of METTL3 from its role in promoting translation, it is likely that m6A normally plays a role in the recruitment or ‘tethering’ of METTL3 complexes to a subset of target mRNAs. It will be interesting to explore whether METTL3 itself or possibly unknown METTL3-complex components can function as m6A-reader proteins to help ‘tether’ METTL3 to specific sites in the mRNA targets. Future experiments combining depletion of METTL3 and its associated factors, PAR-CLIP, ribosome footprinting, and/or quantitative translation initiation sequencing (QTI-seq) (Gao et al., 2015) will provide more insight into the selectivity of METTL3 in regulating m6A and translation. Also, considering our data demonstrating that METTL3-depletion suppresses cancer cell viability future efforts aimed at inhibiting this pathway could lead to the development of new cancer therapies.
EXPERIMENTAL PROCEDURES
Cell culture and cell lines
Human lung cancer cell lines (A549, H1299, H1792), HeLa, and HEK293T cells were cultured with DMEM supplemented with 10% FBS and antibiotics. Human normal skin fibroblast cell line BJ and normal lung fibroblast cell line IMR-90 were cultured in RPMI-1640 medium with 15% FBS and antibiotics. Cells were grown in a 5% CO2 cell culture incubator at 37 °C.
Plasmids and molecular cloning
Human METTL3 and YTHDF1 cDNA was generated by PCR and cloned into NotI and BglII sites of pFLAG-CMV2 expression plasmid. Q5® Site-Directed Mutagenesis Kit (NEB) was used to generate the METTL3 catalytic mutant (aa395–398, DPPW → APPA) (Alarcon et al., 2015b). The FLAG-METTL3 fragment was subcloned into the NheI and SwaI sites of pCDH lentiviral vector for stable over-expression. MS2 ORF was PCR amplified and cloned into the HindIII site of pFLAG-CMV2-METTL3 to generate the MS2-METTL3 expression plasmid. The pGL3c_TK luciferase reporter containing two MS binding sites (FLuc-MS2bs), pFlag-MS2, and pFlag-MS2-GW182 C-term plasmids are generated as previously described (Chang et al., 2012). pcDNA3-FLAG-CBP80 (Oh et al., 2007) and pcDNA3-FLAG-eIF4E(Kim et al., 2009) were described previously. Bicistronic reporter pR/EMCV/F and pR/CrPV/F were kindly provided by Y.K. Kim. 2x MS2 binding site was PCR amplified from the FLuc-MS2bs (Chang et al., 2012) and cloned into the NotI sites of pR/EMCV/F and pR/CrPV/F. pCMV6-Myc-DDK-FTO expression plasmid was purchased from Origene. The pLKO.1 lentiviral shRNA constructs targeting the human METTL3 (shMETTL3-1: RHS3979-201764032, shMETTL3-2: RHS3979-201764033) and GFP (RHS4459) were purchased from Dharmacon. All cloning primers are listed in Supplemental Table 2.
Transfection and siRNA knockdown
Transfection of plasmids was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transient knockdown of target genes by siRNA was performed with Lipofectamine RNAi Max (Invitrogen). The previously confirmed siRNA targeting sequences (Liu et al., 2014; Wang et al., 2014b; Wang et al., 2015) used in this study are: siMETTL3: CTGCAAGTATGTTCACTATGA; siMETTL14: AAGGATGAGTTAATAGCTAAA; siWTAP AAGCTTTGGAGGGCAAGTACA; siYTHDF1: CCGCGTCTAGTTGTTCATGAA; siYTHDF2: AAGGACGTTCCCAATAGCCAA.
Virus production and transduction
For shRNA knockdown, pLKO.1 constructs together with packing and helper plasmids, pLP1, pLP2 and VSVG, were co-transfected into 293T cells. For over-expression, pCDH lentiviral vectors together with delta 8.9 and VSVG were transfected into 293T cells. Viruses were collected at 48 hours and 72 hours after transfection and then used to infect cells with Polybrene (8 μg/ml, Sigma). 48 hours after infection, Puromycin was added to the culture medium to select the infected cells.
Polysome fractionation
Polysome fractionations were performed as described previously (Kim et al., 2009). Briefly, METTL3 depleted or control HeLa cells (four 150-mm culture dishes) were treated with 100 ug/mL cycloheximide (Sigma) for 10 min at 37°C. The n, cells were harvested and 1 mL of cytoplasmic extract was layered onto 11 mL of 10%–50% sucrose gradient and centrifuged at 36,000 rpm in a Beckman SW-41Ti rotor for 2.5h at 4°C. Gradients were fractionated and monitored at absorbance 254nm (Brandel). Collected fractions were then analyzed by western blotting or RT-PCR.
RNA isolation and RT-PCR
Total RNA was extracted from cells or polysome fractions using Trizol (Invitrogen) following the manufacturer’s instructions. 2 μg total RNA was used for reverse transcription using SuperScript® III Reverse Transcriptase (Invitrogen). Quantitative RT–PCR was performed using SYBR® Green PCR Master Mix with the StepOne Real-Time PCR System (Applied Biosystems). Beta-Actin was used as an internal control for the normalization. For RNA samples from polysome fractions, RT–PCRs were carried out using specific oligonucleotides and α-[32P]-dCTP (PerkinElmer) to determine the distributions of target RNAs in the polysome fractions (Kim et al., 2009). All primers used in this study were listed in Supplemental Table 2.
Cell fractionation, co-immunoprecipitation, and Western blot
To separate the cell cytoplasmic and nuclear fractions, cells were first lyzed with low salt buffer (20 mM Hepes pH 7.9, 10% Glycerol, 1.5 mM MgCl2, 0.05 % NP40, protease inhibitors) on ice for 5 minutes and then centrifuged at 4000rpm for 5 minutes at 4°C. Supernatant was collected as cytoplasmic fractions, the pellet was then lyzed with high salt buffer (20 mM Hepes pH 7.9, 10% Glycerol, 1.5 mM MgCl2, 500mM NaCl, 0.05 % NP40, protease inhibitors) to collect the nuclear fractions. Co-immunoprecipitation and Western blot were performed as previously described (Wang et al., 2014a). The following antibodies were used for Western blotting: METTL3 (Proteintech,15073-1-AP); β-Tubulin (Abcam, ab6046); EGFR (Cell Signaling Technology, #2232); TAZ (BD Pharmingen, 560235); MAPKAPK2 (Cell Signaling Technology, #3042); DNMT3A (Abcam, ab13888); β-Actin (Abcam, ab119716); Fibrillarin (Abcam, ab4566); CBP80 (Choe et al., 2012); CTIF (Choe et al., 2012); eIF4E (Cell signaling technology, #2067); eIF3b (Santa Cruz Biotechnology,sc-16377); eIF4AI (Abcam, ab31217); eIF4GI (Cell signaling technology, #2498); FLAG (Sigma, A8592); METTL14 (Sigma, HPA038002); WTAP (Proteintech Group 60188-1-Ig); YTHDF1 (Abcam, ab99080); YTHDF2 (Proteintech, 24744-1-AP).
m6A meRIP-Seq and total RNA cloning and deep sequencing
meRIP-Seq and data analysis were performed as previously described with minor modifications (Dominissini et al., 2013). Briefly, mRNA was first purified from total RNA using PolyATtract® mRNA Isolation Systems (Promega). Then 5 μg mRNA was fragmented and immunoprecipated with anti-m6A antibody (Synaptic Systems, 202003), the immunoprecipated RNA was washed and eluted by competition with N6-Methyladenosine (Sigma-Aldrich, M2780). The purified RNA fragments from m6A meRIP were used for library construction using TruSeq Stranded mRNA Sample Prep Kits (Illumina RS-122-2101) and sequenced with Illumina HiSeq 2000. Reads mapping, m6A peak calling and motif search were performed as described (Dominissini et al., 2013). Analysis of the published PAR-CLIP was performed as previously described (Liu et al., 2014). For deep sequence analysis of RNA expression, 2 μg total RNA was used for the library construction using TruSeq Stranded mRNA Sample Prep Kits (Illumina RS-122-2101) according to the manufacturer’s instructions.
Metabolic labeling
A549 cells were washed twice with PBS and then incubated in methionine- and cysteine-free DMEM (Gibco) medium for 2 hours. Cells were then incubated for 20 min after supplementation with [35S]-methionine ([35S]-Met; 100 mCi/mL; PerkinElmer), washed twice with ice-cold PBS, harvested, and lysed. For the quantitation of [35S]-Met-labeled proteins, cell lysates were subjected to 4–12% Tris-Glycine SDS-PAGE and analyzed by autoradiography. Colloidal blue staining was performed to verify equal amounts of protein loaded.
Tethering assays
Tethering assays were performed as previously described(Chang et al., 2012). Briefly, a firefly luciferase reporter FLuc-MS2bs or bicistronic reporter R/EMCV/F-MS2bs and R/CrPV/F-MS2bs with MS2 binding sites in the 3′ UTR, and a plasmid expressing either MS2 protein or MS2-fusion were transfected with Lipofetamine 2000 (Invitrogen). 48 hours after transfection, the luciferase activities were measured with the dual-luciferase assay kit (Promega). Firefly/Renilla ratios were calculated to determine the roles of MS2-fusion proteins in regulation of luciferase translation.
Cell proliferation, apoptosis, and invasion assays
Cell proliferation, apoptosis, and invasion assays were performed as described previously (Zhang et al., 2014). Briefly, for cell proliferation, 1000 cells were seeded in 96-well plate on day 0. Absorbances at 490nm were measured using CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega) on day 2, day 4 and day 6 to measure the cellular proliferation. The numbers of apoptotic cells were quantified by flow cytometric assays using Annexin V-FITC Apoptosis Detection Kit (BioVision). Cell invasion was measured using BioCoat™ Matrigel™ Invasion Chamber (Corning) following manufacturer’s instructions.
Statistical analysis
Data from q.RT-PCR, apoptosis and invasion were analyzed using Student’s t-test. p<0.05 was considered as statistically significant.
Supplementary Material
Highlights.
METTL3 promotes the translation of important oncogenes such as EGFR and TAZ
METTL3 promotes translation independent of its catalytic activity and m6A readers
METTL3 interacts with translation initiation machinery to enhance translation
METTL3 is required for the growth, survival, and invasion of cancer cells
Acknowledgments
We thank Ronald Mathieu (Boston Children’s Hospital) for help with flow cytometry. S.L. is a Damon Runyon-Sohn Pediatric Fellow supported by the Damon Runyon Cancer Research Foundation (DRSG-7–13). R.I.G was supported by a grant from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386) and a pilot grant from NIGMS (5P01GM099117).
Footnotes
AUTHOR CONTRIBUTIONS
S.L. designed and performed most of the experiments in Figures 1, 6 and part of Figures 2, 5. J.C. designed and performed most of the experiments in Figures 3, 4 and part of Figures 2, 5. P.D. performed all bioinformatic analysis. R.T. cloned the METTL3 human cDNA. S.L., J.C., and R.I.G. analyzed data and wrote the paper with input from P.D. and R.T.
The authors declare no competing financial interests.
ACCESSION NUMEBR
The raw and analyzed deep sequencing data have been submitted to NCBI Gene Expression Omnibus (accession number: GSE76367).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015a;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6- methyladenosine marks primary microRNAs for processing. Nature. 2015b;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6- adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Choe J, Oh N, Park S, Lee YK, Song OK, Locker N, Chi SG, Kim YK. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J Biol Chem. 2012;287:18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. Quantitative profiling of initiating ribosomes in vivo. Nature methods. 2015;12:147–153. doi: 10.1038/nmeth.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. 398e391–392. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilman KL, Leach RA, Tuck MT. Internal 6-methyladenine residues increase the in vitro translation efficiency of dihydrofolate reductase messenger RNA. Int J Biochem Cell Biol. 1996;28:823–829. doi: 10.1016/1357-2725(96)00014-3. [DOI] [PubMed] [Google Scholar]
- Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc Natl Acad Sci U S A. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles MM, Law MH, Stacey SN, Han J, Fang S, Pfeiffer R, Harland M, Macgregor S, Taylor JC, Aben KK, et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45:428–432. 432e421. doi: 10.1038/ng.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20- dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24:1490–1492. doi: 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gregory RI. Methyltransferases modulate RNA stability in embryonic stem cells. Nature cell biology. 2014;16:129–131. doi: 10.1038/ncb2914. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosinedependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh N, Kim KM, Cho H, Choe J, Kim YK. Pioneer round of translation occurs during serum starvation. Biochem Biophys Res Commun. 2007;362:145–151. doi: 10.1016/j.bbrc.2007.07.169. [DOI] [PubMed] [Google Scholar]
- Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5′ terminus. Cell. 1975;4:387–394. doi: 10.1016/0092-8674(75)90159-2. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6- methyladenosine methyltransferase. Cell research. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem J. 1992;288(Pt 1):233–240. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MT, James CB, Kelder B, Kopchick JJ. Elevation of internal 6- methyladenine mRNA methyltransferase activity after cellular transformation. Cancer Lett. 1996;103:107–113. doi: 10.1016/0304-3835(96)04203-6. [DOI] [PubMed] [Google Scholar]
- Wang LN, Wang Y, Lu Y, Yin ZF, Zhang YH, Aslanidi GV, Srivastava A, Ling CQ, Ling C. Pristimerin enhances recombinant adeno-associated virus vectormediated transgene expression in human cell lines in vitro and murine hepatocytes in vivo. Journal of integrative medicine. 2014a;12:20–34. doi: 10.1016/S2095-4964(14)60003-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014b;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6- methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology. 2014c;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in posttranscriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YH, Wang Y, Yusufali AH, Ashby F, Zhang D, Yin ZF, Aslanidi GV, Srivastava A, Ling CQ, Ling C. Cytotoxic genes from traditional Chinese medicine inhibit tumor growth both in vitro and in vivo. Journal of integrative medicine. 2014;12:483–494. doi: 10.1016/s2095-4964(14)60057-1. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014;24:1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.