Abstract
Meiotic clade AAA ATPases, which were initially grouped on the basis of phylogenetic classification of their AAA ATPase cassette, include four relatively well characterized family members, Vps4, spastin, katanin, and fidgetin. These enzymes all function to disassemble specific polymeric protein structures, with Vps4 disassembling the ESCRT-III polymers that are central to the many membrane-remodeling activities of the ESCRT pathway, and spastin, katanin p60 and fidgetin affecting multiple aspects of cellular dynamics by severing microtubules. They share a common domain architecture that features an N-terminal MIT domain followed by a single AAA ATPase cassette. Meiotic clade AAA ATPases function as hexamers that can cycle between the active assembly and inactive monomers/dimers in a regulated process, and appear to disassemble their polymeric substrates by translocating subunits through the central pore of their hexameric ring. Recent studies with Vps4 have shown that nucleotide-induced asymmetry is a requirement for substrate binding to the pore loops, and that recruitment to the protein lattice via MIT domains also relieves auto-inhibition and primes the AAA ATPase cassettes for substrate binding. The most striking, unifying feature of meiotic clade AAA ATPases may be their MIT domain, which is a module that is found in a wide variety of proteins that localize to ESCRT-III polymers. Spastin also displays an adjacent microtubule-binding sequence, and the presence of both ESCRT-III and microtubule binding elements may underlie the recent findings that the ESCRT-III disassembly function of Vps4 and the microtubule-severing function of spastin, and potentially katanin and fidgetin, are highly coordinated.
Keywords: ESCRT, microtubules, Vps4, Spastin, Enzyme mechanism
Graphical abstract
Introduction
ATPases associated with diverse cellular activities (AAA+) are mechanoenzymes that convert chemical energy from ATP hydrolysis into mechanical force that is used to remodel either nucleic acid or protein substrates1. They are characterized by the presence of one or more AAA ATPase cassettes, which comprise a large N-terminal domain that adopts an Additional Strand Conserved E (ASCE) fold and a small helical domain at the C-terminus, the presence of which distinguishes AAA+ proteins from other P-loop NTPases2; 3. The large AAA ATPase domain contains all of the determinants for ATP binding and catalysis4. In addition to the P-loop, which is also referred to as the Walker A motif and contains residues that coordinate β- and γ-phosphates of nucleotides, the large AAA ATPase domain also contains the conserved Walker B motif and sensor-1 residues that function in the binding and hydrolysis of nucleotides and in the sensing of the nucleotide state. AAA+ ATPases function as higher-order oligomers, typically hexamers, that bind ATP at the interface between subunits, where a conserved arginine side chain (arginine finger) from the adjacent protomer complements the active site5.
Within the AAA+ superfamily, members of the AAA family are distinguished by a conserved sequence motif called the second region of homology (SRH)5, which includes the arginine “finger” that senses changes in coordination at the ATP/ADP-binding site. Variations in the SRH, specifically the position of an additional conserved arginine residue relative to the arginine finger, allow further classification into clades. The ‘meiotic clade’ AAA ATPases3 were defined as having the additional arginine directly preceding the arginine finger, and were named based on the meiotic functions that were known for mei-1 and mei-26, the C. elegans homologs of katanin p60 and p80. In addition to katanin p60, the best-characterized clade members are Vps4 and spastin, and to a lesser extent fidgetin. These enzymes function in far more than meiosis, and drive a wide range of biological pathways through their ability to disassemble or sever polymeric protein substrates, which are microtubules in the case of katanin, spastin, and fidgetin, and are ESCRT-III polymers in the case of Vps4. Strikingly, all of these enzymes, with the possible exception of fidgetin, display an N-terminal Microtubule Interacting and Trafficking (MIT) domain. MIT domains do not appear to be present in other AAA+ ATPases, although they are found in many other proteins where they mediate recruitment to ESCRT-III polymers by binding sequences termed MIT interacting motifs (MIM)7 near the C-termini of ESCRT-III subunits. This commonality may coordinate the distinct biochemical activities of Vps4 and the microtubule-severing enzymes by jointly recruiting them to ESCRT-III polymers8. In this review we describe the available structural insights and the mechanistic and functional implications for the meiotic clade AAA ATPases Vps4, spastin, and katanin.
Meiotic AAA ATPase isoforms
The microtubule severing meiotic clade AAA ATPases are found throughout the animal kingdom and in plants and protozoa, whereas Vps4 is also present in yeast and in some but not all archaeal phyla. While members of the meiotic clade of AAA ATPases all contain an N-terminal MIT domain and a single AAA ATPase cassette at the C-terminus (Figure 1), each member has unique features, and some exist in multiple isoforms in higher eukaryotes.
Figure 1. Domain structures of meiotic AAA ATPases and their binding partners.
Domains for which some structural information is available are color coded and labeled. MIT domain, cyan; large AAA ATPase domain (AAA-L), light blue; small AAA ATPase domain (-S), dark blue; β-domain, magenta; C-terminal helix (C), green; MTBD, dark red. Spastin isoform variants hydrophobic domain (HD) and exon 4 (4). The Vps4 binding partner LIP5/Vta1, wheat. The katanin p60 binding partner katanin p80 and WD40-less p80, white.
Spastin exists in four isoforms in mammalian cells8, with the full-length, canonical protein comprising 616 amino acid residues and referred to as M1 spastin. Alternative promoter use produces M87 spastin9; 10, which lacks the first 86 residues and is the most abundant isoform. Alternative splicing of both M1 and M87 spastin produces the other two isoforms8. The first 86 residues of full-length M1 spastin contain a hydrophobic domain that may have the ability to insert into membranes as well as binding sites for proteins that determine localization to centrosomes or the ER. Thus, the M1 and M87 variants differ in their subcellular localization, with M87 spastin, in which these motifs are missing, remaining in a cytoplasmic pool. Each of the four spastin isoforms contains the MIT domain11, which can mediate recruitment to different cellular locations, as well as nuclear localization signals12. A stretch of ~60 residues after the MIT domain and immediately N-terminal to the AAA ATPase cassette of all isoforms is necessary and sufficient for microtubule binding, and is referred to as the microtubule binding domain (MTBD)13.
Katanin is a heterodimer comprising the unrelated p60 and p80 subunits14, with only p60 displaying the MIT domain15 and AAA ATPase cassette16. Katanin p80 does not sever microtubules on its own, but enhances the in vitro ATPase and microtubule-severing activity of p60 by a factor of two16, which suggests that it might stabilize the active conformation of katanin p60. Katanin p80 contains a series of WD40 domains that appear to play a role in localization16, but are not required for the interaction with the p60 subunit, which is mediated by the C-terminal half of the protein. The katanin p80 B1-like protein displays sequence similarity to katanin p80, but lacks the WD40 repeats17. Katanin-like 1 and 2 are distinct proteins that are closely related to katanin p60 but as yet have not been extensively characterized18; 19; 20.
Fidgetin is less characterized than spastin or katanin, but clearly has the ability to sever microtubules21. The presence of an MIT domain seems probable17, but verification will be important because MIT domain sequences are highly variable and can be missed in the absence of structure determination. Fidgetin has two known paralogs in human, known as fidgetin-like (FIGL) 1 and 222.
Mammalian cells possess two highly similar isoforms of Vps4, VPS4A and VPS4B23, whereas a single isoform is found in yeast and archaea. Eukaryotic Vps4 proteins, but not the archaeal homolog or other members of the meiotic clade, have a unique insertion to the AAA ATPase cassette termed the β-domain, which binds the Vps4 activator, LIP5/Vta124; 25. VPS4A and VPS4B can form heteromeric complexes23, but at least in the context of HIV release (see below), the budding phenotype observed upon depletion of both isoforms can be rescued by overexpressing only VPS4B26, indicating that the isoforms may function independently of each other. Because their MIT domains differ in their binding affinity for different ESCRT-III subunits26; 27, it seems possible that VPS4A and VPS4B may function in different branches of the ESCRT pathway.
Substrates and Biological Functions
The microtubule substrates of spastin, katanin, and fidgetin are cytoskeleton components that define intracellular structure and polarity, and organize information flow throughout the eukaryotic cell by comprising the tracks along which a wide variety of cargo can be transported28; 29; 30, including protein or RNA molecules, organelles and vesicles. Microtubules are also central to major structural transitions and motility, for example in the separation of anaphase chromosomes31, and in ciliary and flagellar motility32; 33. They consist of polymers of α- and β-tubulin heterodimers34; 35 (Figure 2A), which display highly similar structures with a two-part β-sheet that comprises six parallel strands with Rossmann fold topology and an additional 4-stranded mixed β-sheet, with a total of 12 α-helices flanking both sides of the β-sheet. Both subunits bind a GTP molecule at the usual site for a Rossmann fold. Heterodimers of α- and β-tubulin interact end-to-end to form filaments that further assemble as hollow tubes of 13 protofilaments with structurally distinct ends (Figure 2B). The minus-end, where α-tubulin is exposed, is often bound to the γ-tubulin ring complex, which can nucleate microtubule assembly and anchors microtubules to the centrosome or other microtubule organizing centers36, from where microtubules typically grow toward the periphery of the cell.
Figure 2. Substrates of meiotic clade AAA ATPases.
(a) Structure of the heterodimer of GTP-bound α-tubulin (green/cyan) and GDP-bound β-tubulin (pink/magenta). The unstructured C-terminal tails that are engaged by the pore loops of spastin and katanin are colored red. (b) Schematic of the microtubule structure. Growing microtubules add GTP-bound tubulin heterodimers (green/blue) to their plus end. Following assembly, β-tubulin hydrolyses ATP to GDP (green/pink). C-terminal tails are shown for some representative subunits. (c) Crystal structure of CHMP3 (PDB ID: 3FRT)70 showing the canonical four helix bundle of the ESCRT-III fold (green). Sequences from helix 5 (red) are preferentially engaged by Vps4 pore loops115. The MIM motif is part of the unstructured C-terminal tail and is shown schematically. (d) ESCRT-III assembles into membrane-bound helical polymers. MIM motifs are exposed on the surface, where they are available to recruit MIT-domain containing proteins, including Vps4.
As cells change and adapt, microtubules are continually being remodeled by a dynamic process of assembly and disassembly that is governed by multiple regulatory processes. The GTP hydrolysis state has a substantial effect on microtubule dynamics37; 38; 39. Both the α- and the β-tubulin subunits are bound with GTP during polymerization. Assembly stimulates GTP-hydrolysis in β-tubulin, which destabilizes the polymer. Consequently, hydrolysis of the GTP in β-tubulin subunits that cap the plus-end of the microtubule can trigger rapid depolymerization. Additional microtubule regulation is provided by a plethora of proteins, including those that associate specifically with the plus-end40 or minus-end41, and the microtubule-severing enzymes katanin, spastin and fidgetin14; 16; 21; 42; 43; 44; 45. This severing activity is possible because α- and β-tubulin display flexible C-terminal tails of ~20 residues that project from the sides of microtubules35 and are accessible for spastin and katanin to bind and translocate in order to destabilize the tubulin polymer14; 43; 46. Microtubule severing can generate seeds for new microtubule growth, remove branching, induce microtubule bundling, or induce rapid depolymerization due to the removal of stabilizing plus-end capping proteins17; 47. The microtubule-severing enzymes thereby perform critical functions in many if not all of the biological processes that depend upon microtubules, including meiosis and mitosis, where they regulate spindle assembly and chromatid separation, cytokinetic abscission, ciliogenesis, cell migration, endosomal tubulation and neuronal morphogenesis (reviewed in17; 47).
The other well-characterized member of the meiotic clade, Vps4, functions to disassemble polymers of ESCRT-III proteins rather than microtubules. Adaptor proteins of the ESCRT (Endosomal Sorting Complexes Required for Transport) pathway recruit cargo and ESCRT-III subunits, which subsequently polymerize to constrict membrane necks followed by a fission event48; 49; 50. Thus far, all of the reported ESCRT pathway-driven processes are “reverse topology” membrane changes, in which membranes are drawn toward the cytoplasmic face50. The most ancient pathway catalyzed by the ESCRT system appears to be the final abscission step of cytokinesis, because this pathway also operates in some archaeal phyla51; 52; 53. The ESCRT pathway drives many additional processes that involve equivalent membrane topology changes in eukaryotic cells, including budding of intraluminal vesicles into the endosome to form multivesicular bodies (MVB) that are then targeted for lysosomal degradation54, exosome release55; 56; 57, shedding of microvesicles58; 59, plasma membrane wound repair60, pruning of axons and dendrites61, clearance of off-pathway nuclear pore complex assembly intermediates62, and nuclear envelope sealing during mitotic spindle disassembly63; 64. HIV and many other viruses exploit the ESCRT pathway to facilitate budding from the plasma membrane or to downregulate host proteins via the endolysosomal pathway65; 66; 67; 68; 69.
The human family of ESCRT-III proteins has 12 known members that include IST1 and charged multivesicular body proteins (CHMPs, also known as chromatin modifying proteins)50, that are grouped into subfamilies CHMP1-7, with two isoforms A and B of CHMP1 and CHMP2, respectively, and three isoforms, A, B and C, of CHMP4. ESCRT-III proteins share a common structure comprised of an N-terminal core and a C-terminal tail (Figure 2C)70. The core forms a four-helix bundle structure in which the first two helices form a long helical hairpin and the third and fourth helices form a short helical hairpin. A fifth helix packs against the long hairpin near its tip. ESCRT-III proteins exist in an inactive, unassembled state, that upon activation appears to undergo a conformational change that results in polymerization71. Specific pairs of ESCRT-III subunits are thought to assemble as double-helices that can coil into spiral domed structures50 (Figure 2D) that bind, distort and constrict the membrane72; 73; 74, with different types of ESCRT-III subunits predominating in distinct pathways68. The C-terminal residues of ESCRT-III subunits contain MIM sequences that are inherently flexible and accessible on the polymer surface, and serve as a signaling hub by binding to a variety of proteins that possess MIT domains, including deubiquitylases, kinases and proteases7. Among the MIT-containing proteins recruited to ESCRT-III polymers is Vps4, which subsequently catalyzes disassembly of the polymer to regenerate free soluble ESCRT-III subunits that become available to mediate subsequent rounds of membrane fission75. Vps4 recruitment occurs shortly before budding, leading to the suggestion that its activity might contribute directly to the fission reaction, for example by remodeling the ESCRT-III polymer to allow a more constricted configuration76; 77; 78. In the absence of Vps4 or upon expression of a dominant negative mutant, membrane fission is inhibited and bud necks cannot be resolved26; 79. Several models have been proposed to explain how ESCRT-III and Vps4 accomplish membrane fission50, and determining the actual mechanism employed in the different ESCRT pathways is a major priority.
ESCRT-III polymers can also recruit spastin, whose MIT domain binds the CHMP1B MIM and is thereby localized to the midbody during cell division80. Disrupting this interaction will cause a delay in cytokinetic abscission. Thus, the ESCRT-III assembly that functions in the final membrane scission step of cytokinesis, also recruits the enzyme that performs the penultimate step of severing the dense array of microtubule bundles that derive from the mitotic spindle and traverse the midbody8; 81. Recent studies have shown that the ESCRT machinery and spastin also cooperate at an earlier stage of eukaryotic cell division, during late anaphase, when reformation of nuclear envelopes is coordinated with mitotic spindle disassembly63, a context in which spastin is recruited by IST1. Similarly, it has recently been established that spastin and the ESCRT pathway both function in endosomal tubulation82. Interestingly, in all these processes, spastin recruitment is independent of microtubule binding and depends solely on the MIM-MIT interaction. Recruitment to the ESCRT-III lattice has not yet been reported for katanin or fidgetin, although the presence of an MIT domain in katanin and probably also in fidgetin make this possibility an attractive hypothesis. It will be interesting to better understand the level of coordination between Vps4 and microtubule-severing enzyme recruitment and activities in these different pathways, with one possibility being that MIT domains have been retained in the microtubule-severing enzymes in order to allow their coordinated recruitment to ESCRT-III filaments.
The mechanism of recruiting the microtubule-severing enzymes specifically to microtubules is less clearly defined. NMR data suggest that there is a direct interaction between the katanin MIT domain and tubulin83, and the MIT domain of Drosophila melanogaster spastin has been reported to bind microtubules with low affinity43. The most definitive experiments indicate that spastin binds microtubules through the ~60 residue MTBD in the linker between the MIT domain and the AAA ATPase cassette. This region is both necessary and sufficient for microtubule binding of the human and C. elegans spastin homologs13; 84, although further characterization will be important because this sequence is poorly conserved. Thus, unraveling the mechanisms of recruitment to microtubules and colocalization to microtubules and ESCRT-III polymers are also important questions for future studies.
MIT domain structure and interactions
First identified in sorting nexin 1585, MIT domains have now been found in many proteins including meiotic clade AAA ATPases11; 15, kinases86; 87; 88; 89; 90, deubiquitylases (AMSH, UBPY)89; 91; 92 and proteases (Calpain-7)11. Although their designation as Microtubule Interacting and Trafficking domains implies interaction with microtubules, MIT domains are now primarily thought of as a domain that functions to target proteins to ESCRT-III polymers. Although, as noted above, their MIT domains may contribute to microtubule binding by the microtubule-severing AAA ATPases, it is clearly established that the MIT domain of spastin binds to the MIM of the ESCRT-III proteins CHMP1B80; 93 and IST163; 82; 86.
The structures of the MIT domains of VPS4A27, VPS4B94; 95, spastin80 and katanin83 are highly similar to each other (Figure 3A). The ~80 residue MIT domain comprises an antiparallel three-helix bundle in which helices 2 and 3 form a coiled-coil. The first helix packs against bulky aromatic/hydrophobic residues of the coiled-coil using a series of conserved, regularly spaced alanines. This unusual combination of a coiled-coil motif and an alanine zipper results in asymmetry of the overall structure, and provides distinct binding surfaces for the MIMs of the many different ESCRT-III subunits.
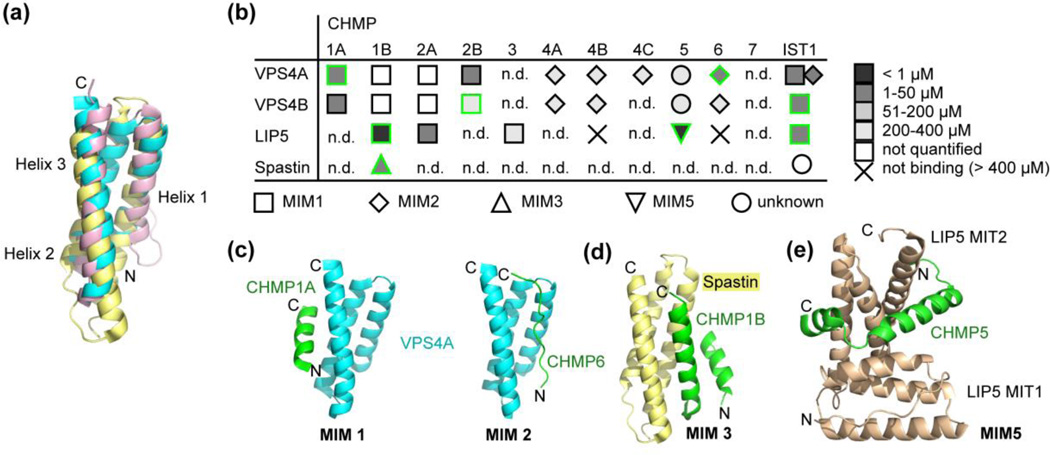
Figure 3. MIM-MIT interactions.
(a) Superposition of the MIT domains of human VPS4A (cyan, PDB ID: 2JQH)27, human spastin (pale yellow, PDB ID: 3EAB)80 and mouse katanin (rose, PDB ID: 2RPA)83. (b) Summary of known MIM-MIT interactions between human ESCRT-III proteins and the MIT domains of VPS4A, VPS4B, LIP5 and spastin26; 27; 79; 80; 86; 87; 93; 94; 96; 98; 142. The type of interaction is indicated by shape. The affinity is indicated by fill color. Green outlines denote interactions that have been characterized structurally. “n.d.”, not determined. No ESCRT-III interactions have been reported for katanin or fidgetin. (c) Structure of the human VPS4A MIT domain in complex with the CHMP1B MIM1 (left, PDB ID: 2JQH)94 and the CHMP6 MIM2 (right, PDB ID: 2K3W)26. (d) Structure of the human spastin MIT domain in complex with the CHMP1B MIM3 (right, PDB ID: 3EAB)80. (e) Structure of MIM5-type binding of the C-terminus of CHMP5 to the tandem MIT domain of LIP5 (PDB ID: 2LXM)96.
Despite the apparent simplicity of the MIT domain structures, their ability to bind the short MIM sequences of ESCRT-III subunits is remarkably versatile. At least five distinct binding modes have been identified for different pairs of MIT domains and MIM peptide motifs involved in the ESCRT pathway96, three of which have thus far been observed for the AAA ATPases (Figure 3B). MIM-MIT interactions have also been visualized between the autophagy-related proteins Atg1 and Atg1390. The MIT domains of both VPS4A and VPS4B can bind Type 1 or Type 2 MIMs26, with the two classes of MIM displaying very different binding geometry (Figure 3BC). MIM1 sequences are found in CHMP1-394 and IST187, and have been visualized in multiple MIT domain complexes (Figure 3B) to reveal a short amphipathic helix binding in the groove between MIT helices 2 and 394; 97; 98; 99. In contrast, MIM2 sequences, which have been identified in CHMP4, CHMP6 and IST1 (IST1 has both MIM1 and MIM2 sequences), bind to Vps4 MIT domains in an extended conformation that lines the groove formed by helices 1 and 326; 53; 100. An MIM2 sequence that can bind the Vps4 MIT domain has also been characterized in the S. cerevisiae protein Vfa1100, although it is not clear whether Vfa1 is a previously unrecognized ESCRT-III subunit. Because the MIM1 and MIM2 binding sites are on opposite sides of the MIT domain, both types of interactions can form at the same time, and simultaneous binding has been demonstrated for the MIM1 and MIM2 sequences of IST1 to the VPS4A MIT domain87. The MIT domain of spastin interacts with CHMP1B through a third binding mode80. Although the MIM3 sequence overlaps with the MIM1 residues that were previously visualized binding to the groove between helices 2 and 3 in other MIT domains, the MIM3 motif occupies the groove between helices 1 and 3 on the spastin MIT domain, where it binds as a long amphipathic helix that is preceded by a short helix that makes additional contacts with MIT helix 1. This finding illustrates that, in addition to the MIM sequences, determinants on the surface of MIT domains of different proteins define the specificity of the interaction. The MIT domain of VPS4A was able to compete with spastin for binding to the CHMP1B MIM3, but it is unclear through which type of binding mode Vps4 interacts with the C-terminal CHMP1B fragment. In addition, spastin can be recruited by IST163.. Notably, the LIP5/Vta1 activator of Vps4 (discussed below) contains a tandem MIT domain, which has the ability to bind several ESCRT-III tails at the same time using an MIM1-type interaction on the first MIT domain or an MIM5 binding mode on the second MIT domain (Figure 3BC), with the MIM5 interaction being the tightest MIM-MIT interaction reported to date (KD = 3 nM)96; 101 and involving two helices of CHMP5 wrapping around MIT2 of LIP5.
AAA ATPase cassette
Vps4 and the microtubule-severing enzymes have a single AAA ATPase cassette that adopts a canonical AAA+ fold102; 103. Crystal structures of human, yeast and archaeal Vps424; 104; 105; 106; 107, human and D. melanogaster Spastin108; 109, and C. elegans Fidgetin-like protein 1 (FIGL-1)110, have revealed details of the domain structure (Figure 4). The large AAA ATPase domain contains 5 α-helices that are arranged on either side of a central 5-stranded parallel β-sheet. Eukaryotic Vps4 contains an additional amino-terminal strand (β’) that packs against the β-sheet in an antiparallel orientation. The corresponding region is disordered in structures of archaeal Vps4, and in spastin and FIGL-1 it forms a short helix that packs against the edge of the central sheet and helix 3. In spastin, this helix contains the final 5 residues of the MTBD, but it is unclear if those specific residues contribute to microtubule binding.
Figure 4. Structure of the AAA ATPase cassette.
Crystal structures of (a) human VPS4B (PDB ID: 1XWI)24, (b) human spastin (PDB ID: 3VFD)109 and (c) human fidgetin-like 1 (FIGL-1, PDB ID: 3D8B)110. Coloring is as in Figure 1. The N-terminal helix (red) in microtubule-severing enzymes lies within the sequence that has been mapped to contain the MTBD in spastin.
The small AAA ATPase domain adopts an antiparallel 4-helix bundle architecture. Unique to eukaryotic Vps4 proteins is the presence of the β-domain, which is inserted into the small ATPase domain following the third helix. This β-domain serves as a binding site for the Vta1/LIP5 activator of eukaryotic Vps424; 25, which does not appear to have a counterpart in archaeal Vps4 or the microtubule-severing AAA ATPases.
Compared to other AAA+ ATPases, the meiotic clade enzymes have an additional C-terminal helix that is connected to the small AAA ATPase domain through a short linker and packs closely against the P-loop and strands of the central β-sheet of the large AAA ATPase domain. The functional role of the C-terminal helix remains poorly understood. Truncation experiments in yeast have suggested a role in oligomerization and consequently in ATP hydrolysis111 although these effects may be indirect.
Models for the active hexamer
AAA ATPases typically function as oligomers, usually hexamers102, and only hydrolyze ATP in the assembled state. Consistent with this, at micromolar concentrations, spastin and katanin both assemble into oligomers that are likely hexameric in the presence of ATP13; 16; 108; 112. Recent biochemical studies have shown that Vps4 also forms a hexamer106, overturning earlier models suggesting a double ring architecture with six or seven subunits in each ring105; 113; 114. Much of the earlier confusion was due to the analysis of a hydrolysis-deficient mutant Vps4 protein that displays aberrant assembly behavior106. Interestingly, in spastin and katanin p60, the equivalent Walker B mutation stabilizes the hexamer. The hexamer model for Vps4 is further supported by the finding that the assembled Vps4 ATPase cassette binds a substrate peptide in its central pore with 6:1 Vps4:peptide stoichiometry115. Determination of a high-resolution structure of an assembled meiotic clade AAA ATPases has been a high priority for many years, although no such structures have been reported to date.
In the absence of an experimentally determined, high-resolution structure of the active hexamer, the leading model is that the meiotic clade enzymes resemble crystal structures of other AAA ATPases. In particular, very similar models of the Vps4 and spastin ATPase hexamers (Figure 5) have been proposed based on the known structures of the p97 D1 ring and the NSF D2 ring, respectively24; 108, both of which were crystallized as symmetric hexamers116; 117; 118; 119; 120. Supporting this p97-like model, mutation of residues in the predicted Vps4 interfaces disrupted higher-order oligomerization and abolished ATPase activity in vitro, and caused failure of Vps4-associated function in cell culture104; 106. Small-angle X-ray scattering data for spastin are consistent with the p97-like model108, as are the dimensions of rings seen in electron micrographs of negatively stained katanin16.
Figure 5. Hexameric models of spastin and Vps4.
Models of p97-like hexamers were generated by superposition of the subunit structures of spastin (left) or VPS4B (right) on the D1 ring of the p97 hexamer (not shown). Hexamerization is favored by the avidity effect of MIT domains binding to the ESCRT-III and tubulin lattices. Spastin and other microtubule-severing enzymes bind microtubules through their MIT domains and microtubule binding domain (MTBD), as indicated by red arrows. The interaction between the LIP5 VSL domain and the VPS4B β-domain was modeled after the crystal structure of the equivalent yeast proteins25.
The model positions the large AAA ATPase domain of each subunit at the center of the ring (Figure 5). Small AAA ATPase domains are located further from the center of the hexamer, and pack against the large subunit of the neighboring protomer. This arrangement positions the arginine finger in the vicinity of the nucleotide binding pocket at the interface between protomers, consistent with its role in complementing the active site4; 5. This arrangement is ideal to transmit conformational changes in response to nucleotide binding and hydrolysis. Importantly, unless all subunits are in the same nucleotide state, the hydrolysis cycle will likely induce conformational changes resulting in asymmetry of the overall structure, which is not accounted for in the current structural model.
The p97-model also places two loops at the central pore of the hexamer. The sequences of both pore loops are highly conserved among AAA+ ATPases that process protein substrates24; 108. Pore loop 1 contains an aromatic-hydrophobic-glycine tripeptide motif, whereas pore loop 2 displays a number of charged residues. The importance of conserved residues in each of the pore loops has been demonstrated by the finding that their substitution disrupts substrate binding in vitro13; 46; 84; 115. Spastin and katanin in which pore loop residues have been mutated still bind to but no longer sever microtubules46; 84, and Vps4 pore loop mutants no longer support the ESCRT-dependent sorting of model cargo proteins or HIV budding24; 104. An important caveat for some of these studies is that mutation of some pore loop residues can diminish hexamerization, indicating that subsequent loss of activity might be an indirect consequence of altered assembly. However, a recent study of Vps4 has identified pore loop mutants that altered substrate binding to the pore but did not destabilize the hexamer115. Similarly, some pore loop mutants of spastin and katanin retain their ability to hydrolyze ATP and bind to microtubules, but no longer function in severing46; 84.
Regulation of Hexamerization
Hexamerization of meiotic clade AAA ATPases is weak and requires high micromolar concentrations in vitro, with an inactive monomeric or dimeric form predominating at lower concentrations. Recruitment to the polymeric substrate presumably increases the local concentration, thereby promoting assembly at precisely the site where activity is needed (Figure 5). Consistent with this idea, the ATPase activity of Vps4 is stimulated when the protein is concentrated on a surface using an affinity tag121, and both katanin and spastin have higher ATPase activity in the presence of microtubules14; 16; 44. The combination of inherently weak hexamerization coupled with avid recruitment to a polymer may thereby provide a mechanism to restrict the ATPase remodeling activity to authentic substrates.
The Vps4 cofactor LIP5/Vta1 stimulates ATP hydrolysis in vitro at least in part by stabilizing the active, hexameric state122; 123. As discussed above, its N-terminal tandem MIT domain binds the MIM motifs of several ESCRT-III proteins, and may thereby further promote association with the ESCRT-III lattice (Figure 5). The tandem MIT domain is followed by an ~100-residue linker that appears to be inherently flexible, followed by a C-terminal VSL domain that is dimeric and binds to the Vps4 β-domain at the periphery of the hexameric ring. LIP5/Vta1 might stabilize the Vps4 hexamer through interactions that bridge adjacent subunits. This idea is supported by the identification of a Vps4 stimulatory element N-terminal to the VSL domain in yeast Vta1124 that directly contacts the small AAA ATPase domain125, although such an arrangement has yet to be visualized. A crystal structure of a VSL complex with the Vps4 small AAA ATPase and β-domain suggests that Vta1 may function to stabilize inter-hexameric association25, although this model is not obviously consistent with the finding that Vta1-Vps4 complexes appear to be stable hexamers106.
The microtubule-severing enzymes lack β-domains and do not appear to possess activators equivalent to LIP5/Vta1, although the non-catalytic p80 subunit of katanin has the ability to stimulate both ATP hydrolysis and microtubule severing by katanin p6016. The mechanism of this stimulation has not yet been determined, but one attractive possibility is that p80 may stabilize the katanin hexamer. Electron micrographs of the katanin p60/p80 complex16 are consistent with this hypothesis, although higher resolution structural information or biochemical data will be necessary to test and advance this model.
Substrate engagement and translocation
Following recruitment by the N-terminal MIT domain and/or linker residues, flexible sequences in the substrates engage with residues of the central pore loops of the ATPase hexamer. This model is supported by mutation of Vps4 pore loop residues and the observation that ESCRT-III-derived peptides bind to Vps4 lacking the MIT domain with 1:6 subunit stoichiometry, which is consistent with one peptide binding to the central pore of the hexamer115. A direct interaction between pore loops and tubulin tails has also been reported for spastin13; 84 and katanin46. It is important to note that the sequence stretches that are preferentially engaged by the pore loops are distinct from the MIM and MTBD sequences that mediate recruitment to the ESCRT-III or tubulin lattice. In the case of Vps4, it has been shown that the AAA ATPase cassette can engage ESCRT-III proteins in the absence of an MIM-MIT interaction, and that this interaction will stimulate ATPase activity, presumably through further stabilization of the active hexamer conformation101; 115; 121. Vps4 and microtubule-severing enzymes all preferentially engage acidic sequence motifs in the C-terminal regions of their respective substrate. When the acidic C-terminal tails of tubulin have been removed, spastin and katanin are still able to bind to microtubules via the MTBD, but their severing activity is blocked14; 46; 126. Vps4 pore loops also bind acidic motifs within their ESCRT-III substrates, and it has therefore been proposed that charge-charge interactions between acidic regions in the substrate and basic pore loop residues define the interaction46; 108; 121. Although acidic motifs are preferentially engaged, it seems likely that the pore loops have the ability to translocate a wide variety of amino acid sequences. Mapping of a minimal binding motif in the C-terminal tail of ESCRT-III subunits showed preferred binding to the ATPase cassette of a segment corresponding to helix 5115; 121. This motif is conserved in all ESCRT-III proteins and is distinct from MIM sequences, which are located C-terminal to helix 5. It is attractive to speculate that polymerization to form ESCRT-III filaments triggers the release of helix 5 from its binding site at the tip of the ESCRT-III core because it has been observed that removing C-terminal segments that include helix 5 promotes assembly70; 71; 127; 128. This model is appealing because it would provide a mechanism to further restrict Vps4 activity to relevant substrates, although direct evidence for that model is currently lacking.
The stoichiometry of a single substrate peptide per hexamer indicates that the bound ATPase conformation is asymmetric115. The importance of asymmetry is also emphasized by the finding that peptide binding to Vps4 is supported by ADP·AlFx and ADP·BeFx, which are able to mimic multiple nucleotide-bound states, but not by ATP, ADP or non-hydrolyzable ATP analogs115. This is an appealing model that is consistent with studies on related or analogous nucleic acid translocases129; 130, protein translocases131; 132; 133; 134, and the F1-ATPase135, which have been inferred to function as asymmetric hexamers in which ATP hydrolysis proceeds progressively through successive positions around the ring as substrate is translocated. In AAA+ ATPases, the nucleotide binding site is located in the cleft between the large and the small AAA ATPase domain, which are connected by a flexible hinge. The nucleotide state is thought to affect the angle between the two domains104; 107, and may also be sensed by the C-terminal helix, which is in close proximity to the P-loop and may also contact the adjacent subunit. Subtle changes due to nucleotide hydrolysis may therefore result in an asymmetric, non-planar overall conformation such as a split washer, which has been seen in structures of AAA+ proteins including the E1 helicase129, the 26S proteasome131; 132, the bacterial enhancer-binding protein NtrC1133 and the membrane fusion protein NSF134. A common feature shared by these structures is the arrangement of the pore loops in a spiraling staircase conformation, which implies an attractive mechanism for the conversion of chemical energy from ATP binding and hydrolysis into mechanical force that translocates the substrate into the central pore.
The extent to which substrates are translocated through the central pore is a fundamental question for AAA+ ATPases. Unfolding and translocation of the entire substrate through the pore has been demonstrated for AAA+ proteins that target their substrate for degradation136, including ClpA and ClpX, which both work with the ClpP protease, and the AAA+ components of the 26S proteasome. Other AAA+ ATPases remodel protein complexes by inducing a conformational change in their substrate that does not involve complete translocation or global unfolding. A recent structural study investigating the mechanism of SNARE disassembly by NSF proposed that ATP hydrolysis results in a shearing or twisting force that causes the three subunits of the post-fusion SNARE complex to unwind and dissociate134. A second example of a AAA+ ATPase that does not trigger global unfolding is RuBisCO activase, which translocates only a short segment of RuBisCO in order to induce a conformational change that releases inhibitory sugar phosphates and allows the enzyme to return to its catalytically active conformation137. A recent study indicates that ESCRT-III substrates are globally unfolded as they are processed by Vps4138. In contrast, it seems unlikely that tubulin subunits are fully unfolded during microtubule severing because tubulin subunits extracted by microtubule-severing enzymes in vitro become available for subsequent rounds of polymerization14, even when in the absence of the type II chaperonin that is required for their initial folding139; 140.
Regulatory role of MIT domains
Recently, a regulatory function for the MIT domain of Vps4 has been described115. A construct lacking the MIT domain bound substrate peptides at the pore loops more tightly than the full-length protein. Moreover, the autoinhibitory effect of MIT domains for binding at the pore loops was eliminated when the MIT domain was engaged with either MIM1 or MIM2 sequences. These observations prompt the model that the MIT domains inhibit substrate engagement and processing until Vps4 is recruited to the site of action by binding of the MIT domain to MIMs on the ESCRT-III lattice, and is an attractive mechanism to prevent premature substrate processing. This is analogous to the finding that activity of the MIT-containing protease calpain-7 is repressed prior to binding of the MIT domain with ESCRT-III141. In the case of Vps4, autoinhibition does not appear to affect oligomerization, suggesting that MIT domains might regulate substrate binding by a mechanism such as obstructing access to the pore or by preventing the formation of the asymmetric binding site that is competent for substrate binding. It is remarkable that both MIM1 and MIM2 sequences can alleviate autoinhibition because they bind to different sites on the MIT domain, and it will be of considerable interest to understand how this is achieved. Another important goal for future studies is to understand how many of the six MIT domains in one Vps4 hexamer need to be engaged by the ESCRT-III lattice in order to activate substrate binding and translocation at the pore loops. Regardless of the mechanistic details, these observations indicate the presence of a powerful autoregulatory mechanism to ensure that Vps4 unfoldase activity, which may be sufficiently permissive to translocate all of the sequence motifs found in an entire ESCRT-III subunit138, and therefore may have the inherent capacity to translocate many if not most protein sequences, is restricted to assembled ESCRT-III lattices. It will be of interest to determine if an analogous mechanism exists for the microtubule-severing enzymes, and if so, if it is mediated by engagement of MIT domains with an ESCRT-III polymer and/or by engagement of the MTBD with microtubules.
Models and open questions
The mechanisms by which Vps4, katanin p60, spastin and fidgetin remodel their distinct ESCRT-III or microtubule lattice substrates appear to be similar (Figure 6). The leading model is that they exist as inactive monomeric or dimeric proteins in the cytoplasm, and assemble into active hexamers upon recruitment and local concentration through binding to their polymeric ESCRT-III or microtubule substrates. The hexamers are competent to engage substrates via loops lining the central pore and to translocate their respective ESCRT-III or microtubule substrates through the central pore in an ATP-driven cycle. In the case of Vps4, MIT binding to MIM sequences promotes ESCRT-III substrate engagement, and a currently unknown mechanism presumably exists to ensure that katanin, spastin, and fidgetin faithfully engage their microtubule substrate.
Figure 6. Model of ATPase assembly and protein lattice disassembly.
Meiotic clade AAA ATPases are monomeric or dimeric in the cytoplasm. The microtubule or ESCRT-III protein polymer (green) is depicted schematically. Recruitment of the ATPase by MIM-MIT interactions or microtubule interactions promotes formation of the active hexamer through increased local concentration. Substrate binding to the central pore loops requires an asymmetric conformation of the hexamer, which is induced by ATP hydrolysis. MIM-MIT interaction releases autoinhibition of substrate engagement at the Vps4 pore loops. Pore loops initially bind acidic motifs in the C-terminal tails of either ESCRT-III or tubulin, and translocation through the central pore results in subunit unfolding and consequent disassembly of the ESCRT-III lattice or severing of microtubules.
Similarities extend to the types of sequences that are initially engaged for substrate processing. Vps4 and microtubule-severing enzymes bind acidic sequences near the C-terminus of their substrate, suggesting that in both cases the direction of translocation is toward the structured N-terminus. Continued translocation will lead to conformational destabilization and consequent disassembly of the protein lattice.
Many important mechanistic questions remain to be resolved. One of the most fundamental is determination of high-resolution structures of active, asymmetric hexamers, and determination of how they engage and translocate substrate through the central pore. Building on this, it will be important to understand the mechanism by which LIP5/Vta1 stabilizes and activates Vps4, and the role of additional cofactors, such as katanin 80, for the microtubule-severing enzymes. Mechanisms of recruitment should be clarified for the microtubule-severing enzymes, including the interplay between MIT and MTBD-mediated recruitment to ESCRT-III and microtubules, and in all cases the mechanisms of activation upon recruitment remain to be determined. There is much more to be done to understand how these fascinating enzymes drive a wide array of essential biological processes.
Highlights.
The N-terminal MIT domain mediates recruitment to substrate.
The ATPase cassette forms an asymmetric hexamer that disassembles protein polymers.
Vps4 functions in membrane fission by disassembling ESCRT-III polymers.
Spastin, katanin and fidgetin sever microtubules in multiple cellular pathways.
Polymers are disassembled by translocating subunits through the hexamer pore.
Acknowledgements
We thank Janet Iwasa for artwork, Wesley Sundquist and Adam Frost for helpful discussions, and John McCullough, Jörg Votteler, Dawn Wenzel, Jack Skalicky and Lauren Williams for comments on the manuscript. This work was supported by NIH P50 GM082545 (C.P.H.). N.M. was also supported by NIH T32 AI055434.
Abbreviations
- AAA
ATPase associated with diverse cellular activities
- ESCRT
Endosomal Sorting Complexes Required for Transport
- MIT
microtubule interacting and trafficking
- MTBD
microtubule binding domain
- EM
electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- 2.Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J Struct Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Wendler P, Ciniawsky S, Kock M, Kube S. Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 6.Mains PE, Kemphues KJ, Sprunger SA, Sulston IA, Wood WB. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 1990;126:593–605. doi: 10.1093/genetics/126.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley JH, Yang D. MIT domainia. Dev Cell. 2008;14:6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Lumb JH, Connell JW, Allison R, Reid E. The AAA ATPase spastin links microtubule severing to membrane modelling. Biochim Biophys Acta. 2012;1823:192–197. doi: 10.1016/j.bbamcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Claudiani P, Riano E, Errico A, Andolfi G, Rugarli EI. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res. 2005;309:358–369. doi: 10.1016/j.yexcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Mancuso G, Rugarli EI. A cryptic promoter in the first exon of the SPG4 gene directs the synthesis of the 60-kDa spastin isoform. BMC Biol. 2008;6:31. doi: 10.1186/1741-7007-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciccarelli FD, Proukakis C, Patel H, Cross H, Azam S, Patton MA, Bork P, Crosby AH. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics. 2003;81:437–441. doi: 10.1016/s0888-7543(03)00011-9. [DOI] [PubMed] [Google Scholar]
- 12.Beetz C, Brodhun M, Moutzouris K, Kiehntopf M, Berndt A, Lehnert D, Deufel T, Bastmeyer M, Schickel J. Identification of nuclear localisation sequences in spastin (SPG4) using a novel Tetra-GFP reporter system. Biochem Biophys Res Commun. 2004;318:1079–1084. doi: 10.1016/j.bbrc.2004.03.195. [DOI] [PubMed] [Google Scholar]
- 13.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 15.Rigden DJ, Liu H, Hayes SD, Urbe S, Clague MJ. Ab initio protein modelling reveals novel human MIT domains. FEBS Lett. 2009;583:872–878. doi: 10.1016/j.febslet.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 17.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonbuchner TM, Rath U, Sharp DJ. KL1 is a novel microtubule severing enzyme that regulates mitotic spindle architecture. Cell Cycle. 2010;9:2403–2411. doi: 10.4161/cc.9.12.11916. [DOI] [PubMed] [Google Scholar]
- 19.Ververis A, Christodoulou A, Christoforou M, Kamilari C, Lederer CW, Santama N. A novel family of katanin-like 2 protein isoforms (KATNAL2), interacting with nucleotide-binding proteins Nubp1 and Nubp2, are key regulators of different MT-based processes in mammalian cells. Cell Mol Life Sci. 2015 doi: 10.1007/s00018-015-1980-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith LB, Milne L, Nelson N, Eddie S, Brown P, Atanassova N, O'Bryan MK, O'Donnell L, Rhodes D, Wells S, Napper D, Nolan P, Lalanne Z, Cheeseman M, Peters J. KATNAL1 regulation of sertoli cell microtubule dynamics is essential for spermiogenesis and male fertility. PLoS Genet. 2012;8:e1002697. doi: 10.1371/journal.pgen.1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the "Pacman-flux" machinery that moves chromosomes. J Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Mahaffey CL, Berube N, Nystuen A, Frankel WN. Functional characterization of fidgetin, an AAA-family protein mutated in fidget mice. Exp Cell Res. 2005;304:50–58. doi: 10.1016/j.yexcr.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Scheuring S, Rohricht RA, Schoning-Burkhardt B, Beyer A, Muller S, Abts HF, Kohrer K. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol. 2001;312:469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 24.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, Sundquist WI. Structural and mechanistic studies of VPS4 proteins. Embo j. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang D, Hurley JH. Structural role of the Vps4-Vta1 interface in ESCRT-III recycling. Structure. 2010;18:976–984. doi: 10.1016/j.str.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 29.Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 31.Duro E, Marston AL. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev. 2015;29:109–122. doi: 10.1101/gad.255554.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haimo LT, Rosenbaum JL. Cilia, flagella, and microtubules. J Cell Biol. 1981;91:125s–130s. doi: 10.1083/jcb.91.3.125s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishikawa T. Cryo-electron tomography of motile cilia and flagella. Cilia. 2015;4:3. doi: 10.1186/s13630-014-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogales E, Wang HW. Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr Opin Struct Biol. 2006;16:221–229. doi: 10.1016/j.sbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 36.Oakley BR, Paolillo V, Zheng Y. gamma-Tubulin complexes in microtubule nucleation and beyond. Mol Biol Cell. 2015;26:2957–2962. doi: 10.1091/mbc.E14-11-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouhard GJ. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Mol Biol Cell. 2015;26:1207–1210. doi: 10.1091/mbc.E13-10-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 39.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 40.Akhmanova A, Steinmetz MO. Microtubule +TIPs at a glance. J Cell Sci. 2010;123:3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- 41.Akhmanova A, Hoogenraad CC. Microtubule minus-end-targeting proteins. Curr Biol. 2015;25:R162–R171. doi: 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 42.Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 44.Salinas S, Carazo-Salas RE, Proukakis C, Cooper JM, Weston AE, Schiavo G, Warner TT. Human spastin has multiple microtubule-related functions. J Neurochem. 2005;95:1411–1420. doi: 10.1111/j.1471-4159.2005.03472.x. [DOI] [PubMed] [Google Scholar]
- 45.Stoppin-Mellet V, Gaillard J, Vantard M. Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem J. 2002;365:337–342. doi: 10.1042/BJ20020689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johjima A, Noi K, Nishikori S, Ogi H, Esaki M, Ogura T. Microtubule severing by katanin p60 AAA+ ATPase requires the C-terminal acidic tails of both alpha- and beta-tubulins and basic amino acid residues in the AAA+ ring pore. J Biol Chem. 2015;290:11762–11770. doi: 10.1074/jbc.M114.614768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125:2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 55.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 56.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 57.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 58.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, Perez F. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 61.Loncle N, Agromayor M, Martin-Serrano J, Williams DW. An ESCRT module is required for neuron pruning. Sci Rep. 2015;5:8461. doi: 10.1038/srep08461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 64.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bieniasz PD. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss ER, Gottlinger H. The role of cellular factors in promoting HIV budding. J Mol Biol. 2011;410:525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amorim NA, da Silva EM, de Castro RO, da Silva-Januario ME, Mendonca LM, Bonifacino JS, da Costa LJ, daSilva LL. Interaction of HIV-1 Nef protein with the host protein Alix promotes lysosomal targeting of CD4 receptor. J Biol Chem. 2014;289:27744–27756. doi: 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, Schoehn G, Weissenhorn W, Sadoul R. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 77.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 80.Yang D, Rismanchi N, Renvoise B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, Seaman MN, Hazan J, Reid E. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J Cell Biol. 2013;202:527–543. doi: 10.1083/jcb.201211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwaya N, Kuwahara Y, Fujiwara Y, Goda N, Tenno T, Akiyama K, Mase S, Tochio H, Ikegami T, Shirakawa M, Hiroaki H. A common substrate recognition mode conserved between katanin p60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J Biol Chem. 2010;285:16822–16829. doi: 10.1074/jbc.M110.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsushita-Ishiodori Y, Yamanaka K, Hashimoto H, Esaki M, Ogura T. Conserved aromatic and basic amino acid residues in the pore region of Caenorhabditis elegans spastin play critical roles in microtubule severing. Genes Cells. 2009;14:925–940. doi: 10.1111/j.1365-2443.2009.01320.x. [DOI] [PubMed] [Google Scholar]
- 85.Phillips SA, Barr VA, Haft DH, Taylor SI, Haft CR. Identification and characterization of SNX15, a novel sorting nexin involved in protein trafficking. J Biol Chem. 2001;276:5074–5084. doi: 10.1074/jbc.M004671200. [DOI] [PubMed] [Google Scholar]
- 86.Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20:1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caballe A, Wenzel DM, Agromayor M, Alam SL, Skalicky JJ, Kloc M, Carlton JG, Labrador L, Sundquist WI, Martin-Serrano J. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. Elife. 2015;4 doi: 10.7554/eLife.06547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 90.Fujioka Y, Suzuki SW, Yamamoto H, Kondo-Kakuta C, Kimura Y, Hirano H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 91.Solomons J, Sabin C, Poudevigne E, Usami Y, Hulsik DL, Macheboeuf P, Hartlieb B, Gottlinger H, Weissenhorn W. Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure. 2011;19:1149–1159. doi: 10.1016/j.str.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 93.Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 94.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 95.Takasu H, Jee JG, Ohno A, Goda N, Fujiwara K, Tochio H, Shirakawa M, Hiroaki H. Structural characterization of the MIT domain from human Vps4b. Biochem Biophys Res Commun. 2005;334:460–465. doi: 10.1016/j.bbrc.2005.06.110. [DOI] [PubMed] [Google Scholar]
- 96.Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, Uter NT, Bajorek M, Myszka DG, Sundquist WI. Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J Biol Chem. 2012;287:43910–43926. doi: 10.1074/jbc.M112.417899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 98.Guo EZ, Xu Z. Distinct mechanisms of recognizing endosomal sorting complex required for transport III (ESCRT-III) protein IST1 by different microtubule interacting and trafficking (MIT) domains. J Biol Chem. 2015;290:8396–8408. doi: 10.1074/jbc.M114.607903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vild CJ, Li Y, Guo EZ, Liu Y, Xu Z. A novel mechanism of regulating the ATPase VPS4 by its cofactor LIP5 and the endosomal sorting complex required for transport (ESCRT)-III protein CHMP5. J Biol Chem. 2015;290:7291–7303. doi: 10.1074/jbc.M114.616730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vild CJ, Xu Z. Vfa1 binds to the N-terminal microtubule-interacting and trafficking (MIT) domain of Vps4 and stimulates its ATPase activity. J Biol Chem. 2014;289:10378–10386. doi: 10.1074/jbc.M113.532960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shim S, Merrill SA, Hanson PI. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell. 2008;19:2661–2672. doi: 10.1091/mbc.E07-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 103.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 104.Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol. 2008;384:878–895. doi: 10.1016/j.jmb.2008.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hartmann C, Chami M, Zachariae U, de Groot BL, Engel A, Grutter MG. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 106.Monroe N, Han H, Gonciarz MD, Eckert DM, Karren MA, Whitby FG, Sundquist WI, Hill CP. The oligomeric state of the active Vps4 AAA ATPase. J Mol Biol. 2014;426:510–525. doi: 10.1016/j.jmb.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiao J, Xia H, Yoshino-Koh K, Zhou J, Xu Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J Mol Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taylor JL, White SR, Lauring B, Kull FJ. Crystal structure of the human spastin AAA domain. J Struct Biol. 2012;179:133–137. doi: 10.1016/j.jsb.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng W, Lin Z, Li W, Lu J, Shen Y, Wang C. Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein. J Biol Chem. 2013;288:29305–29312. doi: 10.1074/jbc.M113.502559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vajjhala PR, Nguyen CH, Landsberg MJ, Kistler C, Gan AL, King GF, Hankamer B, Munn AL. The Vps4 C-terminal helix is a critical determinant for assembly and ATPase activity and has elements conserved in other members of the meiotic clade of AAA ATPases. FEBS J. 2008;275:1427–1449. doi: 10.1111/j.1742-4658.2008.06300.x. [DOI] [PubMed] [Google Scholar]
- 112.Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 113.Landsberg MJ, Vajjhala PR, Rothnagel R, Munn AL, Hankamer B. Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure. 2009;17:427–437. doi: 10.1016/j.str.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 114.Yu Z, Gonciarz MD, Sundquist WI, Hill CP, Jensen GJ. Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. Binding of Substrates to the Central Pore of the Vps4 ATPase Is Autoinhibited by the Microtubule Interacting and Trafficking (MIT) Domain and Activated by MIT Interacting Motifs (MIMs) J Biol Chem. 2015;290:13490–13499. doi: 10.1074/jbc.M115.642355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 117.Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. Embo j. 2004;23:1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huyton T, Pye VE, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6A. J Struct Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 119.Yu RC, Hanson PI, Jahn R, Brunger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 120.Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 121.Merrill SA, Hanson PI. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J Biol Chem. 2010;285:35428–35438. doi: 10.1074/jbc.M110.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc Natl Acad Sci U S A. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Norgan AP, Davies BA, Azmi IF, Schroeder AS, Payne JA, Lynch GM, Xu Z, Katzmann DJ. Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J Biol Chem. 2013;288:26147–26156. doi: 10.1074/jbc.M113.494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davies BA, Norgan AP, Payne JA, Schulz ME, Nichols MD, Tan JA, Xu Z, Katzmann DJ. Vps4 stimulatory element of the cofactor Vta1 contacts the ATPase Vps4 alpha7 and alpha9 to stimulate ATP hydrolysis. J Biol Chem. 2014;289:28707–28718. doi: 10.1074/jbc.M114.580696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eckert T, Le DT, Link S, Friedmann L, Woehlke G. Spastin's microtubule-binding properties and comparison to katanin. PLoS One. 2012;7:e50161. doi: 10.1371/journal.pone.0050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 129.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 130.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Forster F, Unverdorben P, Sledz P, Baumeister W. Unveiling the long-held secrets of the 26S proteasome. Structure. 2013;21:1551–1562. doi: 10.1016/j.str.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 132.Matyskiela ME, Martin A. Design principles of a universal protein degradation machine. J Mol Biol. 2013;425:199–213. doi: 10.1016/j.jmb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sysoeva TA, Chowdhury S, Guo L, Nixon BT. Nucleotide-induced asymmetry within ATPase activator ring drives sigma54-RNAP interaction and ATP hydrolysis. Genes Dev. 2013;27:2500–2511. doi: 10.1101/gad.229385.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 136.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 137.Portis AR, Jr, Li C, Wang D, Salvucci ME. Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot. 2008;59:1597–1604. doi: 10.1093/jxb/erm240. [DOI] [PubMed] [Google Scholar]
- 138.Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo j. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 141.Osako Y, Maemoto Y, Tanaka R, Suzuki H, Shibata H, Maki M. Autolytic activity of human calpain 7 is enhanced by ESCRT-III-related protein IST1 through MIT-MIM interaction. FEBS J. 2010;277:4412–4426. doi: 10.1111/j.1742-4658.2010.07822.x. [DOI] [PubMed] [Google Scholar]
- 142.Howard TL, Stauffer DR, Degnin CR, Hollenberg SM. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]