SUMMARY
Neurons receive input from the outside world or from other neurons through neuronal receptive endings (NREs). Glia envelop NREs to create specialized microenvironments; however, glial functions at these sites are poorly understood. Here we report a molecular mechanism by which glia control NRE shape and associated animal behavior. The C. elegans AMsh glial cell ensheathes NREs of twelve neurons, including the thermosensory neuron AFD. KCC-3, a K/Cl transporter, localizes specifically to a glial microdomain surrounding AFD receptive-ending microvilli, where it regulates K+ and Cl− levels. We find that Cl− ions function as direct inhibitors of an NRE-localized receptor-guanylyl-cyclase, GCY-8, which synthesizes cGMP. High cGMP mediates the effects of glial KCC-3 on AFD shape by antagonizing the actin-regulator WSP-1/NWASP. Components of this pathway are broadly expressed throughout the nervous system, suggesting that ionic regulation of the NRE microenvironment may be a conserved mechanism by which glia control neuron shape and function.
INTRODUCTION
Neurons receive information from the environment or other neurons through dendritic structures termed neuronal receptive-endings (NREs). In the mammalian central nervous system (CNS), postsynaptic neurons receive excitatory inputs at NREs termed spines, actin-rich receptive-endings that protrude from the dendrite shaft. Developmental and experience-dependent remodeling suggests that plasticity of spine morphology may correlate with learning and memory (Bourne and Harris, 2008). Perturbations in spine shape are associated with disorders including epilepsy, dementia, schizophrenia, Huntington’s disease, Alzheimer’s disease, and Fragile X syndrome (Penzes et al., 2011). Spines are often ensheathed by astrocytic glia (Chung et al., 2015). Glial cues are implicated in spine shape control, however, mechanisms by which they regulate spine morphology in vivo are not well understood (Christopherson et al., 2005; Chung et al., 2015; Murai et al., 2003).
Sensory NREs are comprised of microtubule-based cilia or actin-based microvilli, and are also glia-approximated. Sensory NRE shape perturbation leads to sensory deficits and is common in patients with congenital defects such as deafness-blindness Usher’s syndrome, or inherited conditions such as retinal degeneration. Genetic lesions underlying these syndromes affect sensory organ glia or associated neurons/neuron-like cells (Estrada-Cuzcano et al., 2012; Kremer et al., 2006). Glial mediators of sensory NRE shape are not known.
Glia often provide trophic support for neurons, complicating investigation of their roles in NRE shape control in vivo. C. elegans AMsh glia, which envelop neurons of the amphid sense organ, resemble mammalian glia but are dispensable for neuron survival. They are thus an excellent model to study glial control of NRE shape in vivo (Shaham, 2010). The AMsh glial cell ensheathes microvilli NREs of the AFD thermosensory neuron, as well as NREs of eleven other neurons, including the ciliated NRE of the AWC chemosensory neuron (Figure 1A) (Ward et al., 1975). We previously showed that ablation of AMsh glia disrupts AFD microvilli and AWC cilia NREs (Bacaj et al., 2008). Whether this reflect passive or active glial roles was unclear. Furthermore, molecules mediating glial contribution to NRE shape were unknown.
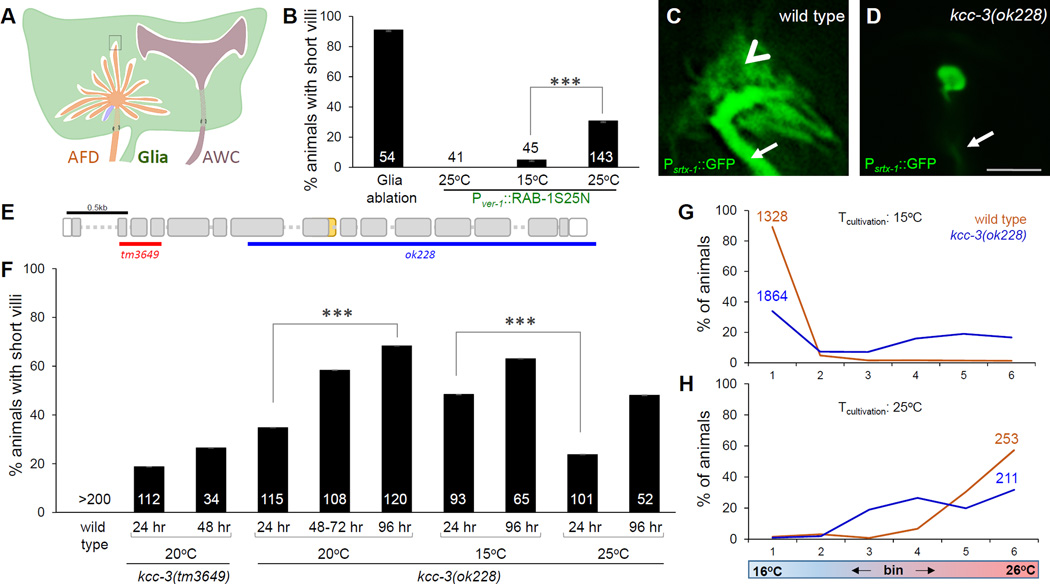
Figure 1. Glial KCC-3 controls AFD neuron receptive-ending shape.
(A) AMsh glia ensheathes multiple neurons. AFD (orange) also has a cilium (blue). Boxed region magnified in Figure 7. (B) Histogram depicting AFD defects in indicated genotypes. Numbers, animals scored. Error bars, SEM. ***p<0.0002. For transgenics, three independent lines were scored. Green text, glia expression; red text, AFD expression. Second bar, non-transgenic siblings (C,D) Fluorescence images of wild-type (C) and kcc-3(ok228) (D) AFD microvilli. Scale bar, 1µm. Arrow, dendritic shaft. Arrowhead, microvilli. (E) kcc-3 gene structure, deletions used are indicated by red and blue bars. Sequences not included in WormBase annotation, orange. (F) Histogram details as in Figure 1B. Time, hours post mid L4 stage. (G, H) Thermotaxis behavior assays for indicated genotype raised at 15°C (G) and 25°C (H). Animals in (G), 24 hours post mid-L4 stage. Animals in (H), 96 hours post mid-L4 stage. N, Number of animals. See also Figure S1 and S2.
AFD NREs consist of microvilli and a single simple cilium (Figure 1A) (Doroquez et al., 2014; Perkins et al., 1986). Biogenesis of C. elegans cilia has been explored (Inglis et al., 2007). However, mutations in genes affecting cilium development perturb AFD-dependent thermosensation only weakly, and do not affect the length, number, or distribution of AFD microvilli (Figure S1A) (Perkins et al., 1986; Tan et al., 2007). Conversely, mutations in the gene ttx-1 block microvilli formation and disrupt temperature sensation, but leave the AFD cilium intact (Procko et al., 2011; Satterlee et al., 2001). Thus, different molecular programs control cilium and microvilli structures, and microvilli are important for AFD-mediated thermosensation. Shape control of microvilli NREs is poorly understood in any system.
Here we report a mechanism by which a sense organ glial cell continuously regulates the shape of a microvilli-based sensory NRE, and its associated animal behavior, by regulating the NRE microenvironment. We find that an AMsh glia-expressed K/Cl co-transporter, KCC-3, regulates AFD NRE shape and C. elegans thermosensory behavior by controlling Cl− levels surrounding AFD NREs. Cl− ions directly inhibit the AFD neuron-specific receptor guanylyl cyclase (rGC) GCY-8 by binding to the conserved S(x)nGPxC motif in its extra-cellular domain. GCY-8 determines cGMP levels within AFD along with the phosphodiesterases PDE-1 and 5. High cGMP levels antagonize the actin regulator WSP-1/WASP, blocking NRE growth.
While KCC-3 affects AFD neuron shape, it is not required for AWC neuron NRE maintenance. KCC-3 localizes to a glial microdomain surrounding the AFD NRE, but not around the AWC NRE. Thus, a single glial cell discriminates between different neurons with which it associates.
Homologous and analogous components of this pathway are expressed throughout the CNS/PNS of many species. Our results suggest that ionic modulation of receptor activity at NREs may be a conserved mechanism by which glia regulate NRE shape and function.
RESULTS
Glia-secreted and/or membrane-bound factors control AFD NRE shape
To determine whether AMsh glia provide merely passive structural support, or whether active signaling promotes AFD NRE shape acquisition, we aimed to determine whether glia-secreted or membrane-bound proteins are required for AFD microvilli shape control. We generated animals containing a dominant-negative version of the ER-Golgi trafficking regulator RAB-1 (RAB-1S25N) under control of the AMsh glia-specific and temperature-sensitive ver-1 promoter. This construct is predicted to block exocytic traffic of secreted and membrane proteins at 25°C (Satoh et al., 1997). Thus, transient incubation at 25°C for 24 hours leaves glia structurally intact, but should impair signaling. Using this tool, we blocked glial secretion in young adults. A pronounced defect in AFD and AWC NRE shape was observed, mimicking the effects of glia ablations (Figure 1B, S2A). By contrast, non-transgenic animals raised at 15°C and transferred as L4 larvae to 25°C, or transgenic animals raised at 15°C (Figure 1B) have normal AFD microvilli. Thus, secreted or membrane-bound glial cues are required continuously to maintain AFD microvilli shape post-development. Glia-dependent dynamic plasticity of sensory structures had not been previously appreciated.
KCC-3 is a regulator of AFD NRE shape and function
To identify the relevant glial cues controlling AFD microvilli shape, we performed candidate mutant and RNA interference screens focused on inactivating glia-enriched genes encoding membrane or secreted proteins (Bacaj et al., 2008). We found that mutations in the kcc-3 gene result in AFD microvilli loss (Figure 1C–F). KCC-3 encodes a predicted K/Cl co-transporter homologous to human solute carrier protein SLC12A4, a protein predicted to require RAB-1-mediated trafficking for membrane localization. A null lesion in the gene, kcc-3(ok228), results in temperature- and age- dependent loss of AFD NREs. 96-hour kcc-3(ok228) adults or kcc-3(ok228) adults grown at 15°C display more pronounced deficits than 24-hour adults or adults grown at 25°C, respectively (Figure 1F). Weaker, but similar defects accompany kcc-3(tm3649) mutants, which harbor a smaller in-frame truncation of the gene (Figure 1E,F), consistent with the severity of gene disruption.
AFD is the primary thermosensory neuron in C. elegans, and its NREs house proteins that mediate thermosensory transduction (Garrity et al., 2010). This transduction apparatus is also required for the ability of C. elegans to remember its temperature of cultivation (Mori and Ohshima, 1995). We found that kcc-3(ok228) animals exhibit deficits in thermotaxis behavior when asked to choose their cultivation temperature on a temperature gradient. These behavior defects mirror the temperature- and age-dependence of kcc-3(ok228) AFD NRE defects (Figures 1G,H; S2B), suggesting a common underlying mechanism by which KCC-3 regulates AFD receptive-ending shape and AFD-mediated animal behavior.
The behavior and NRE shape defects we observed in kcc-3(ok228) mutants are similar to those reported for AMsh glia-ablated animals (Bacaj et al., 2008), suggesting that KCC-3 is a major mediator of the glial effect on AFD.
KCC-3 functions in AMsh glia to regulate AFD NRE shape
To confirm that KCC-3 is expressed in AMsh glia, we examined animals carrying a kcc-3 [5’UTR::gfp::3’UTR] transgene (Tanis et al., 2009). kcc-3 expression is detected exclusively in glia, including AMsh glia, but not in AFD (Figure 2A,B). Expression of kcc-3 coding sequences under the same 5’ UTR, or expression of a kcc-3 cDNA under control of a heterologous AMsh glia-specific promoter rescued kcc-3(ok228) AFD neuron shape defects (Figure 2C). Thus, KCC-3 functions specifically in AMsh glia to control AFD NRE shape.
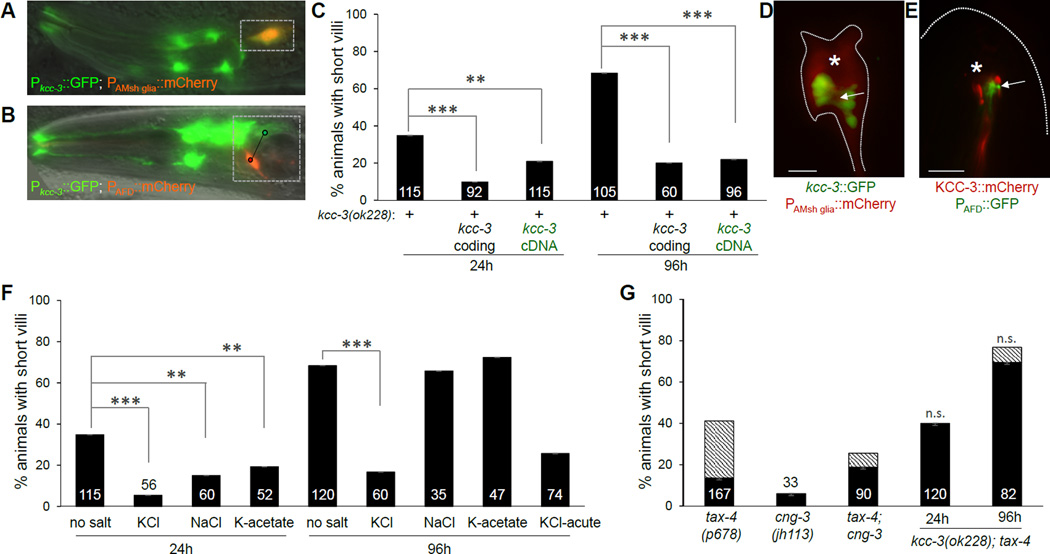
Figure 2. KCC-3 localizes to glial membranes surrounding AFD and regulates KCl homeostasis.
(A,B) Fluorescence images depicting expression of indicated transcriptional reporters. Box, cells of interest. Circles + line in (B) indicate distinct cells. (C) Histogram details as in 1B and 1F. +, presence of kcc-3(ok228) allele. **p=0.009. (D, E) Arrow, AFD receptive-ending region. Asterisk, location of AWC cilium. (F, G) Histogram details as in 1B and 1F. **p<0.02. n.s., not significant compared to age-matched kcc-3(ok228). Hatched bars, animals with disorganized AFD microvilli.
KCC-3 localization is restricted to a glial micro-domain surrounding AFD NREs
AMsh glia ensheath sensory NREs of other neurons, such as AWC, in addition to those of AFD. Furthermore, AMsh glia ablation or exocytosis block with RAB-1S25N disrupts NRE structure of these neurons (Figure S2A) (Bacaj et al., 2008). Thus, a general mechanism may underlie NRE shape control by glia. Surprisingly, however, we found that kcc-3 mutations do not affect AWC NREs (Figure S2A,C,D). To investigate the origin of this specificity, we examined KCC-3 subcellular localization using a genomic fosmid clone containing the kcc-3 locus recombineered with GFP immediately upstream of the kcc-3 stop codon, or a clone in which a kcc-3 cDNA is tagged with mCherry and expressed under an AMsh glia-specific promoter. We found that KCC-3 localizes to the portion of the AMsh glia apical domain in which AFD microvilli are embedded, and is conspicuously absent from the region surrounding AWC or any other amphid neuron (asterisk, Figure 2D,E). Restricted localization, therefore, likely explains the specific effects of KCC-3 on AFD receptive-endings.
Thus, contrary to our initial assumption, the AMsh glial cell employs different molecular mechanisms to control AFD and AWC receptive-ending shape. While single glial cells in other systems also contact multiple neurons, whether they discriminate between these neurons has been a major outstanding question. Our results provide definitive in vivo evidence that they can do so.
Glial KCC-3 regulates AFD NRE shape by regulating K+ and Cl− levels
KCC channels transport K+ and Cl− ions across membranes (Russell, 2000). We wondered, therefore, whether ionic imbalance may underlie AFD defects in kcc-3(ok228) mutant animals. To test this, we compared AFD receptive-ending defects in kcc-3(ok228) mutants cultivated on standard C. elegans growth medium, and those raised on plates supplemented with 150 mM KCl. Remarkably, AFD microvilli morphology in kcc-3(ok228) mutants raised on high KCl was largely normal (Figure 2F). Rescue of the kcc-3 mutant defects could also be achieved acutely, as 72-hour animals transferred to KCl-supplemented plates showed significant rescue within 24 hours (Figure 2F, “KCl-acute”).
To determine whether both K+ and Cl− were required for rescue, we raised kcc-3(ok228) mutants on plates supplemented with 150 mM Na+Cl− or K+Acetate−. While modest rescue was seen in younger animals in both cases, this was not sustained in older ones (Figure 2F).
Thus, K+ and Cl−, regulated by KCC-3, are acutely required to control AFD microvilli shape. Furthermore, the specific localization of KCC-3 to glial membranes around AFD strongly suggests that these ions function in the vicinity of the AFD NRE.
KCC-3 regulates AFD NRE shape through the AFD-specific rGC, GCY-8
AFD neuronal activity requires a cyclic nucleotide gated channel (CNG) composed of the TAX-2 β subunit and the TAX-4 or CNG-3 α subunits (Cho et al., 2004; Coburn and Bargmann, 1996; Komatsu et al., 1996). CNG channels allow cations (Na+, K+, and Ca+) to flow across membranes down their electrochemical gradients. Our finding that medium supplementation with K-acetate fails to rescue kcc-3(ok228) mutants suggested that impaired K+ conductance through these cation channels is unlikely to fully explain the AFD NRE defects of these mutants. To test this directly, we examined tax-4 and cng-3 single mutants, or tax-4; cng-3 double mutants. We found that these animals have only minor defects in AFD microvilli length, with some exhibiting elongated and disorganized microvilli (Figure 2G; hatched bars). Furthermore, mutations in genes functioning in downstream interneurons (ttx-3), or in adaptation of other neurons (egl-4) also have no effect on AFD shape (Figure S1B). Thus, the effects of KCC-3 on AFD shape cannot be entirely explained by reduced CNG channel conduction, and appear independent of downstream circuit dynamics.
We considered the possibility that glial KCC-3 defects could lead to increased K+ and Cl− accumulation in the extracellular space surrounding AFD microvilli, leading to hyperactive signaling through CNG channels. If this were the case, CNG channel mutations should mitigate the AFD shape defects of kcc-3(ok228) mutants. However, we found that tax-4 mutations did not suppress kcc-3(ok228) defects at all (Figure 2G). Our data, therefore, suggest that neuronal activity is not the primary effector of KCC-3 for AFD microvilli shape control. We therefore sought another neuronal effector.
CNG channels are activated by cGMP, synthesized by receptor guanylyl cyclases (rGCs). rGCs are Type I transmembrane proteins with extracellular (ECD), transmembrane (TM), kinase-homology (KHD), hinge (H), and guanylyl cyclase catalytic (CAT) domains (Figure 3A). The AFD neuron expresses the rGCs GCY-8, -18, and -23, which function redundantly to regulate thermotaxis behavior (Inada et al., 2006; Yu et al., 1997). We examined gcy-8(tm949), gcy-18(nj38), and gcy-23(ok797) single loss-of-function mutants and found no defects in AFD receptive-ending shape. Remarkably, however, the gcy-8(tm949) lesion, predicted to eliminate GCY-8 cyclase activity, strongly mitigates the AFD microvilli defects of kcc-3(ok228) mutants (Figures 3B, S3). This effect is largely specific to gcy-8, as the gcy-23(ok797) loss-of function allele only weakly rescues the AFD NRE defects of kcc-3(ok228) mutants (Figure 3B). Intriguingly, a genomic fosmid, in which GFP sequences are fused just upstream of the gcy-8 stop codon, is expressed specifically in AFD, and GFP fluorescence is localized to AFD microvilli (Figure S4A), consistent with previous studies.
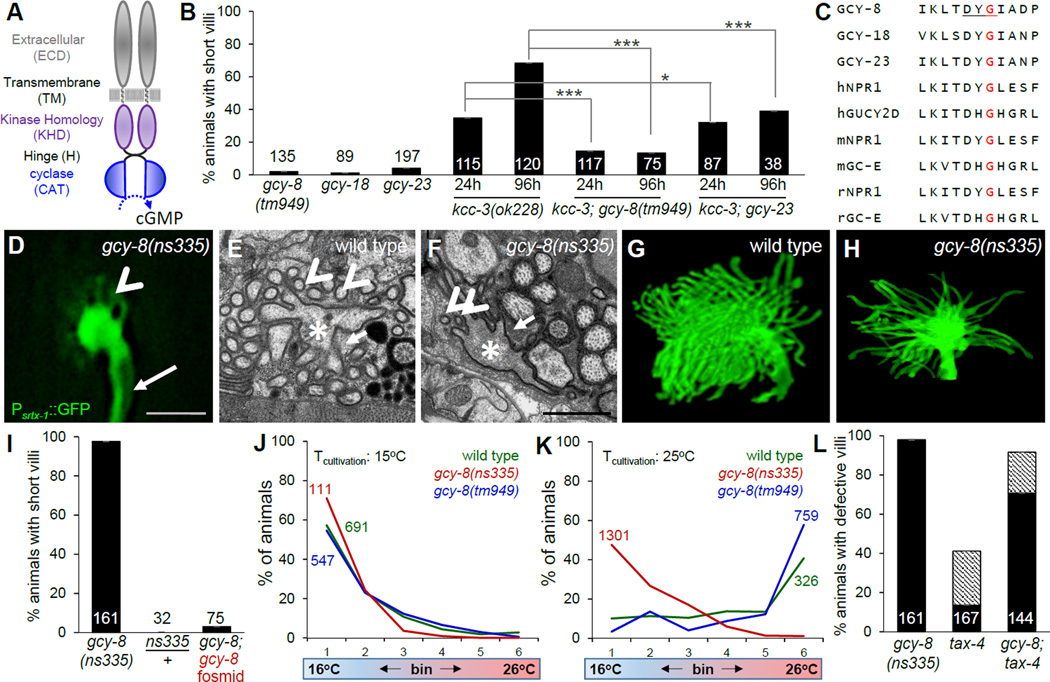
Figure 3. The neuronal rGC GCY-8 functions downstream of glial KCC-3.
(A) GCY-8 domain structure. (B) Histogram details as in 1B and 1F. *p=0.34. (C) Partial alignment of GCY-8. Red, conserved glycine mutated in gcy-8(ns335). (D,F,H) Fluorescence, TEM, and FIB-SEM reconstruction, respectively, of gcy-8(ns335) AFD receptive-ending. (E, G) TEM and FIB-SEM reconstruction, respectively, of wild-type AFD receptive-ending. (D) Arrow, dendritic shaft. Arrowhead, short microvilli. (E,F) Arrowheads, microvilli. Asterisk, dendritic shaft. Arrow, glial cytosol. Scale bar, 1µm. (I) Histogram details as in 1B. (J, K) Thermotaxis assays as in 1G. Animals raised at 15°C (J) or 25°C (K). (L) Histogram as in 1B. Hatched, animals with disorganized microvilli. See also Figure S3, S4 and Movies 1–4.
Together, our studies suggest that GCY-8 is a key effector of KCC-3, and that loss of KCC-3 results in an increase in GCY-8 activity.
Increased GCY-8 activity, through KHD inactivation, blocks AFD NRE extension
The hypothesis that increased GCY-8 activity mediates the effects of glial kcc-3 lesions was serendipitously corroborated by results of a genetic screen we conducted seeking mutants with defective AFD microvilli. From 10,800 F2 progeny examined (3.5 genomes), we recovered six relevant mutants. Genetic mapping, whole-genome sequencing, and transformation rescue studies revealed that one of these, ns335, has a causal lesion in gcy-8. The gcy-8(ns335) mutation is predicted to cause a G707E change in a highly conserved D(F/H/Y)G motif within the kinase homology domain (Figures 3A,C; S3A). This lesion has not been previously described in other rGCs.
Fluorescence microscopy revealed that gcy-8(ns335) mutants have fewer and shorter AFD microvilli (Figure 3D), and we confirmed this using serial-section transmission electron microscopy (TEM) (Figure 3E,F) and focused ion beam scanning EM (FIB-SEM)(Figure 3G,H; Movies S1–S4). Using TEM, we found that while wild-type AFD neurons have 43 ± 2 microvilli (n=6), gcy-8(ns335) mutants have 12 ± 1 (n=4) microvilli. SEM image analysis showed maximal microvilli length of 2.5 µm for wild-type animals, but only 1.5 µm for gcy-8(ns335).
gcy-8(ns335)/+ heterozygotes do not show AFD NRE abnormalities, and the defects of gcy-8(ns335) homozygotes are rescued by expression of wild-type genomic sequences (Figure 3I), demonstrating that ns335 is a recessive allele. However, gcy-8(tm949) mutants, lacking the GCY-8 cyclase domain, have no effect on AFD microvilli shape, and suppress kcc-3(ok228) mutant defects (Figure 3B). Thus, gcy-8(ns335) is unlikely to be a loss-of-function allele.
Supporting this idea, while gcy-8(tm949) mutants have near normal thermotaxis behavior on a linear thermal gradient (Wasserman et al., 2011), gcy-8(ns335) mutants accumulate at low temperatures (15°C) regardless of their cultivation temperature (Figure 3J,K). Furthermore, unlike gcy-8(tm949), the gcy-8(ns335) allele does not suppress the AFD NRE shape defects of kcc-3(ok228) (Figure S4B). Thus, gcy-8(ns335) appears to be a gain of GCY-8 function. The recessive nature of gcy-8(ns335) could be explained by the observation that rGC dimerization is essential for function (Potter, 2011). Formation of heterodimeric mutant/wild-type GCY-8 complexes in gcy-8(ns335)/+ heterozygotes would be predicted to reduce cyclase activity. Indeed, expression of a cyclase-dead GCY-8D976A protein in AFD can rescue gcy-8(ns335) microvilli defects (Figure 4A), presumably by promoting non-productive dimer formation as is the case for equivalent mutations in other rGCs (Thompson and Garbers, 1995).
Figure 4. Excess GCY-8 and cGMP cause loss of AFD receptive-ending.
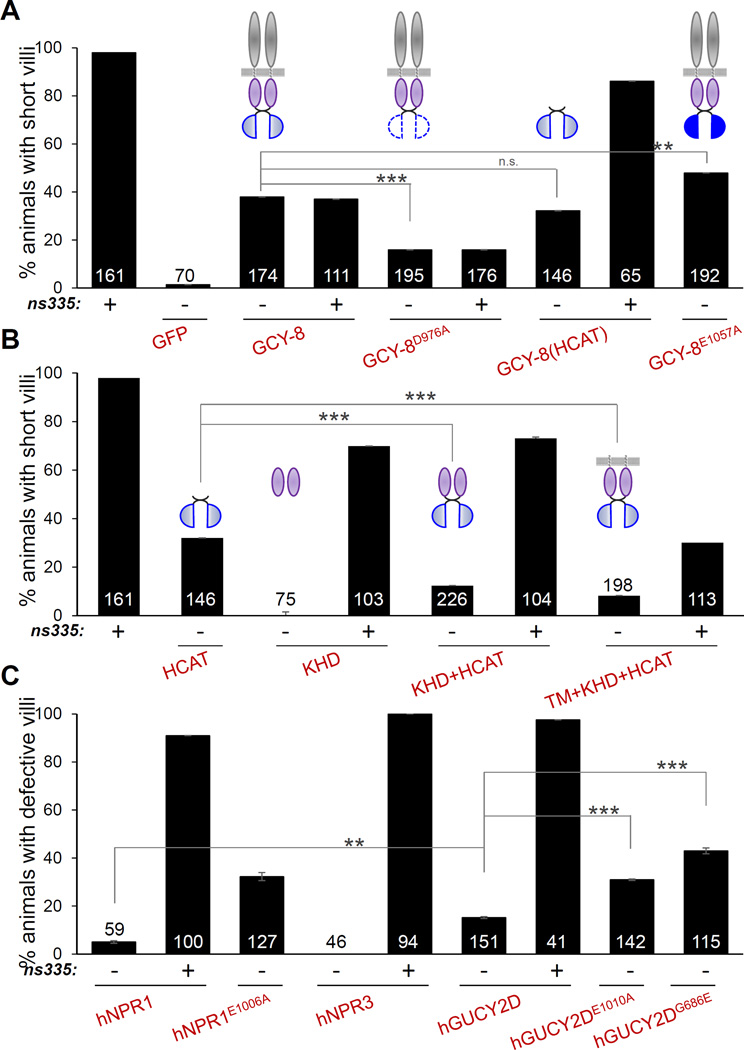
Histogram details are as in Figure 1B. +, −, presence or absence, respectively, of gcy-8(ns335). Diagrams depict overexpressed GCY-8 protein. White-blue, weak-strong predicted cyclase activity (A). **p<0.02. n.s., not significant.
The gcy-8(ns335) mutation results in a G707E substitution in a conserved D(F/H/Y)G motif in the kinase-homology domain (KHD) of GCY-8 (Figures 3A,C; S3A). Deletion of the KHD of the human rGC NPR1 enhances cyclase activity in cell-culture experiments (Koller and Goeddel, 1992). Thus, the gcy-8(ns335) mutation may block the inhibitory effect of the KHD domain on cyclase activity. To understand the role of the KHD, we first developed an in vivo assay for GCY-8 function. Specifically, we found that overexpression of a GCY-8 cDNA in AFD neurons of otherwise wild-type animals using an AFD promoter::gcy-8 cDNA plasmid injected at high copy promotes microvilli loss (Figure 4A). Thus, increased GCY-8 activity leads to AFD NRE loss. Furthermore, expression of full-length GCY-8 containing an E1057A mutation, whose equivalent hyper-activates other rGCs (Wedel et al., 1997), weakly but significantly exaggerated the effects of wild-type GCY-8 overexpression (Figure 4A). We also generated wild-type animals expressing the cyclase-dead GCY-8D976A protein. This single amino-acid substitution curtailed the ability of GCY-8 overexpression to block microvilli growth (Figure 4A).
To test the effect of the KHD domain on cyclase activity, we first showed that overexpressing a fragment of GCY-8 consisting only of the HCAT domain was sufficient to elicit AFD microvilli defects (Figure 4A). Similar constructs for other rGCs have constitutive cyclase activity in cell culture studies (Thompson and Garbers, 1995). As expected if gcy-8(ns335) is a gain-of-function allele, expression of the hyper-activated GCY-8 HCAT fragment fails to rescue gcy-8(ns335) defects (Figure 4A). However, AFD microvilli NRE loss was abrogated if the HCAT domain was fused to the KHD domain, with or without the transmembrane domain (KHD-HCAT or TM-KHD-HCAT) (Figure 4B). Of note, while the TM-KHD-HCAT protein rescues gcy-8(ns335), the KHD-HCAT protein fragment does so significantly less efficiently (Figure 4B), suggesting that membrane localization of GCY-8 is important for its dimerization and cyclase activity regulation. Our results suggest that elevated cyclase activity, effected by inhibition of the KHD domain, blocks AFD NRE extension.
Our findings also suggest that the conserved DFG motif, mutated in gcy-8(ns335) animals, and present in the KHD of all rGCs is likely the relevant element mediating cyclase inhibition by the KHD domain in vivo.
GCY-8 functions downstream of KCl
As is the case for kcc-3 mutants, the effect of gcy-8(ns335) on AFD microvilli structure is largely independent of AFD neuron function, as tax-4 mutations fail to suppress gcy-8(ns335) AFD microvilli defects (Figure 3L). Importantly, dietary supplementation with KCl does not rescue gcy-8(ns335) NRE shape defects (Figure S4B).
Thus, rescue of kcc-3(ok228) NRE shape defects by exogenous KCl is not due to non-specific effects on neuron morphology. Furthermore, GCY-8 functions downstream of KCC-3 and KCl, as would be predicted for a KCC-3 effector.
GCY-8 has a ligand-independent basal cyclase activity inhibited by the DFG motif
Our finding that glial kcc-3 mutant defects can be overcome by inactivating GCY-8 in AFD neurons suggests that GCY-8 may have constitutive cyclase activity that is inhibited by KCC-3. Consistent with this idea, we found that overexpression of either GCY-8 or the orphan human retinal rGC GUCY2D, previously shown to have basal activity (Duda et al., 1996; Shyjan et al., 1992), promotes AFD microvilli loss in wild-type animals (Figure 4C). However, expression of the human rGC NPR-1, which requires ANP peptide for activity, or the rGC scavenger receptor, NPR-3, lacking a cyclase domain, had negligible effects on AFD shape (Figure 4C).
To directly test whether GCY-8 has basal activity, we generated stably-integrated HEK293T cell lines expressing wild-type or mutant versions of hNPR-1 or GCY-8 proteins, and assayed these for steady-state cGMP levels (Figure 5A,B). We draw three important conclusions from these studies. First, while hNPR1 has essentially no basal activity, as previously reported, GCY-8-expressing cells show significant activity, as predicted by our genetic data. Second, consistent with our in vivo studies, GCY-8G707E, which affects the conserved DFG motif in the KHD domain, enhances this basal cyclase activity, similar to or greater than an activating lesion previously described in other rGCs (GCY-8E1057A). Third, the GCY-8G707E-analogous mutation in hNPR1 (hNPR1G680E) also shows increased cyclase activity. Thus, GCY-8 has basal cyclase activity and the glycine residue of the DFG motif is important for cyclase inhibition across rGCs.
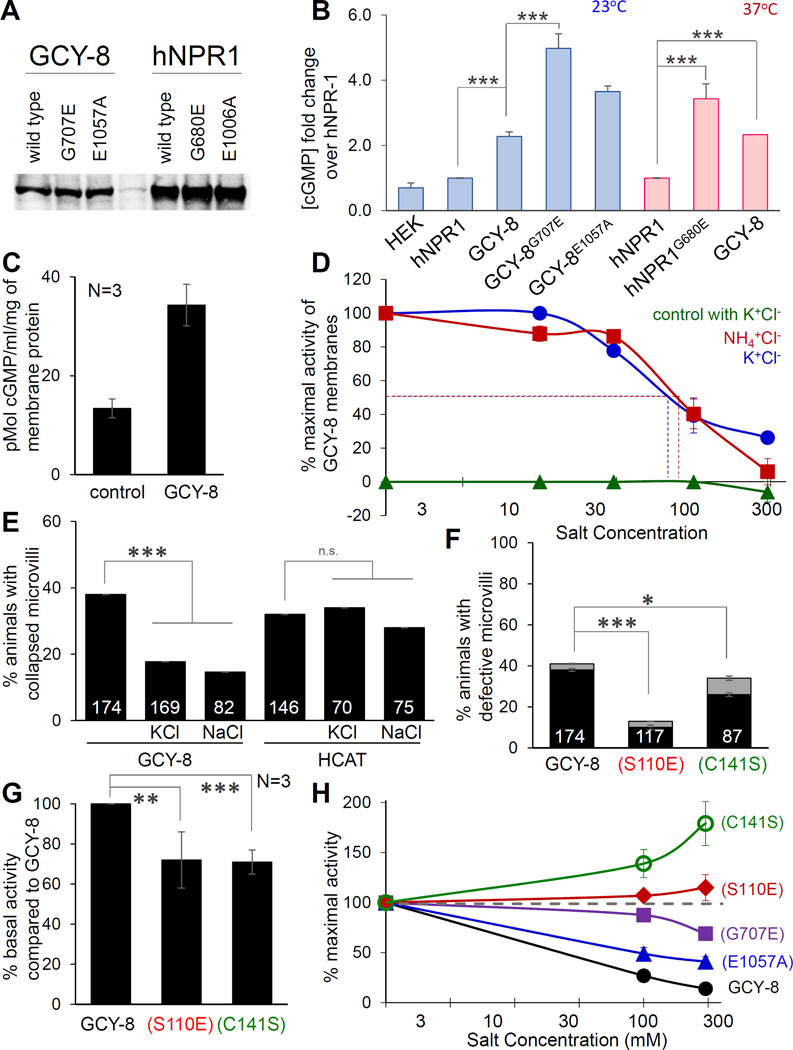
Figure 5. Chloride inhibits GCY-8 cyclase basal activity.
(A) Western blot showing expression of indicated rGC proteins in HEK239T cells. (B) Histogram showing steady-state cGMP levels in HEK239T cells expressing the indicated constructs. Error bars, SEM of 3–4 experiments. ***p<0.0008. (C) cGMP measurements in membrane fractions of control HEK293T cells or GCY-8-expressing HEK293T cells. (D) Inhibition of GCY-8 membrane fraction guanylyl cyclase activity by chloride. Dotted lines, half-maximal effect. X-axis is plotted on a log10 scale. (E, F) Histogram details as in Figure 1B. (F) Mutations engineered in full-length GCY-8 noted in color, see also Figure S5C.*=p<0.03 (G) Histogram details as in Figure 5C. Mutations noted as in Figure 5F. ***p=0.004; **p=0.01 (H) Details as in Figure 5D and 5F. Activity of each protein normalized to basal activity in 0mM NH4+Cl− (dashed grey line). See also Figure S5.
Cl− ions inhibit GCY-8
Our data raise the possibility that KCC-3 may inhibit GCY-8 through the action of K+ or Cl−. To test whether this effect is direct and which ion can inhibit GCY-8 activity, we performed reconstituted membrane fraction assays using the stably integrated HEK293T cell lines described above. As with the whole cell assays, we found a 2.7-fold increase in cGMP production in GCY-8 containing fractions (Figure 5C). Strikingly, we found that Cl− ions, but not K+ or other anions, are potent inhibitors of GCY-8 cyclase activity and cGMP production (Figures 5D, S5A), with an IC50 of ~60mM. This IC50 is within the physiological range for extracellular Cl− concentrations in many settings (Tora et al., 2015), suggesting that changes in extracellular Cl−, as would be predicted to occur in kcc-3 mutants, should indeed affect GCY-8 activity. Consistent with these in vitro results, we found that exogenous supplementation with Cl− mitigates the over-expression defects of GCY-8 in vivo (Figure 5E). Also, previous studies suggest that cGMP levels may be reduced at lower temperatures to allow CNG channels in AFD to close as part of the thermosensory response (Garrity et al., 2010). Thus, defects in inhibition of cGMP production by GCY-8 should be more pronounced at 15°C, which is exactly what we see in null kcc-3 mutants (Figure 1F). Taken together, our results support a model in which KCC-3 regulates extracellular Cl− levels, which, in turn, directly influence GCY-8 guanylyl cyclase activity.
Cl− inhibits GCY-8 by binding to a functionally conserved Cl− binding motif in its ECD
Crystal structures of the rat rGC NPR1 reveal a Cl− in the ECD domain within a pocket defined by the motif S(x)nGPxC (van den Akker et al., 2000; Ogawa et al., 2010). A conserved S(x)nGPxC motif is present in the ECD of GCY-8 (Figure S5B,C), but not in GCY-23, mutations in which suppress kcc-3 lesions only modestly (Figures 3B, S5C). The S(x)nGPxC motif is structurally conserved in the ECD of metabotropic glutamate receptors (mGluRs), and Cl− is an orthosteric ligand for mGluRs (DiRaddo et al., 2015; Tora et al., 2015). Of note, the IC50 for Cl− inhibition of GCY-8 is identical to the Cl− EC50 measured for mGluR2 activity, consistent with a possible role for the GCY-8 S(x)nGPxC ECD motif in mediating the effects of Cl−.
Supporting this idea, exogenous in vivo supplementation with Cl− salts does not suppress the over-expression defects of the HCAT fragment lacking the ECD, but suppresses defect of full-length GCY-8 overexpression (Figure 5E). Furthermore, mutating the conserved Cl− binding domain serine of mGluRs to glutamate mimics the effects of Cl− (Dutzler, 2003; Tora et al., 2015). We found that a similar mutation in GCY-8 (GCY-8S110E) inhibits the ability of GCY-8 to promote NRE involution in vivo (Figure 5F), suggesting that this motif may indeed be functionally conserved. To confirm this, we expressed GCY-8S110E in HEK293T cells and measured basal guanylyl cyclase activity in reconstituted membrane fractions. GCY-8S110E had reduced basal activity compared to GCY-8, as expected of a mutation that mimics binding of an inhibitory Cl− ligand (Figure 5G). Furthermore, GCY-8S110E was insensitive to increasing levels of Cl−, unlike full length GCY-8 (Figure 5H). By contrast, the GCY-8G707E and GCY-8E1057A activated proteins that possess intact ECDs retain Cl− inhibition, but with dampened efficacy compared to full-length GCY-8 (Figure 5H).
Changing the conserved cysteine of the S(x)nGPxC motif to serine enhances Cl− coordination in mGluRs by introducing a second hydroxyl moiety into the Cl− binding pocket (Figure S5B, C) (Tora et al., 2015). We found that the same may be true for the S(x)nGPxC motif of GCY-8. GCY-8C141S slightly reduces the ability of GCY-8 to prevent NRE extension in vivo (Figure 5F), and shows reduced basal activity compared to GCY-8 (Figure 5G) and reduced Clsensitivity in vitro (Figure 5H).
Taken together, these results suggest that Cl− ions directly inhibit GCY-8 activity by binding to the conserved S(x)nGPxC motif in the extra-cellular domain of GCY-8.
Excess cGMP blocks AFD NRE growth
Our studies suggest the hypothesis that increased cGMP production by GCY-8 promotes AFD NRE disappearance. To examine the involvement of cGMP in AFD NRE shape control, we reasoned that GCY-8-independent manipulations that alter cGMP levels within AFD should also alter microvilli morphology. Therefore, we expressed cyclase-activated forms of human NPR-1 (hNPR-1E1006A) and human GUCY2D (hGUCY2DE1010A, a retinal rGC) in the AFD neuron (Wedel et al., 1997). As shown in Figure 4C, transgenic animals display defects in microvilli extension, suggesting that excess cGMP produced by these heterologous guanylyl cyclases indeed blocks AFD NRE growth. Similarly, hGUCY2D protein containing a lesion homologous to the GCY-8G707E lesion (hGUCY2DG686E), also significantly shortens AFD receptive-ending microvilli (Figure 4C).
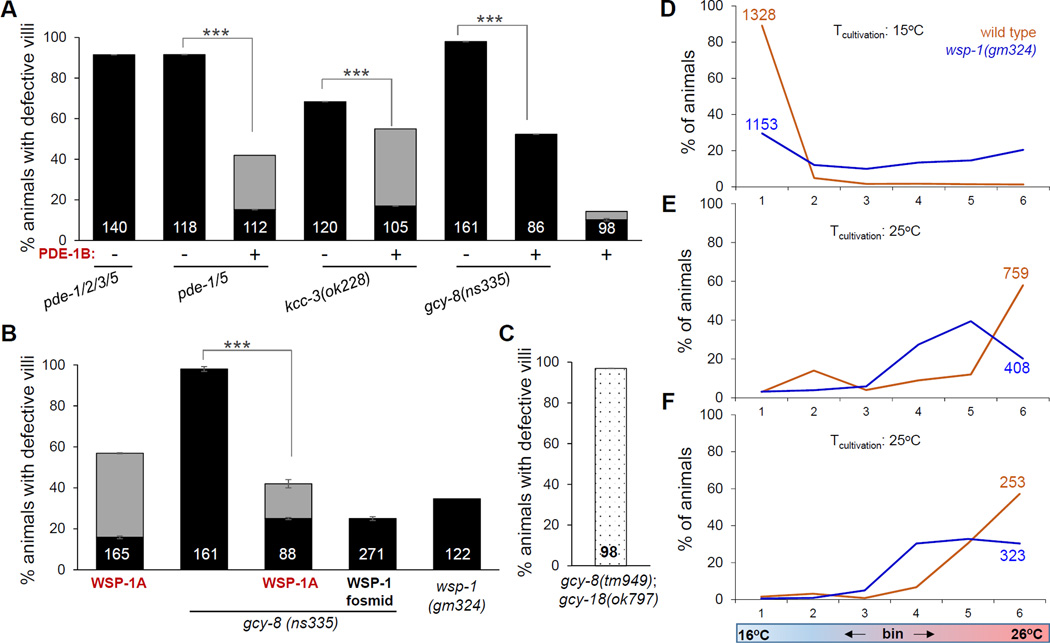
As an alternate means of modulating cGMP levels, we examined the consequences of changing the activities of phosphodiesterase (PDE) genes, encoding proteins that degrade cGMP, on AFD receptive-ending morphology. C. elegans has four such genes (pde-1, -2, -3, -5) (Liu et al., 2010), two of which (pde-2, and -5) are known to be expressed in AFD (Wang et al., 2013). While a pde-1 gene reporter containing limited 5’ regulatory sequences fused to GFP was not previously detected in AFD (Wang et al., 2013), we found that a fosmid derived from genomic DNA surrounding the locus, and recombineered to introduce GFP coding sequences just upstream of the pde-1 stop codon, is expressed in the cell (Figure S6C). Thus, at least 3 PDEs are expressed in AFD.
A quadruple mutant inactivating all cGMP PDE genes showed complete loss of AFD microvilli (Figure 6A). A pde-5 pde-1 double mutant also displayed a fully penetrant loss of AFD receptive-endings (Figure 6A). Single, double, or triple combinations of mutations in the other cGMP PDE genes or in the cAMP-specific PDE gene, pde-4, had no effect on AFD receptive-ending shape (Figure S6A). Thus, PDE-1 and PDE-5 are the only PDEs required for AFD microvilli extension, and appear to function redundantly. We conclude that PDE-1 and PDE-5 likely act cell autonomously in the AFD neuron to regulate receptive-ending shape, and that excess cGMP blocks microvilli extension.
Figure 6. The actin regulator WSP-1 regulates AFD shape downstream of GCY-8.
(A–C) Histogram details as in Figure 1B. (A)+/−, presence or absence, respectively of PDE-1B over-expression (B) Gray, animals with extended microvilli. (C) Dotted, animals with slightly shorter, but not absent, microvilli. (D–F) Thermotaxis assays as in Figure 1G. 24 hours post mid-L4 stage animals raised at 15°C (D) or 25°C (E), and 96 hours post mid-L4 stage animals raised at 25°C (F). See also Figure S6 and S7.
To test whether increasing PDE activity reciprocally promotes AFD microvilli elongation, we examined pde-5 pde-1 double mutants expressing a cDNA derived from the pde-1B mRNA isoform, in AFD (Figure S6B). This transgene strongly rescues the AFD defects of the double mutant (Figure 6A), supporting the idea that PDE-1 and PDE-5 exercise interchangeable cell-autonomous activities required for AFD morphogenesis. Furthermore, PDE-1B overexpression restores AFD NRE microvilli to kcc-3(ok228) and gcy-8(ns335) mutants, consistent with the idea that high cGMP levels block microvilli elongation in these mutants (Figure 6A). Importantly, we noticed that in some animals overexpressing PDE-1B, microvilli are longer than in the wild type, and are sometimes misshapen (Figures 6A, gray bars; S6D,E). Our results suggest, therefore, that cGMP levels are sufficient to dictate the extent of microvilli elongation.
cGMP antagonizes the actin regulator WSP-1 to control AFD receptive-ending shape
Since the effects of cGMP on AFD NRE shape are largely independent of CNG channels, we sought to identify a relevant mediator for shape determination. From a screen of candidate effectors, we found that overexpression in otherwise wild-type AFD neurons of a cDNA corresponding to the gene wsp-1, encoding the well-studied actin regulator NWASP, results in elongated AFD microvilli (Figures 6B, S7A,B), suggesting that WSP-1 promotes microvilli formation. Strikingly, WSP-1 overexpression restores microvilli to gcy-8(ns335) mutants (Figure 6B). A similar result is obtained using a genomic clone carrying the wsp-1 gene (Figure 6B). These observations suggest that high cGMP inhibits microvilli growth by antagonizing WSP-1. Supporting the idea that WSP-1 is normally required for AFD NRE growth, wsp-1(gm324) mutants homozygous for a loss-of-function mutation perturbing the wsp-1A mRNA isoform are defective in AFD microvilli elongation. Moreover, wsp-1(gm324) animals also show defects in thermotaxis behavior at all temperatures and ages tested (Figure 6D–F). Another allele, wsp-1(gk208630), which only affects the wsp-1B isoform, does not affect AFD NREs (Figure S7C,D).
We note that wild-type animals over-expressing WSP-1A occasionally display shorter microvilli (Figure 6B). One explanation for this may be that high levels of WSP-1 target free actin to new filament ends, preventing elongation of existing filaments (Smith et al., 2013a). Consistent with this hypothesis, double mutants between gcy-8(tm949) and gcy-23(ok797) null alleles show a modest reduction in microvilli length, as do some animals overexpressing PDE-1B (Figure 6A,C).
The finding that wsp-1(gm324) animals exhibit a weaker NRE defect than gcy-8(ns335) mutants suggests that a second actin polymerizing factor also functions downstream of cGMP. Importantly, however, the ability of WSP-1 overexpression alone to promote AFD microvilli elongation in wild type animals, and rescue the defects of gcy-8(ns335), demonstrates that actin nucleation in AFD actin-rich microvilli is likely the key step that is being regulated by the glia-neuron interactions we uncovered. We therefore conclude that cGMP antagonizes WSP-1 activity, which likely regulates nucleation of actin filaments in AFD NREs. These results also imply that unexpectedly, we have found that cGMP regulates at least two independent effectors – CNG channels and WSP-1, with different functions in the AFD neuron, and WSP-1 is the key downstream effector for regulation of AFD NRE shape.
DISCUSSION
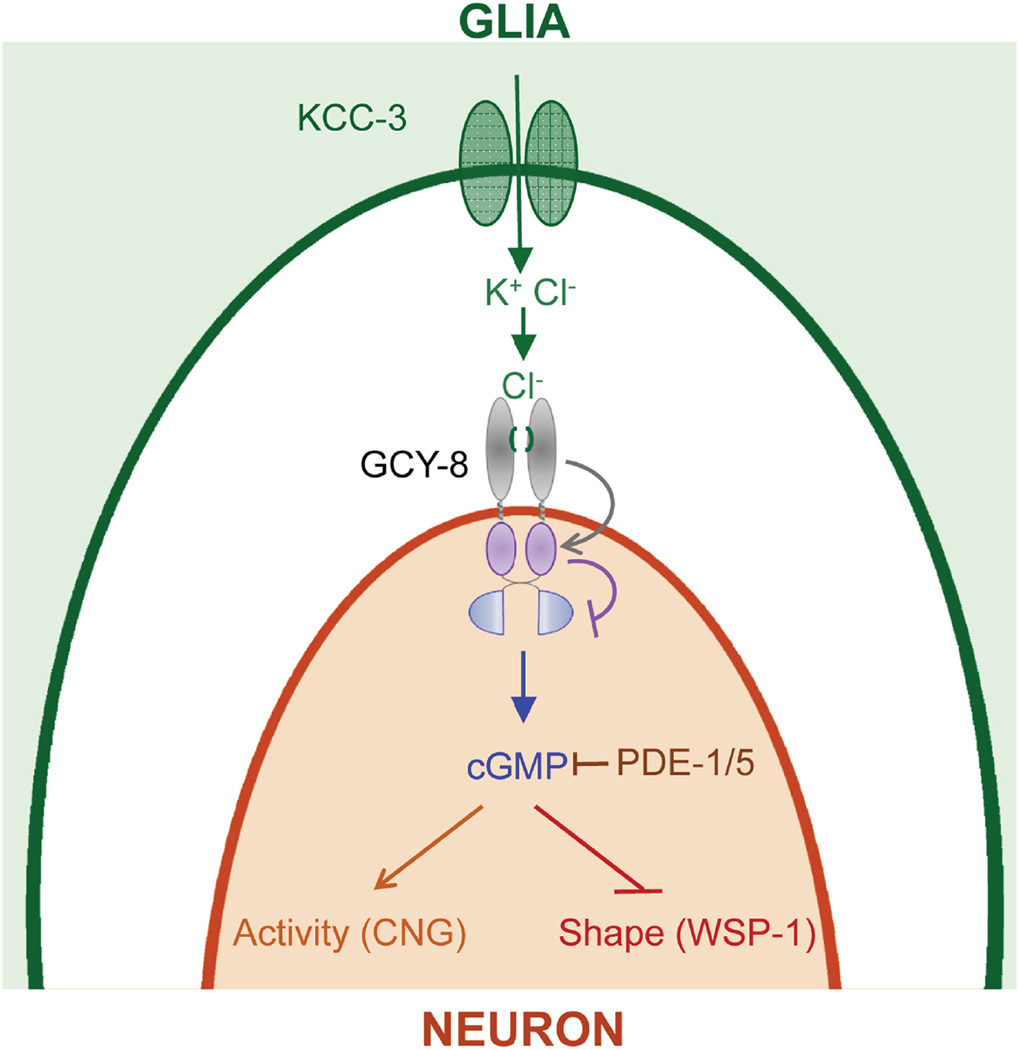
Our studies identify a glial regulator of sensory NRE shape, and reveal that glia can dictate NRE shape by controlling the NRE microenvironment. Our findings support a model (Figure 7) in which the glial K/Cl transporter KCC-3 localizes around and controls Cl− levels surrounding AFD microvilli. Cl− inhibits the AFD-specific rGC GCY-8, which has a conserved Cl− binding structural motif. GCY-8, along with PDE-1 and PDE-5, modulate neuron cGMP levels, and cGMP antagonizes WSP-1, which promotes NRE elongation, presumably through actin nucleation. Importantly, our studies reveal that glia continuously maintain AFD NRE shape, presumably by regulation of the microenvironment.
Figure 7. Model for AFD neuron receptive-ending shape control by AMsh glia.
Magnified view of boxed region in Figure 1A. Glial KCC-3 regulates chloride in the extracellular milieu around AFD receptive-ending. Chloride inhibits GCY-8 by binding the S(x)nGPxC motif in its ECD. GCY-8, PDE-1 and PDE-5 control cGMP levels, which antagonizes the actin cytoskeleton through WSP-1.
Glia discriminate among NREs
Our findings that KCC-3 localizes around AFD NREs and is selectively required for their function, demonstrate that a single glial cell discriminates between neurons with which it associates by targeting regulators to specific neuron-contact sites. Mammalian astrocytes can associate with 100,000 NREs (Chung et al., 2015), and Ca2+ fluxes in astrocytes have localized features (Khakh and Sofroniew, 2015). We suggest that the ability to discriminate between associated neurons may be a universal property of glia. Understanding how KCC-3 localization is achieved may provide a molecular handle on targeted control of neurons by glia.
Glia control sensory and synaptic NRE shape and function
How actin-based microvilli NREs are formed and maintained has not been explored. Our results will likely apply to many such sensory structures. Indeed, glia/glia-like cells regulate the ionic milieu of diverse actin-based sensory NREs, and human disease mutations affect these cells (Estrada-Cuzcano et al., 2012; Hamel, 2007). KCC and NKCC co-transporters are expressed in glia of the ear, retina, and in Schwann cells (Boettger et al., 2002; Kettenmann and Verkhratsky, 2008), and their disruption can lead to NRE degeneration (Gallemore et al., 1997; Strauss, 2005). KCC3 loss in humans leads to sensory neuropathy (Kahle et al., 2015).
rGCs are also widely expressed in sensory and other neuron types, and retinal GUCY2D and Grueneberg ganglion GC-G have basal guanylyl cyclase activities similar to GCY-8 (Chao et al., 2015; Shyjan et al., 1992). Increased GUCY2D activity leads to defects in photoreceptor outer segment shape in Leber’s Congenital Amaurosis with some patient mutations mapping to the ECD (Perrault et al., 2000). These effects are reminiscent of the effects of GCY-8G707E on AFD NREs.
Our work may also present a paradigm for understanding glial effects on spine morphology in the CNS. Astrocytic KCC channels regulate K/Cl levels in the CNS (Kettenmann and Verkhratsky, 2008). Disruption of K/Cl levels contributes to neuronal dysfunction in Huntington’s disease and epilepsy models (Kahle et al., 2015; Tong et al., 2014), and to defects in spine shape (Murmu et al., 2015).
AMsh glia form a bounded compartment around AFD NREs, which, as in synapses, allows tight control of the NRE milieu. A role for extracellular Cl− in modulating spine-localized metabotropic glutamate receptors has been explored (Tora et al., 2015). Our studies suggest that in addition to providing a diffusion barrier, glia may actively control synaptic Cl− levels to modulate spine activity and shape. Supporting this proposition, mGluRs (Class III GPCRs) and rGCs adopt a LIVBP (Leu/IsoLeu/Val binding periplasmic protein-like) fold in the extracellular domain (Acher et al., 2011). The S(x)nGPxC Cl− binding motif is conserved across nervous system LIVBP fold receptors (Acher et al., 2011).
Glial control of thermosensation
Electrophysiological studies of AFD neurons suggest that high/low cGMP opens/closes CNG channels upon warming/cooling (Garrity et al., 2010; Ramot et al., 2008) relative to the cultivation temperature. Thus, within the operating range of the thermosensory apparatus, a basal level of cGMP must exist that can be modulated up or down. That GCY-8 has basal activity supports this idea, as does our observation that kcc-3 mutants exhibit more pronounced thermosensory deficits at low temperature, where higher GCY-8 inhibition may be required.
We previously showed that expression of the VER-1 receptor tyrosine kinase in AMsh glia is temperature dependent and independent of the AFD neuron (Procko et al., 2011). It is possible that modulation of AMsh glia-dependent properties, such as KCC-3 activity, by temperature may also form part of the thermosensory apparatus.
Control of rGC activity
rGCs are prevalent receptors and are important therapeutic targets (Potter, 2011). Our data highlight unexpected aspects of rGC structure and function. That neuronal rGCs (e.g. GCY-8, GUCY2D) have basal activities but non-neuronal rGCs (e.g. NPR-1 and NPR-2) do not, suggests the possibility of a fundamental dichotomy. The identification of Cl− as a GCY-8 inhibitor raises the possibility that rGCs with basal activity, for which activating ligands have not been identified, may instead be regulated by inhibitory extracellular cues. Our results suggest that the relationship between the ECD and KHD may be inverted in rGCs with basal activity vs. ligand-activated non-neuronal rGCs.
That Cl− ions influence GCY-8 activity also suggests that other ion may control rGCs. A recent study exploring rGCs in K+ and I− sensation in C. elegans revealed that stimulus specificity tracks with receptors ECDs (Smith et al., 2013b). Thus, rGCs may sense different ions through their ECD.
Finally, we show that a conserved glycine residue in the KHD has inhibitory roles in GCY-8 and also in human NPR1. The D(F/H/Y)G motif containing this amino acid is conserved in all kinases and pseudo-kinases and forms part of the activation loop. Thus, this domain may facilitate ATP binding, previously suggested to affect rGC activity (Goraczniak et al., 1992).
EXPERIMENTAL PROCEDURES
C. elegans methods
Standard culturing and germ-line transformation methods were used (Brenner, 1974; Mello and Fire, 1995). Mutants, transgenes, genetic methods, RNAi methods and thermotaxis assays are described in Supplemental Methods.
Plasmids
Plasmid construction details are provided in Supplemental Methods.
Microscopy and Image Processing
Images were collected on an Axioplan 2 microscope (Zeiss) with 63x/1.4 NA objective (Zeiss) and dual-band filter set (Chroma, Set 51019). Some images were collected on a DeltaVision Core imaging system (Applied Precision) with a PlanApo 603/1.42 NA or UPLSApo 1003/1.40 NA oil-immersion objective and a Photometrics CoolSnap HQ camera (Roper Scientific). Images were de-convolved using ImageJ. Electron microscopy methods are described in Supplemental Methods.
cGMP ELISAs
ELISAs were performed using the Direct cGMP kit (Enzo Life Sciences, ADI-900-014). 5×105 cells were grown on poly-D-Lysine coated plates (Corning BioCoat, 354414) for 12 hours, rinsed in D-PBS and incubated in serum free medium supplemented with 0.5 mM IBMX at 23°C or 37°C with 5% C02 for one hour. Cells were lysed in 0.1 M HCl + 0.1% Triton X-100 and assayed for cGMP concentration as per manufacturer’s protocol (Enzo Life Sciences) (Guo et al., 2007, 2009). Optical density measurements were performed on a BioTek Synergy NEO using the Gen5 Data Analysis software. All GCY-8 experiments were performed in biological quadruplicate and NPR-1 experiments in biological triplicate. Importantly, the assay measures steady-state accumulation of cGMP (production minus degradation), and is always an underestimate of cGMP production. Membrane assays were carried out as follows: Cells were cultured in growth medium to ~95% confluency and washed in 50 mM NH4Ac and 200 mM sucrose pH 7 (plus 1 mM IBMX and protease inhibitors). Cells were passed through a 26 gauge needle several times and sonicated for 10 seconds. After centrifugation at 2000 rpm for 5 min, membrane fraction was prepared by centrifugation at 100,000 g for 1.5 h at 4°C. The membrane pellet was resuspended in 50 mM NH4Ac, 200 mM Sucrose pH 7, 1 mM DTT, 10 mM MgAc2 and 1 mM MnSO4. Membrane preparations were treated with KCl, KAc, or NH4Cl. 5 mM GTP was added to start the cyclase reaction. After 30 min at 30°C, membrane preparations were lysed in 0.2 M HCl and assayed for cGMP.
Supplementary Material
Acknowledgments
We thank Menachem Katz, Piali Sengupta, Cori Bargmann, Jessica Tanis, and Michael Koelle for reagents; Tarakhovsky lab members for help with cell culture; and Cori Bargmann and Shaham lab members for comments. William J. Rice at the Simons Electron Microscopy Center (NYSBC) helped with the FIB/SEM imaging. AS was an American Cancer Society postdoctoral fellow (PF-13-083-01-DDC), an NIH-T32 institutional postdoctoral fellow (5T32CA967334), and a Murray Foundation fellow. This work was supported in part by NIH grants NS081490 and HD078703 to S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
AS and SS designed experiments and wrote the manuscript. AS performed experiments and analyses except for electron microscopy, performed by YL, and membrane fraction cGMP assays, performed by BL and XYH. CF and JF assisted AS in construct cloning.
REFERENCES
- Acher FC, Selvam C, Pin J-P, Goudet C, Bertrand H-O. A Critical Pocket Close to the Glutamate Binding Site of mGlu Receptors Opens New Possibilities for Agonist Design. Neuropharmacology. 2011;60:102–107. doi: 10.1016/j.neuropharm.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S. Glia are essential for sensory organ function in C. elegans. Science. 2008;322:744–747. doi: 10.1126/science.1163074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger T, Hübner Ca, Maier H, Rust MB, Beck FX, Jentsch TJ. Deafness and renal tubular acidosis in mice lacking the K-Cl co-transporter Kcc4. Nature. 2002;416:874–878. doi: 10.1038/416874a. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Chen C, Lin Y, Breer H, Fleischer J. Receptor guanylyl cyclase-G is a novel thermosensory protein activated by cool temperatures. 2015;34:294–307. doi: 10.15252/embj.201489652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Choi KY, Park CS. A new putative cyclic nucleotide-gated channel gene, cng-3, is critical for thermotolerance in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2004;325:525–531. doi: 10.1016/j.bbrc.2004.10.060. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CCa, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres Ba. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Chung W-S, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease? Nat. Neurosci. 2015;18:1539–1545. doi: 10.1038/nn.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife. 2014;3:e01948. doi: 10.7554/eLife.01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T, Goraczniak R, Surgucheva I, Rudnicka-Nawrot M, Gorczyca Wa, Palczewski K, Sitaramayya A, Baehr W, Sharma RK. Calcium modulation of bovine photoreceptor guanylate cyclase. Biochemistry. 1996;35:8478–8482. doi: 10.1021/bi960752z. [DOI] [PubMed] [Google Scholar]
- Dutzler R. Gating the Selectivity Filter in ClC Chloride Channels. Science (80-.) 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- Estrada-Cuzcano A, Roepman R, Cremers FPM, Hollander AIDen, Mans Da. Non-syndromic retinal ciliopathies: Translating gene discovery into therapy. Hum. Mol. Genet. 2012;21 doi: 10.1093/hmg/dds298. [DOI] [PubMed] [Google Scholar]
- Gallemore RP, Hughes Ba, Miller SS. Retinal pigment epithelial transport mechanisms and their contributions to the electroretinogram. Prog. Retin. Eye Res. 1997;16:509–566. [Google Scholar]
- Garrity Pa, Goodman MB, Samuel AD, Sengupta P. Running hot and cold: behavioral strategies, neural circuits, and the molecular machinery for thermotaxis in C. elegans and Drosophila. Genes Dev. 2010;24:2365–2382. doi: 10.1101/gad.1953710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraczniak RM, Duda T, Sharma RK. A structural motif that defines the ATP-regulatory module of guanylate cyclase in atrial natriuretic factor signalling. Biochem. J. 1992;282(Pt 2):533–537. doi: 10.1042/bj2820533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Tan YC, Wang D, Madhusoodanan KS, Zheng Y, Maack T, Zhang JJ, Huang XY. A Rac-cGMP Signaling Pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Zhang JJ, Huang XY. Stimulation of guanylyl cyclase-D by bicarbonate. Biochemistry. 2009;48:4417–4422. doi: 10.1021/bi900441v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel CP. Cone rod dystrophies. Orphanet J. Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H, Ito H, Satterlee J, Sengupta P, Matsumoto K, Mori I. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics. 2006;172:2239–2252. doi: 10.1534/genetics.105.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007:1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Khanna AR, Alper SL, Adragna NC, Lauf PK, Sun D, Delpire E. K-Cl cotransporters, cell volume homeostasis, and neurological disease. Trends Mol. Med. 2015;21:513–523. doi: 10.1016/j.molmed.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Verkhratsky A. Neuroglia: the 150 years after. Trends Neurosci. 2008;31:653–659. doi: 10.1016/j.tins.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Khakh BS, Sofroniew MV. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015;18:942–952. doi: 10.1038/nn.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller KJ, Goeddel DV. Molecular biology of the natriuretic peptides and their receptors. Circulation. 1992;86:1081–1088. doi: 10.1161/01.cir.86.4.1081. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- Kremer H, van Wijk E, Märker T, Wolfrum U, Roepman R. Usher syndrome: Molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet. 2006;15:262–270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- Liu J, Ward A, Gao J, Dong Y, Nishio N, Inada H, Kang L, Yu Y, Ma D, Xu T, et al. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat. Neurosci. 2010;13:715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995 doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat. Neurosci. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- Murmu RP, Li W, Szepesi Z, Li J-Y. Altered Sensory Experience Exacerbates Stable Dendritic Spine and Synapse Loss in a Mouse Model of Huntington’s Disease. J. Neurosci. 2015;35:287–298. doi: 10.1523/JNEUROSCI.0244-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones Ka, VanLeeuwen J-E, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins La, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S, Ghazi I, Ducroq D, Souied E, Leowski C, Bonnemaison M, Dufier JL, Munnich a, et al. Spectrum of retGC1 mutations in Leber’s congenital amaurosis. Eur. J. Hum. Genet. 2000;8:578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- Potter LR. Regulation and therapeutic targeting of peptide-activated receptor guanylyl cyclases. Pharmacol. Ther. 2011;130:71–82. doi: 10.1016/j.pharmthera.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Lu Y, Shaham S. Glia delimit shape changes of sensory neuron receptive endings in C. elegans. Development. 2011;138:1371–1381. doi: 10.1242/dev.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot D, MacInnis BL, Goodman MB. Bidirectional temperature-sensing by a single thermosensory neuron in C. elegans. Nat. Neurosci. 2008;11:908–915. doi: 10.1038/nn.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JM. Sodium-Potassium-Chloride Cotransport. 2000;80:211–276. doi: 10.1152/physrev.2000.80.1.211. [DOI] [PubMed] [Google Scholar]
- Satoh a, Tokunaga F, Kawamura S, Ozaki K. In situ inhibition of vesicle transport and protein processing in the dominant negative Rab1 mutant of Drosophila. J. Cell Sci. 1997;110(Pt 2):2943–2953. doi: 10.1242/jcs.110.23.2943. [DOI] [PubMed] [Google Scholar]
- Satterlee JS, Sasakura H, Kuhara A, Berkeley M, Mori I, Sengupta P. Specification of thermosensory neuron fate in C. elegans requires ttx-1, a homolog of otd/Otx. Neuron. 2001;31:943–956. doi: 10.1016/s0896-6273(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Shaham S. Chemosensory organs as models of neuronal synapses. Nat. Rev. Neurosci. 2010;11:212–217. doi: 10.1038/nrn2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyjan aW, de Sauvage FJ, Gillett Na, Goeddel DV, Lowe DG. Molecular cloning of a retina-specific membrane guanylyl cyclase. Neuron. 1992;9:727–737. doi: 10.1016/0896-6273(92)90035-c. [DOI] [PubMed] [Google Scholar]
- Smith Ba, Padrick SB, Doolittle LK, Daugherty-Clarke K, Corrêa IR, Xu MQ, Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife. 2013a;2013:1–25. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HK, Luo L, O’Halloran D, Guo D, Huang X-Y, Samuel ADT, Hobert O. Defining specificity determinants of cGMP mediated gustatory sensory transduction in Caenorhabditis elegans. Genetics. 2013b;194:885–901. doi: 10.1534/genetics.113.152660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The Retinal Pigment Epithelium in Visual Function. Physiol. Rev. 2005:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Tan PL, Barr T, Inglis PN, Mitsuma N, Huang SM, Garcia-Gonzalez MA, Bradley BA, Coforio S, Albrecht PJ, Watnick T, et al. Loss of Bardet Biedl syndrome proteins causes defects in peripheral sensory innervation and function. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17524–17529. doi: 10.1073/pnas.0706618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis JE, Bellemer A, Moresco JJ, Forbush B, Koelle MR. The potassium chloride cotransporter KCC-2 coordinates development of inhibitory neurotransmission and synapse structure in Caenorhabditis elegans. J. Neurosci. 2009;29:9943–9954. doi: 10.1523/JNEUROSCI.1989-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DK, Garbers D. Dominant negative mutation of the guanylyl cyclase-A receptor. J. Biol. Chem. 1995 doi: 10.1074/jbc.270.1.425. [DOI] [PubMed] [Google Scholar]
- Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, Anderson Ma, Mody I, Olsen ML, Sofroniew MV, et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat. Neurosci. 2014;17:694–703. doi: 10.1038/nn.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tora AS, Rovira X, Dione I, Bertrand H-O, Brabet I, De Koninck Y, Doyon N, Pin J-P, Acher F, Goudet C. Allosteric modulation of metabotropic glutamate receptors by chloride ions. FASEB J. 2015;29:4174–4188. doi: 10.1096/fj.14-269746. [DOI] [PubMed] [Google Scholar]
- Wang D, O’Halloran D, Goodman MB. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J. Gen. Physiol. 2013;142:437–449. doi: 10.1085/jgp.201310959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J. Comp. Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Wasserman SM, Beverly M, Bell HW, Sengupta P. Regulation of response properties and operating range of the AFD thermosensory neurons by cGMP signaling. Curr. Biol. 2011;21:353–362. doi: 10.1016/j.cub.2011.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel BJ, Foster DC, Miller DE, Garbers DL. A mutation of the atrial natriuretic peptide (guanylyl cyclase-A) receptor results in a constitutively hyperactive enzyme. Proc. Natl. Acad. Sci. U. S. A. 1997;94:459–462. doi: 10.1073/pnas.94.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.