Abstract
MicroRNA expression profiling in human liver progenitor cells following hepatocytic differentiation identified miR-122 and miR-194 as the microRNAs most strongly upregulated during hepatocytic differentiation of progenitor cells. MiR-194 was also highly upregulated following hepatocytic differentiation of human embryonic stem cells (hESCs). Overexpression of miR-194 in progenitor cells accelerated their differentiation into hepatocytes, as measured by morphological features such as canaliculi and expression of hepatocytic markers. Overexpression of miR-194 in hESCs induced their spontaneous differentiation, a phenotype accompanied with accelerated loss of the pluripotent factors OCT4 and NANOG and decrease in mesoderm marker HAND1 expression. We then identified YAP1 as a direct target of miR-194. Inhibition of YAP1 strongly induced hepatocytic differentiation of progenitor cells and YAP1 over expression reversed the miR-194-induced hepatocytic differentiation of progenitor cells. In conclusion, we identified miR-194 as a potent inducer of hepatocytic differentiation of progenitor cells and further identified YAP1 as a mediator of miR-194's effects on hepatocytic differentiation and liver progenitor cell fate.
Keywords: miR-194, differentiation, hepatocytes, progenitor cells, YAP1
Introduction
Liver diseases, which are associated with liver function loss, are a common cause of mortality worldwide. Orthotopic liver transplantation has been the only effective treatment for patients with end-stage liver diseases [1]. However, this method of treatment is limited by the shortage of donor organs. Thus, the development of alternative methods is necessary for treating life-threatening liver diseases. Alternative methods such as cell transplantation and other cell-based therapies have great potential for improving the lives of patients with liver failure [2]. Establishing a readily available source of stem or liver progenitor cell-derived hepatocytes would be invaluable for cell-based liver development therapies. Hepatic progenitor cells that reside in the liver have been shown to have the capacity to restore liver functions [3] and non-hepatocyte sources such as embryonic stem cells (ESCs) have been proposed as an alternate for generation of hepatocytes [4]. These reports have demonstrated their ability to express a hepatocyte-like phenotype under specific in vitro growth conditions and in animal models.
MicroRNAs (miRNAs) are small non-coding RNAs between 21–25 nucleotides long that can silence cognate target genes by specifically binding and cleaving messenger RNAs or by inhibiting their translation [5]. The interaction between a miRNA and its target mRNA does not require perfect complementarity. Hence, a single miRNA has the potential to regulate multiple target mRNAs [6]. More than 2500 unique mature human miRNAs have been identified so far (http://microrna.sanger.ac.uk/sequences/). It is estimated that more than one-third of human protein-coding genes are subjected to regulation by miRNAs [7]. MiRNAs are involved in a variety of biological processes, including developmental timing, embryogenesis, organogenesis, and differentiation of stem cells and progenitor cells [8]. Spectrums of miRNA expression profiling in human embryonic stem cells (hESCs) and ESC-derived embryoid bodies have been well described [9, 10]. In addition, there are several reports showing the importance of specific miRNAs during hematopoiesis [11], neuronal differentiation [12] and skin stem cell differentiation [13]. MiRNAs have also been recognized as key regulators in liver development. For instance, miR-30a is required for bile duct development in zebrafish [14]. MiR-23b cluster miRNAs (miR-23b, 27b, and 24-1), repress bile duct gene expression in fetal hepatocytes [15]. MiR-122, the most abundant miRNA in the liver accounting for approximately 70% of total miRNAs [16], and is required for proper progression of hepatocyte differentiation [17-19].
In the present study, we wished to identify miRNAs other than miR-122 that regulate hepatocytic differentiation. To that end, we used two cell models: the HepaRG cells that display potent hepatocytic differentiation-inducible properties sharing similar features with liver progenitor cells [20-22] and the pluripotent human embryonic stem cell line H9 [23, 24].
Materials and Methods
Cell Culture and Hepatocytic Differentiation
HepaRG cells were cultured in William's E medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Sigma), 100 units/mL penicillin, 100 μg/mL streptomycin (Invitrogen), 5 μg/mL insulin (Sigma), and 5 × 10-5 mol/L hydrocortisone hemisuccinate (Sigma). To induce HepaRG differentiation, a two-step procedure was used as previously described [20-22]. Briefly, cells (1.5 × 105) were maintained for two weeks in complete medium. Then, the culture medium was supplemented with 1% DMSO (Sigma) and 20 ng/mL epidermal growth factor (EGF; Peprotech) for two additional weeks. The medium was renewed every 2 or 3 days. Cells were harvested at 2, 14, and 28 days after seeding. Cell culture pictures were taken using a phase-contrast microscope (Leica) and bile canaliculi (refringent area) at the intersection of two or three hepatocyte-like cells were counted [20]. The hESC line WA-09 (H9) was cultured on hESC qualified Matrigel (BD Biosciences) in mTeSR1 media (Stemcell Technologies). The medium was changed daily, and cells were passaged every 4–6 days with 1 mg/ml Dispase (Stemcell Technologies). For directed differentiation of hESCs toward a hepatocyte fate, the hESCs were cultured in differentiation medium as described previously [23, 24]. Briefly, cultured hESCs were disassociated with Accutase (Stemcell Technologies) and plated on matrigel in mTeSR1 with 10uM ROCK inhibitor Y-27632 (Stemgent) at 90% confluency. Differentiation was initiated by culture for 2 days with 100 ng/ml Activin A (R&D Systems), 10 ng/ml BMP4 (R&D Systems) and 20 ng/ml FGF2 (Peprotech) followed by 3 days with only 100 ng/ml Activin A in RPMI 1640 medium (Invitrogen) supplemented with B27 minus Insulin (Invitrogen) under ambient oxygen / 5% CO2, 5 days with 20 ng/ml BMP4 (Peprotech) / 10 ng/ml FGF2 (Invitrogen) in RPMI/B27 under 4% O2 / 5% CO2, then 5 days with 20 ng/ml HGF (Peprotech) in RPMI/B27 under 4% O2 / 5% CO2, and finally for 5 days with 20 ng/ml Oncostatin-M (R&D Systems) in Hepatocyte Culture Media (Lonza) supplemented with SingleQuots (without EGF) in ambient oxygen / 5% CO2.
RNA isolation, MicroRNA Expression Profiling and Quantitative PCR
Total RNA was isolated using miRNeasy extraction Kit (Qiagen). The GeneChip miRNA 1.0 array (Affymetrix) was used for miRNA expression profiling using total RNA from HepaRG cells at the proliferative (day 2) and differentiated (day 28) stages. Total RNA (1 μg) from each sample collected from three independent differentiation experiments was labeled with biotin using the FlashTag Biotin RNA Labeling Kit (Genisphere, Hatfield, PA). Array hybridization, washing, and scanning of the slides were carried out according to Affymetrix's recommendations. Data were extracted from the images, quantile-normalized, summarized (median polish), and log2-transformed with the miRNA QC software from Affymetrix. To quantify levels of mature microRNAs, total RNA was reverse-transcribed using the TaqMan microRNA reverse transcription kit (Applied Biosystems) and reaction was performed using TaqMan primers in a universal PCR master mix in a ViiA7 Real-Time PCR System (Applied Biosystems). To quantify gene expression, RNA samples were submitted to reverse transcription and real-time PCR using primers listed in Supporting Information Table S1. cDNA was amplified using the CFX Connect Real-Time System (Bio-Rad) and iTaq SYBR Green Supermix (Bio-Rad) and data were analyzed using Bio-Rad CFX Manager software (version 2.1). The relative quantification of miRNA or mRNA expression was calculated using the 2−ΔΔCt method.
MiRNA Target Gene Prediction
For prediction of target genes of miR-194, we used the miRWalk website (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), a comprehensive database that provides predicted as well as validated miRNA-binding site information on miRNAs for human, mouse and rat [25]. Ten established miRNA-target prediction programs (miRWalk, RNA22, miRanda, miRDB, TargetScan, RNAhybrid, PITA, PICTAR 4, PICTAR 5, and Diana-microT) were used. The p-values were automatically calculated by the miRWalk algorithm using a probability distribution of random matches of a subsequence in the given sequence.
Establishment of MiR-194 Expressing Cell Lines
HepaRG cells (1.5 × 105) were seeded in a 60 mm dish and the following day, were infected with empty or miR-194 lentiviral particles (SMARTchoice Lentiviral Human CMV/TurboGFP shMIMIC; Thermo Scientific) at a multiplicity of infection of 5. At 48 h post-infection, GFP fluorescent cells were detected by fluorescent microscopy and puromycin (Invitrogen) was added to the medium (1 μg/ml). The medium was changed every 2 days, and puromycin-resistant cells were selected and expanded. hESCs were maintained as feeder-free cultures on hESC qualified Matrigel (BD Biosciences) in mTeSR1. The day before infection, hESCs were plated into 6-well dishes (Matrigel coated) at 30∼40% confluency. hESCs were infected with empty or miR-194 lentiviral particles (SMARTchoice Lentiviral Human CMV/TurboGFP shMIMIC; Thermo Scientific) by spin innoculation at 2,500 rpm at 37 °C for 90 min, and incubated at 37 °C overnight prior to fresh medium replacement. Medium was changed daily and at 48 h post-infection, puromycin (Invitrogen) was added to the medium (1 μg/ml). After 2-3 days, puromycin selected cells were plated into Matrigel coated dishes at limiting cell dilutions and incubated with higher concentration of puromycin (2-5 μg/ml). Single cells with GFP fluorescent that grew into individual colonies were removed from the dish by cell scraping and transferred into a new Matrigel coated plate. Each cell line was then expanded and maintained in mTeSR1 with puromycin. Empty or miR-194 hESC clones were passaged every 4–6 days with 1 mg/ml Dispase (Stemcell Technologies). For undirected differentiation, cells were incubated with MEF conditioned medium composed of DMEM/F12 supplemented with 20% knockout serum replacement, 1 mM L-glutamine, 1% nonessential amino acids (Invitrogen), and 0.1 mM 2-mercaptoethanol (Sigma) (without additional FGF). Samples were collected every 5 days for 20 days.
Lentivirus Transduction and Cell Infection
293T human embryonic kidney (HEK) cells were maintained in DMEM medium (Invitrogen) supplemented with 10% FBS (Sigma), 100 units/mL penicillin, 100 μg/mL streptomycin (Invitrogen). Cells (3.5 × 106) were plated in 10 cm dishes and co-transfected the next day with lentiviral vectors (pGIPZ-YAP1 shRNA, pGIPZ-YAP1 ORF, or Empty control; GE healthcare), pCMV-ΔR8.2 and pCMV-VSV-G (ratio: 1:1:0.1) using Lipofectamine 2000 transfection reagent (Invitrogen). After 16h, 10 ml of fresh culture medium was added. Supernatants containing lentiviral particles, harvested at 48 h and 72 h, were passed through a 0.45 μm filter, centrifuged in 4000 × g for 5 min to remove cellular debris and stored at −80 °C. For transduction of cells with the YAP1 shRNA and YAP1 ORF encoding lentiviral particles, cells (2 × 105) were plated in 60 mm dishes and transduced with lentivirus-containing supernatant in the presence of polybrene (1 μg/ml). Each cell line was then expanded and maintained in medium with puromycin (1 μg/ml) and/or blasticidin (5 μg/ml).
Western Blot Analysis
Cells were washed with ice-cold PBS and cell lysates were prepared in RIPA buffer (Thermo Scientific) containing protease inhibitors (Thermo Scientific). Cell lysates were passed through 1 mL needle syringe to facilitate the disruption of the cell membranes and centrifuged at 14,000 rpm for 15 min at 4°C. Equal amounts of proteins from the supernatants were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with 5% skim milk in TBST and incubated with the following antibodies: anti-Albumin (R&D Systems), anti-Actin (Cell signaling), anti-α-tubulin (Santa Cruz), anti-OCT4 (Santa Cruz), anti-YAP1 (Novus Biologicals) and anti-phospho-YAP1 (Cell Signaling). The ClarityTM western ECL substrate kit (Bio-Rad) was used to detect bound antibodies. The signal was detected using ChemiDocTM XRS+ imaging system (Bio-Rad).
Transient Transfection and Luciferase Assay
For transient transfection, the mirVana™ negative control, mirVana™ miRNA mimic (miR-194-5p) and mirVana™ miRNA inhibitor (antisense miR-194-5p inhibitor) were purchased from Ambion Inc. HepaRG Cells were transfected with miR-194 or negative control using Lipofectamine™ 2000 (Invitrogen). After 6 h of transfection, media was replaced with fresh William's E medium (Invitrogen) containing 10% FBS (Sigma). Cells were seeded into 24-well plates one day before transfection, and then co-transfected with 200 ng of pEZX-MT06 vectors, which harbored YAP1 3′UTR construct (GeneCopoeia), and miR-194 (50 nM or 100 nM), antisense miR-194 inhibitor (50 nM or 100 nM) or 100 nM of negative control. Luciferase assay was performed 48 h after transfection by the Luc-Pair miR luciferase assay kit (GeneCopoeia). For each sample, firefly luciferase activity was normalized to Renilla luciferase activity.
Immunofluorescence Staining
Cells were washed with PBS, fixed with 4% paraformaldehyde solution for 10 min at room temperature, rinsed with PBS, and permeabilized with 0.2% Triton X-100 for 15 min. After blocking unspecific binding sites with 4% bovine serum albumin in PBS, cells were washed three times, incubated overnight at 4°C with ALB (R&D Systems), Ki-67 (Abcam) and SOX2 (R&D Systems) antibodies (1:100), and treated with Alexa Fluor 488, Alexa Fluor 555 or Alexa Fluor 594-conjugated secondary antibodies (1:500, Invitrogen) for 1 h at room temperature. Cells were then washed with PBS, counterstained with DAPI (Sigma) to visualize nuclei (1:5000), and mounted using Gel/Mount solution (Biomeda). All fluorescence images were obtained using a confocal microscope (Zeiss) and analyzed using AxioVision software.
Enzyme-Linked Immunosorbent Assay (ELISA) for Albumin Secretion
During hepatocytic differentiation, the culture medium was replaced with fresh medium every 2 days, and supernatants were collected 24 h after replacing the medium. Albumin secretion in cultured medium was determined using a human albumin ELISA kit (Abcam) according to the manufacturer's instructions. Briefly, each standard (serially diluted), and samples were loaded into the wells and incubated for 1 h followed by the addition of a biotinylated human albumin detection antibody. The streptavidin-peroxidase substrate solution was added for 30 min and the wells were incubated with the chromogen substrate for 20 min. Color reaction was stopped by the stop solution. Absorbance was measured at 450 nm using a microplate reader (Molecular Devices). The amount of albumin released was normalized to the total number of cells in each well.
Measurement of CYP3A4 activity
To check the induction of cytochrome P450 activities upon hepatocytic differentiation, we used the P450-Glo™ CYP3A4 Assay (Luciferin-PFBE) Kit (Promega). Medium was changed with the appropriate luminogenic CYP substrates (Luciferin-PFBE for CYP3A4). Cells were incubated at 37°C for 4 h, and then the supernatants were mixed with equal amount of Detection Reagent, according to the manufacturer's instructions. Luminescence was measured using GloMax 96 microplate luminometer (Promega). Cell number was calculated using CellTiter-Glo Luminescent Cell Viability Assays (Promega) to normalize P450-Glo assay values to cell counts.
Statistical Analysis
Data are expressed as mean ± SEM. Statistical comparisons were made between two groups with Student's t-test. A value of p < 0.05 was considered significant unless otherwise described.
Results
Upregulation of MiR-194 upon Hepatocytic Differentiation of HepaRG Liver Progenitor Cells
The human liver progenitor HepaRG cells were induced to differentiate into morphologically and functionally mature hepatocyte-like cells. Two days after seeding, HepaRG cells showed typical epithelial morphology. The emergence of two cell types, hepatocyte-like and biliary-like cells, was observed at 14 days while morphologically distinct hepatocyte-like cells were seen at day 28 (Fig. 1A). Differentiated HepaRG cells showed features of normal hepatocytes with refractile cellular borders, clearly delineated nuclei and tridimensional polarization with the appearance of refringent circular canaliculi. The differentiation status was validated by measuring the expression of hepatocyte markers, hepatocyte nuclear factor 4 alpha (HNF4A), aldolase B (ALDOB), and Cytochrome P450 3A4 (CYP3A4). HNF4A, ALDOB, and CYP3A4 expression was significantly upregulated upon hepatocytic differentiation of the HepaRG cells (Fig. 1B). HNF4A mRNA showed an early increase at day 14 (48.4 fold) and further increase at day 28 (584.8 fold). ALDOB showed a similar pattern of upregulation (2.4 and 15.8 fold at day 14 and day 28, respectively) while CYP3A4, a marker of late hepatocytic maturation, strongly increased at day 28 (162.5 fold). Differentiation was also evaluated morphologically by determining the accumulation of bile canaliculi at the intersection of three hepatocyte-like cells (Fig. 1C). While no canaliculi were detected at the proliferative stage, an average of 4,257 canaliculi/cm2 was detected at day 14, increasing to 9,109 canaliculi/cm2 at day 28. To identify miRNAs that are regulated upon hepatocytic differentiation of HepaRG cells, we performed miRNA expression profiling analysis using RNA extracted from HepaRG cells at the proliferative (day 2) and differentiated (day 28) stages. Finally, functional assays such as albumin (ALB) secretion and CYP3A4 activity, were performed to further confirm the hepatocyte differentiation status of the HepaRG cells at day 28 (Fig. 1D). Expression of 52 miRNAs was changed by at least two-fold upon hepatocytic differentiation of HepaRG cells (Supporting Information Table S2). Among them, the hepatocyte specific miRNA miR-122, was the most strongly upregulated miRNA (∼506.7-fold; p=0.005), therefore validating the differentiation status of the cells and the miRNA profiling approach. The second most strongly upregulated microRNA was miR-194 (∼459.8-fold; p=0.008). Increased expression of miR-122 and miR-194 upon hepatocytic differentiation of HepaRG cells was further confirmed by qRT-PCR analysis (Fig. 1E). MiR-122 increased by 35.1 fold (p=0.034) and 1,859.8-fold (p=0.001) at day 14 and day 28, respectively and miR-194 increased by 7.2-fold (p=0.003) and 50.4-fold (p<0.0001) at day 14 and day 28, respectively.
Figure 1.
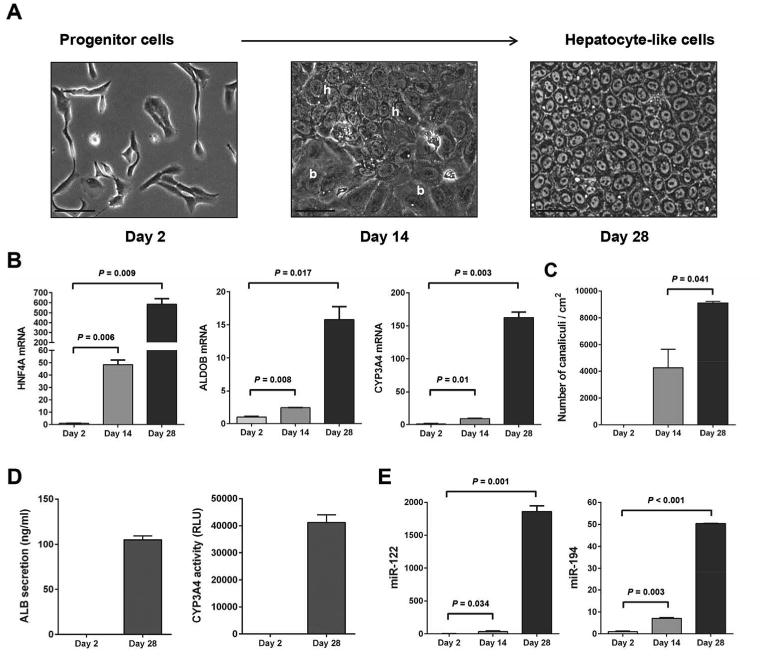
Upregulation of miR-194 upon hepatocytic differentiation of HepaRG liver progenitor cells. HepaRG cells were induced to differentiate to hepatocyte-like cells as described in Materials and Methods. (A): Pictures were taken at different phases of the culture: progenitor cells (day 2), intermediate (Day 14), and hepatocyte-like cells (day 28). h, hepatocyte-like areas; b, biliary-like areas. Bars, 50 μm. (B): Levels of HNF4A, ALDOB and CYP3A4 mRNAs during hepatocytic differentiation of HepaRG cells measured by quantitative PCR and shown as fold-changes and SEM, at day 14 and day 28 relative to day 2, in two independent experiments. (C): Mean bile canaliculi densities from three randomly selected fields in two independent experiments. (D): Graphs show quantification of secreted human albumin and CYP3A4 activity in HepaRG cells at day 2 and day 28 of the hepatocytic differentiation protocol. (E): Levels of miR-122 and miR-194 during hepatocytic differentiation of HepaRG cells measured by quantitative PCR and shown as fold-changes and SEM, at day 14 and day 28 relative to day 2, in two independent experiments.
Upregulation of MiR-194 upon Directed Hepatocytic Differentiation of hESCs
To generalize our finding, we generated hepatocyte-like cells from H9 human embryonic stem cells (hESCs). The generation of hepatocyte-like cells from hESCs followed five distinct stages: pluripotent hESCs (day 0), definitive endoderm cells (day 5), hepatic progenitor cells (day 10), immature hepatocytes (day 15), and mature hepatocytes (day 20) (Fig. 2A). As for the HepaRG cell differentiation, the extent of hepatocytic differentiation of hESCs was evaluated by measuring the expression of the hepatocyte markers HNF4A, ALDOB and CYP3A4 (Fig. 2B). The expression of HNF4A was strongly increased at day 10 corresponding to hepatic progenitor cells (1,191.5-fold) and further increased at day 15 corresponding to hepatocytes (1,931.2-fold). ALDOB expression also increased at day 10, the liver progenitor cell stage (7.2-fold) and further increased at day 15 in hepatocytes (10.7-fold). CYP3A4 specifically increased at day 20 corresponding to mature hepatocytes (400.1-fold). The expression pattern of these three hepatocyte markers is therefore similar to the pattern observed during the differentiation of HepaRG cells. To further confirm the hepatocytic differentiation of hESCs, protein expression levels of the stem cell marker OCT4 and of ALB were measured by western blot analysis (Fig. 2C). OCT4 was strongly expressed at day 0 corresponding to embryonic stem cells but was not detected at the other stages. ALB protein was detected at day 15 and further increased at day 20 in hepatocyte-like cells. Consistently, a similar expression pattern of stem cell marker SOX2 and ALB was observed using immunofluorescence staining (Fig. 2D). The hepatocyte differentiation status was further confirmed by functional assays including ALB secretion and CYP3A4 activity (Fig. 2E). We then measured miR-194 expression in these hESCs at different stages of differentiation (Fig. 2F). MiR-194 was upregulated at day 10 (35.8-fold) in hepatic progenitors, and further increased during hepatocytic differentiation (145.4-fold) and maturation (184.5-fold), confirming that miR-194 expression is associated with hepatocytic differentiation.
Figure 2.
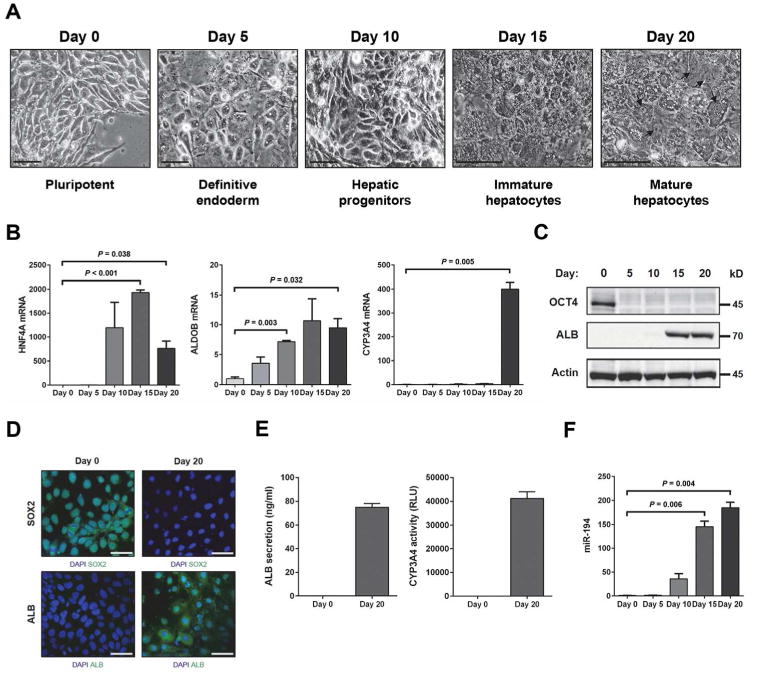
Upregulation of miR-194 upon directed hepatocytic differentiation of hESCs. Differentiation of hESCs toward a hepatocyte fate was performed as described in Materials and Methods. (A): Images showing sequential morphological changes during differentiation from hESCs (pluripotent hESCs + ROCK inhibitor: day 0) to mature hepatocytes (day 20) through definitive endoderm (day 5), hepatic progenitor cells (day 10) and immature hepatocytes (day 15). Hepatocyte morphology including binucleated cells (black arrow). Bars, 50 μm. (B): Quantitative PCR was performed to measure the levels of hepatocyte markers (HNF4A, ALDOB, and CYP3A4) and miR-194 during the hepatocyte differentiation process of hESCs. The graphs show fold-changes and SEM relative to Day 0 in two independent experiments. (C): Western blotting was performed to measure the levels of OCT4 and ALB during the hepatocyte differentiation process of hESCs. Expression of α-tubulin was used as a protein loading control. (D): Immunofluorescence staining of SOX2 (green color, upper panel) and ALB (green color, lower panel) in hESCs at day 0 and day 20 of the hepatocytic differentiation protocol. Nuclei are stained with DAPI blue. Scale bar, 50 μm. (E): Graphs show quantification of secreted human albumin and CYP3A4 activity in hESCs at day 0 and day 20 of the hepatocytic differentiation protocol. (F): Level of miR-194 during hepatocytic differentiation of hESCs measured by quantitative PCR. The graphs show fold-changes and SEM relative to Day 0 in two independent experiments.
Overexpression of MiR-194 Accelerates Hepatocytic Differentiation of Liver Progenitor HepaRG Cells
To investigate whether miR-194 has the capacity to promote hepatocytic differentiation, we established HepaRG cell lines overexpressing miR-194 (HepaRG-miR-194 #1 and HepaRG-miR-194 #2) and control empty cell lines (HepaRG-empty #1 and HepaRG-empty #2). Overexpression of miR-194 in the HepaRG-miR-194 cell lines compared to HepaRG-empty cell lines was confirmed by qRT-PCR analysis (Fig. 3A). Whereas no morphologic differences were noticed between proliferative cell lines, striking differences were observed at day 14 of the differentiation process with miR-194-overexpressing HepaRG cell lines exhibiting increased morphological features of hepatocytes at day 14 compared to empty control cell lines (Fig. 3B). Indeed, even before the addition of differentiation medium, a significantly greater density of bile canaliculi was observed in miR-194 overexpressing cells with 17,086 canaliculi/cm2 compared to 5,525 canaliculi/cm2 in control cells (Fig. 3C). The expression of the hepatocyte markers HNF4A, ALDOB AND ALB and of the hepatocyte-specific microRNA miR-122, was also significantly higher in miR-194 over expressing HepaRG cells compared to control cells (2.8, 2.4, 2.1 and 4.2 fold, respectively) (Fig. 3D). Significantly higher ALB secretion and CYP3A4 activity were also observed in miR-194 over expressing HepaRG cells compared to control cells (Fig. 3E), further confirming with functional assays the increased hepatocytic differentiation resulting from miR-194 over expression (Fig. 3E). This effect on differentiation was concomitant to a reduction in proliferation. Decreased Ki-67 expression was observed at day 14 and day 28 in miR-194 overexpressing HepaRG cells compared to control cells (Supporting Information Fig. S1A). These results were further validated in immunofluorescence staining analysis (Supporting Information Fig. S1B).
Figure 3.
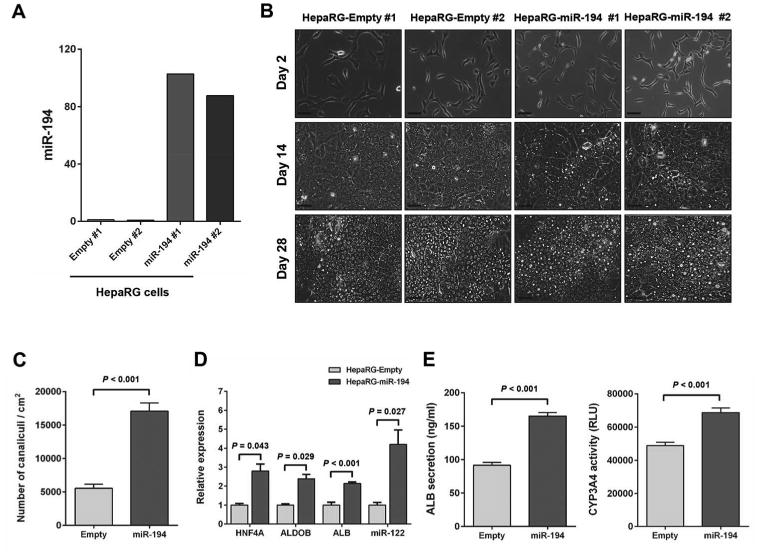
Overexpression of miR-194 accelerates hepatocytic differentiation of HepaRG cells. (A): Expression levels of miR-194 determined by quantitative PCR in HepaRG-empty and HepaRG-miR-194 cell lines. (B): Images of HepaRG-empty and HepaRG-miR-194 cell lines were taken at different phases of the culture: progenitor cells (day 2), intermediate (day 14), and hepatocyte-like cells (day 28). Bars, 50 μm. (C): Mean bile canaliculi densities assessed from three randomly selected fields in two independent experiments at day 14 of hepatocytic differentiation in HepaRG-empty and HepaRG-miR-194 cell lines. (D): Levels of HNF4A, ALDOB, ALB and miR-122 at day 14 of the differentiation process in HepaRG-miR-194 cell lines compared to HepaRG-empty cell lines. (E): Graphs show quantification of secreted human albumin and CYP3A4 activity from HepaRG-empty and HepaRG-miR-194 cell lines at day 28 of hepatocytic differentiation. The results were presented as means ± SEM from two independent experiments (P < 0.05).
Overexpression of MiR-194 Induces Spontaneous Differentiation and accelerates hepatocytic differentiation of hESCs
To assess the capacity for miR-194 to promote hepatic cell fate from a more primitive cell state, we generated hESC cell lines over expressing miR-194 (hESC-miR-194 #1 and hESC-miR-194 #2) and empty control cell lines (hESC-empty #1 and hESC-empty #2). Over expression of miR-194 in the hESC-miR-194 cell lines was confirmed by qRT-PCR analysis (Fig. 4A). Remarkable morphological differences were observed in miR-194-overexpressing hESC cell lines compared to empty control cells when cultured under pluripotency maintaining conditions (mTeSR1) (Fig. 4B). MiR-194-overexpressing hESC cell lines exhibited increased spontaneous differentiation. To further assess the effect of miR-194 over expression on hESC fate and lineage preference, three approaches were used: serial passages under pluripotency maintaining conditions, undirected differentiation and directed hepatocytic differentiation. In serial passages, pluripotency markers OCT4 and NANOG expression levels were significantly decreased in miR-194-overexpressing hESC cell lines compared to empty control cell lines in early passages (Fig. 4C). For undirected differentiation, cells were switched to complete MEF conditioned media minus the FGF2 required to maintain pluripotency to permit spontaneous differentiation. MiR-194-overexpressing hESC cells displayed significantly more pronounced decrease in expression of OCT4 and NANOG during the first 10 days of culture (Fig. 4D, left and middle panels). In addition, while the mesoderm marker HAND1 strongly increased at day 10 in the control empty cells, HAND1 induction was significantly repressed in miR-194 over expression hESC clones (Fig. 4D, right panel). During directed differentiation of hESCs toward a hepatocyte fate, miR-194-overexpressing hESC cells showed a significantly accelerated decrease in the expression of OCT4 and NANOG compared to control cells (Fig. 4E, left and middle panels). Inversely, HNF4A and ALB expression increase during the differentiation process was significantly accelerated in miR-194-overexpressing hESC clones (Fig. 4E, right panel). Taken together, these findings show that increased miR-194 level disrupts hESC pluripotency, specifically inhibits the mesoderm formation of hESCs and accelerates the differentiation towards a hepatocyte fate.
Figure 4.
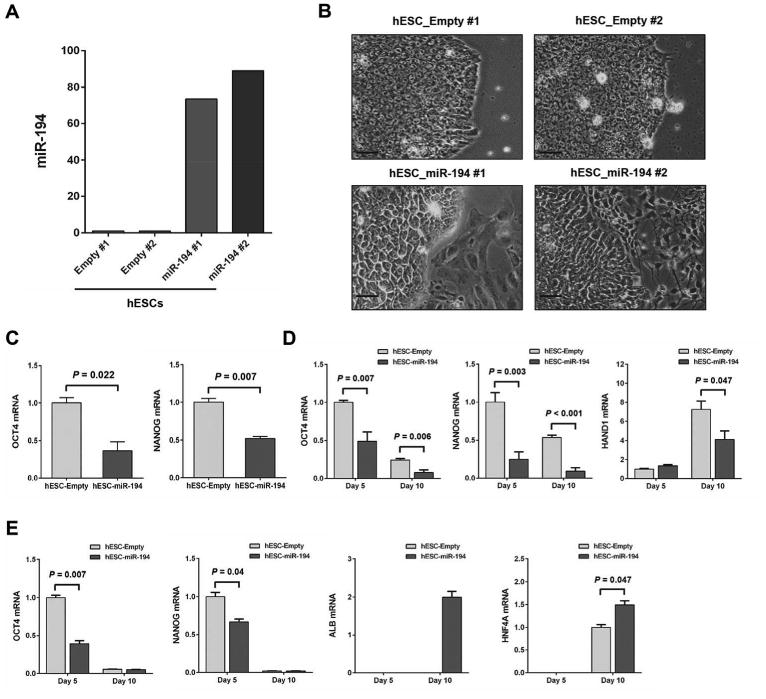
Overexpression of miR-194 induces spontaneous differentiation in hESCs and accelerates hepatocyte differentiation. (A): Expression levels of miR-194 determined by quantitative PCR in hESC-empty and hESC-miR-194 clones. (B): Images showing morphological differences between hESC-empty and hESC-miR-194 clones. Bars, 50 μm. (C): Expression levels of OCT4 and NANOG at an early passage (passage 4) of hESC-empty and hESC-miR-194 clones. (D): Expression levels of OCT4, NANOG and HAND1 during undirected differentiation of hESC-empty and hESC-miR-194 clones, shown as fold-changes and SEM relative to mean level at day 5 in hESC-empty clones in two independent experiments. (E): Expression of OCT4, NANOG, ALB and HNF4A during directed differentiation of hESC-empty and hESC-miR-194 clones, shown as fold-changes and SEM relative to mean level at day 5 in hESC-empty clones in two independent experiments.
Identification of MiR-194 Target Genes with Functional Relevance to Hepatocyte Differentiation
To identify the molecular mechanisms of miR-194-induced hepatocytic differentiation of liver progenitor cells, the program miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) was used to select genes predicted to be targets of miR-194 by at least two prediction algorithms. We then integrated this gene list with a list of genes that we previously reported to be downregulated upon HepaRG hepatocytic differentiation [22]. This integrated analysis identified 319 genes that are not only predicted to be targeted by miR-194 but that are also significantly downregulated upon hepatocytic differentiation of HepaRG cells (Supporting Information Table S3). We selected 17 genes for their known association with cell differentiation, for further validation. Among the 17 selected genes, 16 genes were validated as miR-194 targets as they were significantly downregulated upon miR-194 over expression in both HepaRG cells and hESCs. These genes included BICD2, CENPF, ECT2, ELOVL6, FGFR3, GALNT7, HDAC2, IGF1R, NOTCH2, PRC1, RACGAP1, RAP2B, REV3L, RHEB, THBS1 and YAP1 (Supporting Information Table S4). We further validated YAP1 as a direct target of miR-194. Conserved miR-194 target sites are present in the 3′UTR of human YAP1 (Fig. 5A). Expression of YAP1 mRNA and protein was reduced in mi 194 over expressing HepaRG cell lines compared to control cells (Supporting Information Table S4 and Fig. 5B). Expression of YAP1 mRNA and protein was also decreased in miR-194 over expressing hESCs at day 5 and day 10 (Supporting Information Table S4 and Fig. 5C). The effect of miR-194 over expression on YAP1 mRNA and protein was further validated by transient transfection of miR-194 in HepaRG cell lines (Fig. 5D). To determine whether miR-194 directly targets YAP1, we introduced the luciferase expression vector containing the 3′UTR of YAP1 and miR-194 into HepaRG cells and then measured the effects of miR-194 on luciferase enzyme activity. Co-transfection with miR-194 significantly decreased the luciferase activity of the reporter containing the YAP1 3′UTR in HepaRG cells, but did not affect the luciferase activity with negative control miRNA (Fig. 5E, left panel). Similar results were also observed in miR-194 over expressing HepaRG clones (Fig. 5E, right panel). To further confirm that miR-194 directly targets YAP1, we used antisense oligonucleotide to silence miR-194 in HepaRG cells. Inhibition of miR-194 expression by antisense miR-194 inhibitor was confirmed by qRT-PCR (Fig. 5F). Expression of YAP1 mRNA and protein was increased in HepaRG cells transfected with antisense miR-194 inhibitor (Fig. 5G). Co-transfection of the luciferase expression vector containing the 3′UTR of YAP1 and antisense miR-194 inhibitor significantly increased the luciferase activity of the reporter (Fig. 5H). Downregulation of YAP1 mRNA and protein during hepatocytic differentiation of HepaRG cells and hESCs was also confirmed by qRT-PCR and by western blot analysis (Supporting Information Fig. S2). Taken together, these data demonstrate that miR-194 negatively regulates YAP1 expression by directly binding to YAP1 3′UTR sequence.
Figure 5.
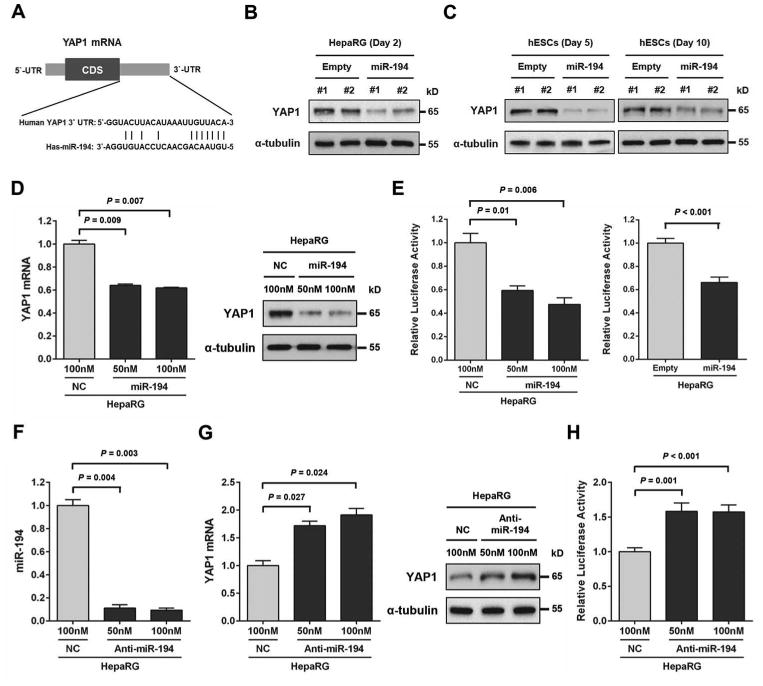
YAP1 is a direct target of miR-194. (A): Putative miR-194 binding sites within the human YAP1-3′UTR are shown in complementary pairing. (B): YAP1 protein expression levels determined by western blot analysis at day 2 in HepaRG-empty and HepaRG-miR-194 cell lines. Expression of α-tubulin was used as a protein loading control. (C): YAP1 protein expression levels determined by western blot analysis at day 5 and day 10 in hESC-empty and hESC-miR-194 cell lines. (D): HepaRG cells were transiently transfected with negative control miRNA (NC; 100 nM) or miR-194 (50 nM or 100 nM) for 48 h. YAP1 mRNA and protein expression levels determined respectively by quantitative PCR and by western blot analysis in HepaRG cells transfected with negative control miRNA or miR-194. (E) Left panel: Cells were transfected with YAP1-3′UTR reporter constructs (200 ng) in the presence or absence of negative control miRNA (100 nM) or miR-194 (50 nM or 100 nM). Effects of miR-194 on the reporter constructs were determined at 48 h after transfection. Right panel: HepaRG-empty and HepaRG-miR-194 cell lines were transfected with YAP1-3′UTR reporter constructs (200 ng). Relative luciferase activities were measured and calculated as the ratio of firefly/renilla activities in the cells, and normalized to those of the negative control miRNA. The results were presented as means ± SEM from three independent experiments (P < 0.05). (F): HepaRG cells were transiently transfected with negative control miRNA (NC; 100 nM) or antisense miR-194 inhibitor (50 nM or 100 nM) for 48 h. MiR-194 levels determined by quantitative PCR in in HepaRG cells transfected with negative control miRNA or antisense miR-194 inhibitor. (G): YAP1 mRNA and protein expression levels determined respectively by quantitative PCR and by western blot analysis in HepaRG cells transfected with negative control miRNA or antisense miR-194 inhibitor. (H): Cells were transfected with YAP1-3′UTR reporter constructs (200 ng) in the presence or absence of negative control miRNA (100 nM) or antisense miR-194 inhibitor (50 nM or 100 nM). Effects of antisense miR-194 inhibitor on the reporter constructs were determined at 48 h after transfection. Relative luciferase activities were measured and calculated as the ratio of firefly/renilla activities in the cells, and normalized to those of the negative control miRNA. The results were presented as means ± SEM from three independent experiments (P < 0.05).
YAP1 Inhibition Induces Hepatocytic Differentiation and Its Overexpression Suppresses MiR-194-Induced Hepatocytic Differentiation in HepaRG Cells
YAP1 is an important transcription factor regulating stem cell fate [26]. Thus, to determine whether YAP1 mediates miR-194-induced hepatocytic differentiation of progenitor cells, we first established using shRNA-containing lentivirus, HepaRG cell lines with reduced YAP1 expression (HepaRG-YAP1 KD #1 and HepaRG-YAP1 KD #2). Reduction of YAP1 expression was confirmed by qRT-PCR and western blot analysis (Fig. 6A). A marked increase of bile canaliculi density was observed in HepaRG-YAP1 KD cells (11,968 canaliculi/cm2) at day 14 as compared to control cells (3,899 canaliculi/cm2) (Fig. 6B). Similar results were observed at day 28. HepaRG YAP1 KD cells also displayed significantly increased expression of HNF4A at day 14 and of HNF4A, ALDOB, CYP3A4 and ALB at day 28, confirming the effect of YAP1 inhibition on hepatocytic differentiation (Fig. 6C). To further evaluate whether YAP1 inhibition mediates miR-194 induced hepatocytic differentiation of HepaRG cells, we overexpressed YAP1 in the miR-194-overexpressing HepaRG cell lines (HepaRG-miR-194-YAP1 #1 and HepaRG-miR-194-YAP1 #2) and generated two empty control cell lines (HepaRG-miR-194-empty #1 and HepaRG-miR-194-empty #2). YAP1 over expression didn’t affect the levels of miR-194 in these cell lines (Fig. 7A). YAP1 over expression was confirmed in HepaRG-miR-194-YAP1 cell lines by qRT-PCR and western blot analysis (Fig. 7B). At day 14 and day 28, a significant decrease in bile canaliculi density was observed in YAP1-overexpressing miR-194 cells compared to miR-194 empty control cells (Fig. 7C). Expression of the hepatocyte markers HNF4A, ALDOB and ALB was also significantly reduced in YAP1-overexpressing miR-194 HepaRG cells at day 14 and day 28 (Fig. 7D). These results demonstrate that YAP1 over expression reverses the effects of miR-194 on hepatocytic differentiation of HepaRG cells, suggesting that YAP1 is the main mediator of miR-194 induced hepatocytic differentiation of progenitor cells.
Figure 6.
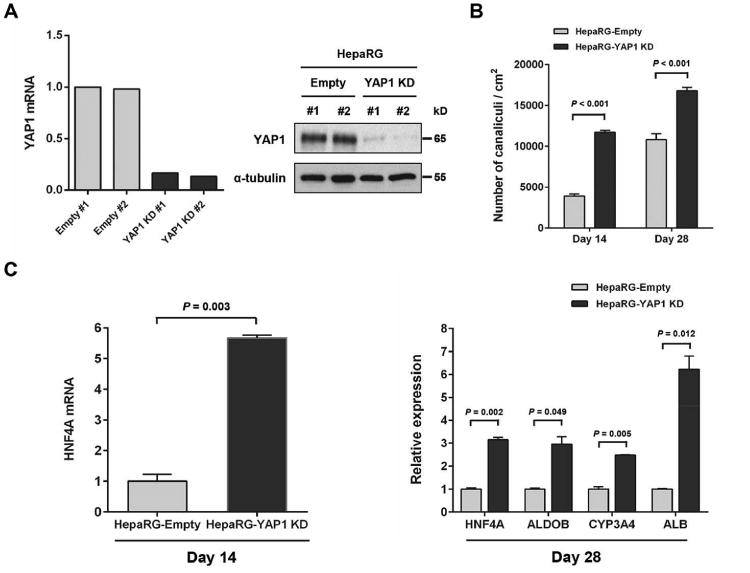
YAP1 inhibition induces hepatocytic differentiation in HepaRG cells. (A): YAP1 mRNA and protein expression levels determined respectively by quantitative PCR (left panel) and western blot analysis (right panel) in HepaRG-empty and HepaRG-YAP1 KD cell lines. α-tubulin was used to control protein loading. (B): Mean bile canaliculi densities of HepaRG-empty and HepaRG-YAP1 KD cell lines assessed from three randomly selected fields in two independent experiments. (C): Relative expression levels of HNF4A at day 14 and of HNF4A, ALDOB, CYP3A4 and ALB at day 28 of hepatocytic differentiation process in HepaRG-YAP1 KD compared to HepaRG-empty cell lines.
Figure 7.
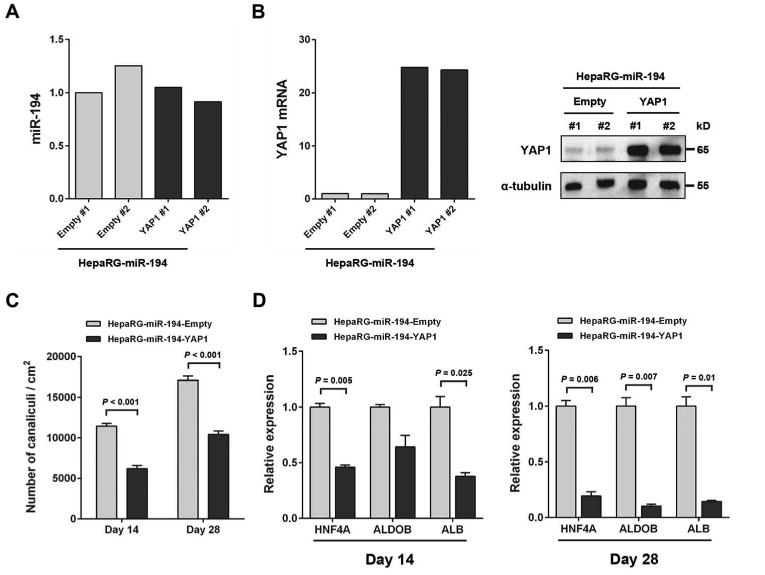
Overexpression of YAP1 suppresses miR-194-induced hepatocytic differentiation in HepaRG cells. (A): MiR-194 levels determined by quantitative PCR in HepaRG-empty, HepaRG-miR-194-empty and HepaRG-miR-194-YAP1 celle lines. (B): YAP1 mRNA and protein expression levels determined respectively by quantitative PCR and by western-blot analysis in HepaRG-empty, HepaRG-miR-194-empty and HepaRG-miR-194-YAP1 cell lines. The expression of α-tubulin was used to control protein loading. (C): Mean bile canaliculi densities of HepaRG-miR-194-empty and HepaRG-miR-194-YAP1 cell lines assessed from three randomly selected fields in two independent experiments. (D): Relative mRNA expression levels of HNF4A, ALDOB and ALB measured by quantitative PCR at day 14 and day 28 of the differentiation process in HepaRG-miR-194-YAP1 cell lines compared to HepaRG-miR-194-empty cell lines.
Discussion
In recent years, miRNAs have been shown to play essential roles in stem cell maintenance, cell differentiation, and organ development [11-13]. In particular, several miRNAs have been reported to modulate liver cell fate and development in health and disease [14, 15, 17, 27-30]. Together, these studies suggest that miRNAs can be key regulators of hepatocytic differentiation. In the present study, we report that miR-194 is the second most highly upregulated miRNA after miR-122 upon hepatocytic differentiation of liver progenitor cells and induces spontaneous differentiation of hESCs.
Recent studies have shown that miR-194 induces intestinal epithelial cell differentiation [31] and is a key mediator of chondrogenic differentiation of adipose-derived stem cells [32]. In addition, miR-194 suppresses cell migration and metastasis in breast, gastric, lung, and liver cancers [33-36]. However, the function of miR-194 in the regulation of hepatocytic differentiation remained unknown. We showed that miR-194 gradually increased in HepaRG liver progenitor cells and hESCs during differentiation into hepatocytes, and exogenous expression of miR-194 promoted the hepatocytic differentiation of HepaRG cells, a phenotype associated with increased expression of hepatocyte markers HNF4A, ALDOB and CYP3A4. Our study also revealed that miR-194 over expression induces the spontaneous differentiation of hESCs as shown by the decrease in the expression of pluripotency markers OCT4 and NANOG in serial passage and undirected differentiation experiments. The liver is derived from the endoderm that the innermost germ layer formed during gastrulation [37]. While we did not observe changes in expression of the endoderm markers FOXA2 and SOX17 upon miR-194 over expression in hESCs during undirected differentiation (data not shown), expression level of the mesoderm marker HAND1 was strongly repressed, suggesting that miR-194 may be involved in suppressing alternative cell fates during hepatocytic differentiation. Overexpression of miR-194 promoted hepatocyte differentiation in HepaRG progenitor cells, but alone was unable to induce a hepatic fate from pluripotent hESCs indicating that its role in hepatocyte differentiation may be cell state specific and dependent on molecular context.
To study the molecular mechanism by which miR-194 regulates hepatocyte differentiation of progenitor cells, we identified 319 predicted miR-194 target genes that are downregulated during HepaRG cell differentiation using the target prediction tool miRWalk [25] and our previously reported gene profiling data [22]. We confirmed for 16 genes that miR-194 over expression results in their decrease in both HepaRG cells and hESCs. These 16 genes included IGF1R that has been previously reported as a target of miR-194 [33]. They included also YAP1, a downstream mediator of Hippo signaling that plays an important role in cell fate determination including differentiation [26]. We showed that forced expression of YAP1 markedly reversed the effects of miR-194 on hepatocytic differentiation. Conversely, inhibition of YAP1 in HepaRG cells strongly induced hepatocytic differentiation. Thus, these results suggest that YAP1 is a negative regulator of hepatocytic differentiation in liver progenitor cells and that YAP1 may be the main mediator of the regulatory effect of miR-194 on hepatocytic differentiation.
Because the Hippo pathway is a key regulator of liver size and tumorigenesis and YAP transcription activity is inhibited by phosphorylation [38, 39], we also measured YAP1 phosphorylation upon hepatocytic differentiation of HepaRG cells and hESCs (Supporting Information Fig. S2). YAP1 was completely or in majority dephosphorylated in proliferative hepaRG cells and hESCs, indicative of high levels of activated YAP1 in these cells. During hepatocytic differentiation of HepaRG cells, YAP1 phosphorylation transiently increased, suggesting that in addition to the strong miR-194-induced downregulation of YAP1 upon hepatocytic differentiation, the remaining YAP protein is further deactivated by phosphorylation. In hESCs, YAP1 phosphorylation followed YAP1 expression, suggesting that no additional regulation by YAP1 kinases was involved in the differentiation process of these cells. A recent study identified a complex network of kinases regulating YAP1 phosphorylation [39] and future studies should investigate their role in hepatocyte differentiation.
A number of additional genes we validated as regulated by miR-194 in both HepaRG cells and hESCs included ECT2, FGFR3, GALNT7, HDAC2, NOTCH2, RACGAP1, RHEB and THBS1 have been shown to participate in cell differentiation processes and liver regeneration [40-47]. Their roles in liver cell fate should be further evaluated.
Conclusion
Our study demonstrates that miR-194 is increased during hepatocytic differentiation of HepaRG cells and hESCs, and that miR-194 enhances hepatic differentiation of progenitor cells by targeting YAP1. This is the first study demonstrating the critical role of miR-194 in hepatocytic differentiation, liver progenitor cell fate and hESCs pluripotency. Our findings support the need to evaluate the utility of targeting miR-194 in a wide array of liver diseases associated with dysregulated liver stem cell fate and abnormal differentiation state of hepatocytes.
Supplementary Material
Acknowledgments
This work was supported in part by the MD Anderson Cancer Center Support Grant CA016672, through a Multidisciplinary Research Program Award and by a fellowship to RMC through a training grant T32-CA009299.
List of abbreviation
- miRNA
microRNA
- mRNA
messenger RNA
- shRNA
small hairpin RNA
- qRT-PCR
quantitative real-time polymerase chain reaction
- ORF
open reading frame
- GFP
green fluorescent protein
- hESCs
human embryonic stem cells
- MEF-CM
mouse embryonic fibroblast-conditioned media
- FGF
fibroblast growth factor
- HNF4A
hepatocyte nuclear factor 4 alpha
- ALDOB
fructose-bisphosphate aldolase B
- CYP3A4
cytochrome P450 3A4
- ALB
albumin
- YAP1
yes-associated protein 1
Footnotes
Author contribution: KHJ initiated the study, performed experiments, analyzed the data, and drafted the manuscript. KHJ, RLM, CZ and NU conducted the in vitro analyses. MCB and RLM critically revised the manuscript. LB supervised the project, advised with regards to the experimental design, and edited the manuscript.
Disclosure of Potential Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Adam R, Hoti E. Liver transplantation: the current situation. Semin Liver Dis. 2009;29:3–18. doi: 10.1055/s-0029-1192052. [DOI] [PubMed] [Google Scholar]
- 2.Yu Y, Fisher JE, Lillegard JB, et al. Cell therapies for liver diseases. Liver Transpl. 2012;18:9–21. doi: 10.1002/lt.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Foster M, Al-Dhalimy M, et al. The origin and liver repopulating capacity of murine oval cells. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay DC, Zhao D, Fletcher J, et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 9.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Lakshmipathy U, Love B, Goff LA, et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 12.Krichevsky AM, Sonntag KC, Isacson O, et al. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi R, Poy MN, Stoffel M, et al. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hand NJ, Master ZR, Eauclaire SF, et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogler CE, Levoci L, Ader T, et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 17.Laudadio I, Manfroid I, Achouri Y, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Doddapaneni R, Chawla YK, Das A, et al. Overexpression of microRNA-122 enhances in vitro hepatic differentiation of fetal liver-derived stem/progenitor cells. J Cell Biochem. 2013;114:1575–1583. doi: 10.1002/jcb.24499. [DOI] [PubMed] [Google Scholar]
- 19.Deng XG, Qiu RL, Wu YH, et al. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver Int. 2014;34:281–295. doi: 10.1111/liv.12239. [DOI] [PubMed] [Google Scholar]
- 20.Parent R, Marion MJ, Furio L, et al. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology. 2004;126:1147–1156. doi: 10.1053/j.gastro.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Cerec V, Glaise D, Garnier D, et al. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology. 2007;45:957–967. doi: 10.1002/hep.21536. [DOI] [PubMed] [Google Scholar]
- 22.Parent R, Beretta L. Translational control plays a prominent role in the hepatocytic differentiation of HepaRG liver progenitor cells. Genome Biol. 2008;9:R19. doi: 10.1186/gb-2008-9-1-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai J, DeLaForest A, Fisher J, et al. StemBook. Cambridge MA: 2012 Uri Ben-Davi and Nissim Benvenisty.; 2008. Protocol for directed differentiation of human pluripotent stem cells toward a hepatocyte fate. [PubMed] [Google Scholar]
- 24.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dweep H, Sticht C, Pandey P, et al. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Lian I, Kim J, Okazawa H, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzur G, Israel A, Levy A, et al. Comprehensive gene and microRNA expression profiling reveals a role for microRNAs in human liver development. PLoS One. 2009;4:e7511. doi: 10.1371/journal.pone.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung CJ, Iyengar S, Blahnik KR, et al. Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PLoS One. 2011;6:e27740. doi: 10.1371/journal.pone.0027740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gailhouste L, Gomez-Santos L, Hagiwara K, et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- 30.Mobus S, Yang D, Yuan Q, et al. MicroRNA-199a-5p inhibition enhances the liver repopulation ability of human embryonic stem cell-derived hepatic cells. J Hepatol. 2015;62:101–110. doi: 10.1016/j.jhep.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Hino K, Tsuchiya K, Fukao T, et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–1442. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Kang Y, Liao WM, et al. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7:e31861. doi: 10.1371/journal.pone.0031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Z, Fu X, Chen X, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le XF, Almeida MI, Mao W, et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One. 2012;7:e41170. doi: 10.1371/journal.pone.0041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Ying X, Chen H, et al. MicroRNA-194 Inhibits the Epithelial-Mesenchymal Transition in Gastric Cancer Cells by Targeting FoxM1. Dig Dis Sci. 2014 doi: 10.1007/s10620-014-3159-6. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Liu T, Fang O, et al. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27kip1. Oncogene. 2014;33:1506–1514. doi: 10.1038/onc.2013.108. [DOI] [PubMed] [Google Scholar]
- 37.North TE, Goessling W. Endoderm specification, liver development, and regeneration. Methods Cell Biol. 2011;101:205–223. doi: 10.1016/B978-0-12-387036-0.00010-4. [DOI] [PubMed] [Google Scholar]
- 38.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng Z, Moroishi T, Mottier-Pavie V, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakata H, Rubin JS, Taylor WG, et al. A Rho-specific exchange factor Ect2 is induced from S to M phases in regenerating mouse liver. Hepatology. 2000;32:193–199. doi: 10.1053/jhep.2000.8271. [DOI] [PubMed] [Google Scholar]
- 41.Simsa-Maziel S, Monsonego-Ornan E. Interleukin-1beta promotes proliferation and inhibits differentiation of chondrocytes through a mechanism involving down-regulation of FGFR-3 and p21. Endocrinology. 2012;153:2296–2310. doi: 10.1210/en.2011-1756. [DOI] [PubMed] [Google Scholar]
- 42.Kahai S, Lee SC, Lee DY, et al. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4:e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye F, Chen Y, Hoang T, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeliazkova P, Jors S, Lee M, et al. Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology. 2013;57:2469–2479. doi: 10.1002/hep.26254. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien RN, Shen Z, Tachikawa K, et al. Quantitative proteome analysis of pluripotent cells by iTRAQ mass tagging reveals post-transcriptional regulation of proteins required for ES cell self-renewal. Mol Cell Proteomics. 2010;9:2238–2251. doi: 10.1074/mcp.M110.000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lafourcade CA, Lin TV, Feliciano DM, et al. Rheb activation in subventricular zone progenitors leads to heterotopia, ectopic neuronal differentiation, and rapamycin-sensitive olfactory micronodules and dendrite hypertrophy of newborn neurons. J Neurosci. 2013;33:2419–2431. doi: 10.1523/JNEUROSCI.1840-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH, Bhang DH, Beede A, et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


