Abstract
We previously reported a mutation in the cholesterol biosynthesis gene, hydroxysteroid (17-beta) dehydrogenase 7 (Hsd17b7rudolph), that results in striking embryonic forebrain dysgenesis. Here we describe abnormal patterns of neuroprogenitor proliferation in the mutant forebrain, namely, a decrease in mitotic cells within the ventricular zone (VZ) and an increase through the remainder of the cortex by E11.5. Further evidence suggests mutant cells undergo abnormal interkinetic nuclear migration (IKNM). Furthermore, intermediate progenitors are increased at the expense of apical progenitors by E12.5, and post-mitotic neurons are expanded by E14.5. In vitro primary neuron culture further supports our model of accelerated cortical differentiation in the mutant. Combined administration of a statin and dietary cholesterol in utero achieved partial reversal of multiple developmental abnormalities in the Hsd17b7rudolph embryo, including the forebrain. These results suggest that abnormally increased levels of specific cholesterol precursors in the Hsd17b7rudolph embryo cause cortical dysgenesis by altering patterns of neurogenesis.
Keywords: cortical neurogenesis, cholesterol biosynthesis, cell fate, neural progenitors, forebrain, neurogenesis, mouse, Hsd17b7, statin, development
Introduction
Cholesterol homeostasis is critical for human health and is implicated in a number of disorders from cardiovascular disease in adults to severe developmental abnormalities in the embryo (Porter and Herman, 2011). The developing embryo is especially susceptible to disruptions in cholesterol homeostasis, because the placental and blood-brain barriers can limit the transport of cholesterol between physiologic compartments. Consistent with a high requirement for in situ synthesis of cholesterol in the brain (Jurevics et al., 1997; Orth and Bellosta, 2012; Tint et al., 2006), a common feature of genetic disorders of cholesterol biosynthesis are central nervous system (CNS) abnormalities. Because the brain is especially sensitive to perturbations in cholesterol metabolism, it is important to understand the mechanism(s) by which genetic or acquired (e.g., drug inhibition) deficiencies in enzymes of cholesterol biosynthesis affect cholesterol production and utilization during forebrain development.
Hsd17b7 (hydroxysteroid 17-beta dehydrogenase 7) encodes a 3-keto-steroid reductase that converts 4-methylzymosterone to 4α-methylzymosterol and zymosterone to zymosterol during post-squalene cholesterol biosynthesis (Marijanovic et al., 2003). Null mutations in mouse Hsd17b7 lead to early embryonic death, precluding studies on the role of Hsd17b7 in brain development (Jokela et al., 2010; Shehu et al., 2008). The Rudolph mutation is a hypomorphic allele of Hsd17b7 (Hsd17b7rud) which, when carried in the homozygous state, allows survival to mid-organogenesis with widespread CNS anomalies (Stottmann et al., 2011). The CNS phenotype of the Hsd17brud mutation is unique among known mutants in the cholesterol biosynthetic enzymatic pathway and provides a novel tool for studying the relationship between cholesterol metabolism and mammalian neural development.
Consistent with reduced activity of Hsd17b7, sterol analysis of the Hsdb17b7rud mutant revealed increased levels of cholesterol intermediates upstream of the Hsd17b7 enzyme, decreased level of downstream intermediates, and an accumulation of unidentified sterols not detected in control embryos. There is also an approximate 20% decrease in cholesterol in Hsd17b7rud brains (Stottmann et al., 2011). It is not entirely clear if the defects in mouse models of cholesterol metabolism result from a decreased level of cholesterol or increased levels of cholesterol precursors and related metabolites. In the case of the Dhcr7 mouse, for example, there is a 67% reduction of brain cholesterol content but no prenatal lethality or forebrain abnormalities in homozygous null mutants (Fitzky et al., 2001). However, a common feature among all mouse mutants with CNS defects is the accumulation of intermediates upstream of each targeted enzyme. In a study by Cunningham et al. (2015), a neural progenitor-specific (GFAP-Cre) ablation of the cholesterol biosynthesis enzyme NSDHL, (NAD(P) dependent steroid dehydrogenase-like), a component of the sterol demethylase complex that also includes HSD17B7, resulted in progressive loss of cortical and hippocampal neurons, marked cortical neuronal apoptosis, and postnatal death with increased levels of several sterol intermediates. In vitro assays provided evidence that reducing the levels of 4-methylsterols improved neural development (Cunningham et al., 2015). We therefore hypothesized that cholesterol deficiency is not the primary cause of the Hsdb17b7rud mutant phenotype and that reducing the high levels of cholesterol intermediates upstream of the mutant Hsd17b7 enzyme would improve neural development in the Hsd17b7rud mouse.
In the present study, we determined with in vivo and in vitro studies that the distinctive phenotype of the Hsdb17b7rud mutant forebrain is caused by precocious differentiation and migration of neuronal progenitors. We also demonstrate that manipulation of the cholesterol biosynthesis pathway through combined pharmacological (simvastatin) and dietary (cholesterol) treatments can ameliorate multiple abnormalities in the Hsd17b7rud mutants, including improving forebrain development.
Results
Apical polarity is disrupted in the Hsd17b7rud cortex
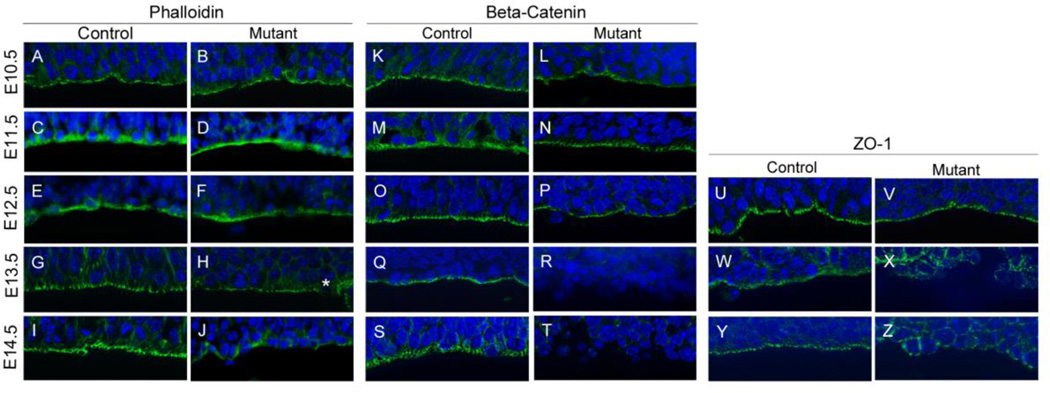
Our previous study of the Hsd17b7rud mutant cortex found disorganized cortical tissue and cellular rosettes at embryonic day (E) 15.5 (Stottmann et al., 2011). To address our hypothesis that the cortical defects were caused by disrupted tissue polarity, we examined markers of apical polarity in Hsd17b7rud mutants from E10.5 to E14.5. At E10.5 through E12.5, both phalloidin and β-catenin, markers for apical adherens junctions, were present along the ventricular zone (VZ) in the appropriate apically-restricted domains in Hsd17b7rud and control embryos at E10.5 through E12.5 (Fig. 1A–F, K–P). However, by E13.5, while both apical markers appeared continuous along the ventricular surface of control embryos, there are minor disruptions in phalloidin staining at the ventricular surface of the mutant (Fig. 1G,H, asterisk) and β-catenin is completely lost along the VZ, failing to form a continuous line of adherens junctions (Fig. 1G,H,Q,R). By E14.5, large sections of the mutant ventricular surface are entirely devoid of both markers (Fig. 1I,J,S,T). In addition to phalloidin and β-catenin staining, staining for the marker of apical junctions, ZO-1, was measured. At E12.5, ZO-1 was restricted to the ventricular surface in both the Hsd17b7rud and control forebrain (Fig. 1U,V). However, altered ZO-1 immunoreactivity was evident by E13.5 and continued through E14.5, while proper alignment of the tight junctions was lost in the Hsd17b7rud cortex (Fig. 1W–Z). Hsd17b7 is not absolutely required to maintain epithelial polarity, however, as siRNA knockdown in an IMCD cell 3-D spheroid assay did not alter spheroid formation (Fig. S1). Thus, the displaced neurons we observe in mutant cortical tissue likely arise through another mechanism.
Figure 1. Apical polarity is lost in the Hsd17b7rud cortex.
Adherens junctions markers phalloidin (A–J), β-catenin (K–T) and ZO-1 (U–Z) are shown for both control and mutant at E10.5 (A,B,K,L), E11.5 (C,D,M,N), E12.5 (E,F,O,P,U,V), E13.5 (G,H,Q,R,W,X) and E14.5 (I,J,S,T,Y,Z). Disruptions in marker distribution are first noted at E13.5 (e.g., asterisk in H). n=3 per embryonic stage for all experiments.
Patterns of proliferation and interkinetic nuclear migration are abnormal in the Hsd17b7rud cortex
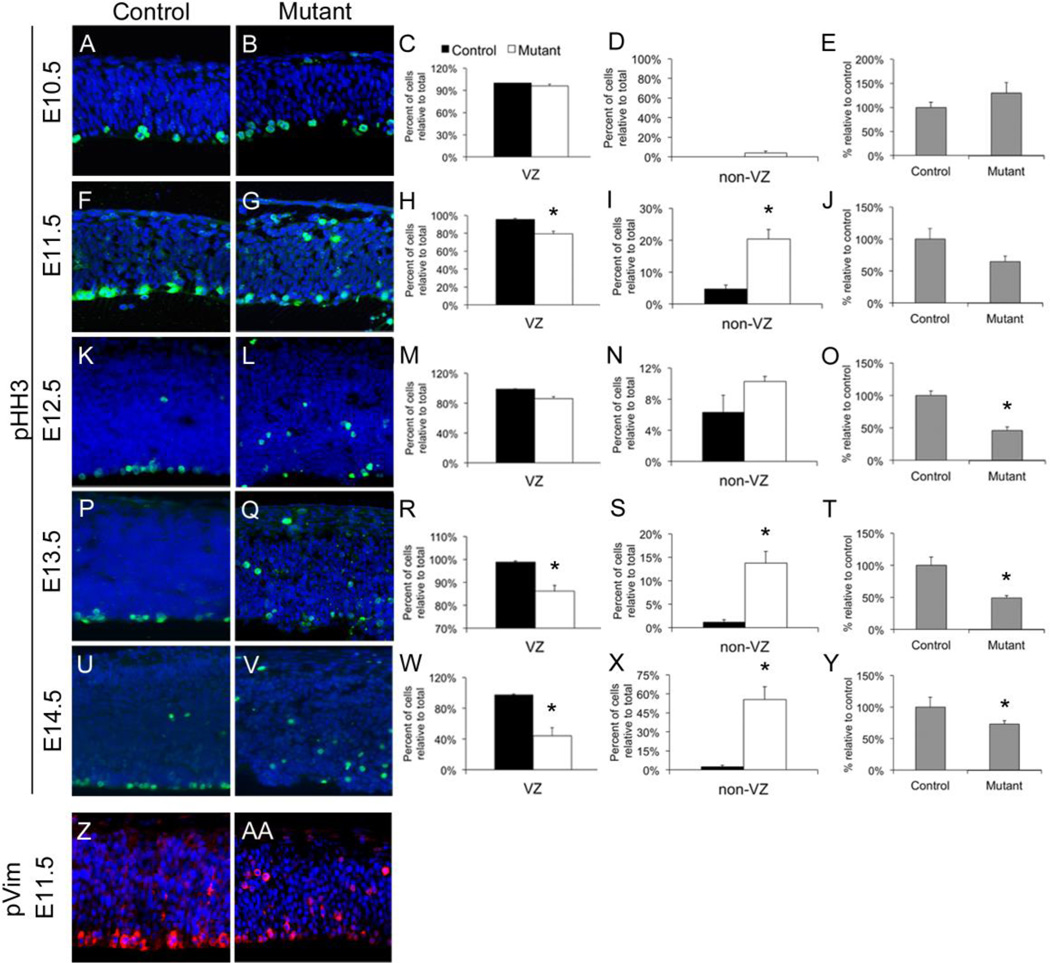
We previously observed decreased proliferation in the Hsd17b7rud cortex at E14.5, which could explain the severe loss of cortical tissue during embryonic brain development (Stottmann et al., 2011). We therefore performed a detailed analysis of neurogenesis in the forebrain to further evaluate this phenomenon. We first examined phospho-Histone H3 (pHH3) localization with immunohistochemistry (IHC) to mark mitotic cells and found that, at E10.5, both the restricted localization of mitotic cells to the VZ and the total number of pHH3-positive cells appeared to be similar in control and mutant cortices (Fig. 2A–D). At E11.5, however, there was a significant reduction of pHH3-positive cells at the VZ in the Hsd17b7rud mutants (96% in control vs. 80% in mutant), with a significant number of cells dividing distant from the VZ in the mutant cortex (4% in control vs. 20% in mutant; Fig. 2F–I). These patterns continued at E12.5, with an overall reduction in proliferation within the mutant cortex (Fig. 2K–N). By E13.5 to 14.5, there were significantly fewer mitotic cells left at the ventricular surface (98% control vs. 86% mutant at E13.5, and 98% control vs. 44% mutant at E14.5; Fig. 2P–S,U–X). In addition to changes in proliferation patterns, there was also a significant reduction in overall proliferation from E12.5 to E14.5 in the mutant relative to the control (Fig. 2O,T,Y). Thus, abnormal patterns and levels of proliferation are evident well before changes in polarity are seen.
Figure 2. Abnormal proliferation and interkinetic nuclear migration (IKNM) in the Hsd17b7rud cortex.
(A–Y) Mitotic activity, as measured by pHH3 (green), is shown from E10.5 through E14.5 (A,B,F,G,K,L,P,Q,U,V) and quantified for each stage (C-E,H-J,M-O,R-T,W-Y). Separate quantification is shown at each stage for mitotic activity in the VZ (C,H,M,R,W), outside the VZ (D,I,N,S,X) and the overall proliferation in each (E,J,O,T,Y). Staining for phosphorylated vimentin (pVim, red) shows dividing radial glia localized along the VZ in the E11.5 control (Z) compared to the Hsd17b7rud mutant (AA).
The ectopic proliferation we see in Hsd17b7rud mutants during the neurogenic phase of development could be due to either increased division of basal progenitors (to produce neurons) or displacement of dividing apical progenitors (radial glia). During early forebrain development, apical progenitor nuclei move up and down radial fibers in coordination with the cell cycle in a process called “interkinetic nuclear migration” (IKNM) (Spear and Erickson, 2012), in which nuclei move basally during G1-S phases and apically during S-G2 phase (Kosodo et al., 2011). During M-phase, apical progenitors align along the VZ and either divide “symmetrically” to maintain the progenitor pool or “asymmetrically” to generate daughter progenitor cells and neurons (Noctor et al., 2004). In contrast, basal progenitor cells (found in the subventricular zone) are unable to self renew in the mouse and divide to generate two post-mitotic neurons (Haubensak et al., 2004). Because phosphorylated Vimentin (pVim) is an intermediate filament protein specific to mitotically active radial glia (Kamei et al., 1998), we used immunohistochemistry (IHC) for pVim to determine if the ectopic proliferation in the Hsd17b7rud mutants is due to displaced apical progenitors, and we found that, whereas the pVim staining was restricted to cells along the VZ surface in the control forebrain, as expected, staining for pVim in the Hsd17b7rud mutants identified the ectopic cells as dividing radial glia (Fig. 2Z,AA). These results indicate that the apical progenitor cells are undergoing improper IKNM and, as a result, dividing and differentiating in locations removed from the VZ instead of renewing the stem cell pool, as would true apical progenitors.
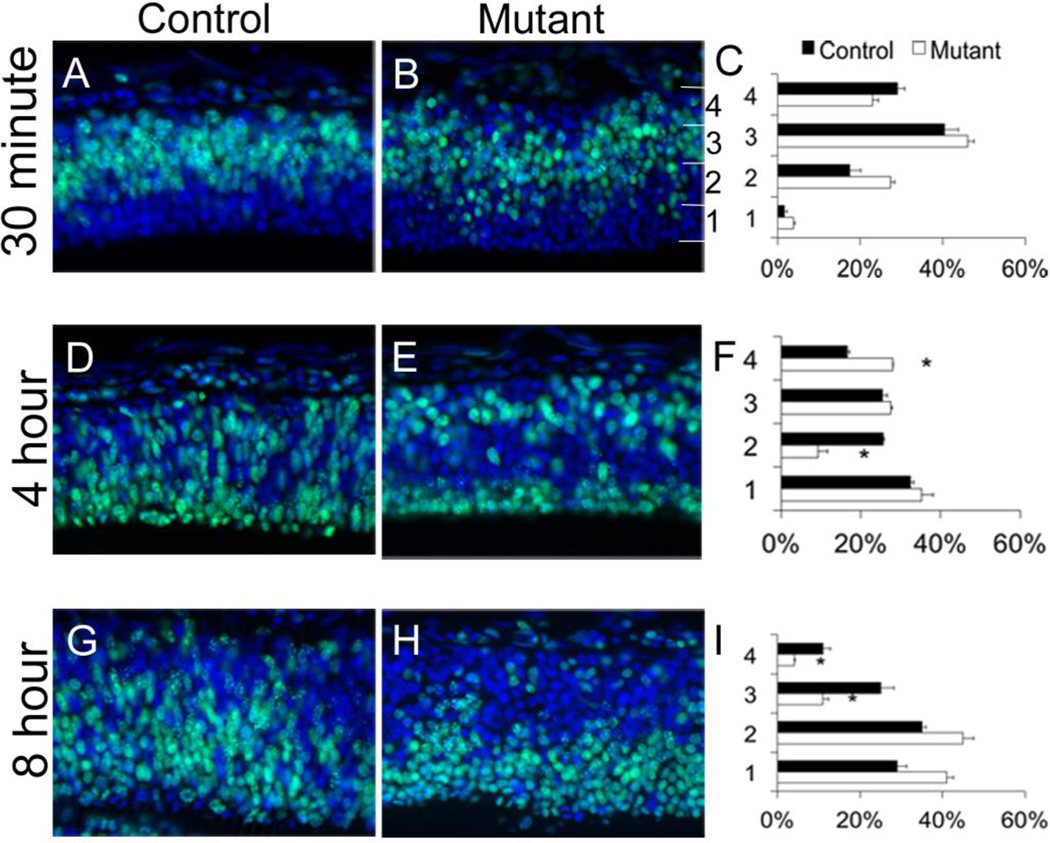
To directly assess IKNM, we tracked the migratory patterns of apical progenitor nuclei in the E11.5 cortex via pulse-chase analysis using EdU, which labels cells in S-phase at the height of IKNM (Buck et al., 2008). We tracked the progress of IKNM by labeling cells at S-phase and analyzing at; 1) 30 minutes, to show the initial locations of the progenitor population; 2) 4 hours, to track basal to apical progress as cells migrate to the VZ; and 3) 8 hours, when nuclei should be resuming apical to basal movement during the IKNM cycle. At 30-minutes post-injection, we found a distinct band of EdU-positive cells in the mid-cortex of the control, as expected (Fig. 3A), with the EdU-positive cells in the mutant appearing only slightly more dispersed (Fig. 3A–C). By 4-hours post injection, most wild-type cells were near the VZ, reflecting the expected gradient of cells moving apically towards the VZ. In the mutant cortex most EdU-positive cells were at the extreme edges of the cortex, with a significant reduction in the number of EdU-positive cells in the mid-cortex (P<0.05, Fig. 3D–F, bin 2,4). At 8 hours, most wild-type cells were at mid-cortex, indicating apical to basal progression (Fig. 3G–I), whereas, in the mutant cortex, most EdU-positive cells were located in the lower cortical bins, with significantly fewer cells present in the two outermost regions (bins 3,4). Similar to the previous finding that cells with improper IKNM showing a more rounded shape (Pacary et al., 2013), we found EdU-positive cells in the Hsd17b7rud cortex to be rounded and less densely packed than cells in control cortex. These results suggest that loss of Hsd17b7 activity induces abnormal neural progenitor IKNM in the neurogenic forebrain, leading to abnormal patterns of differentiation.
Figure 3. EdU patterns are abnormal in Hsd17b7rud cortex.
EdU pulse-chase analyses measured IKNM at 30-minutes (A–C), 4 hours (D–F) and 8 hours (G–I) post-injection. The number of cells in each bin was calculated for each stage (C,F,I). n=3 for each stage and genotype, data shown are the mean±S.E.M., *P<0.05.
Neurogenesis is accelerated in the Hsd17b7rud cortex
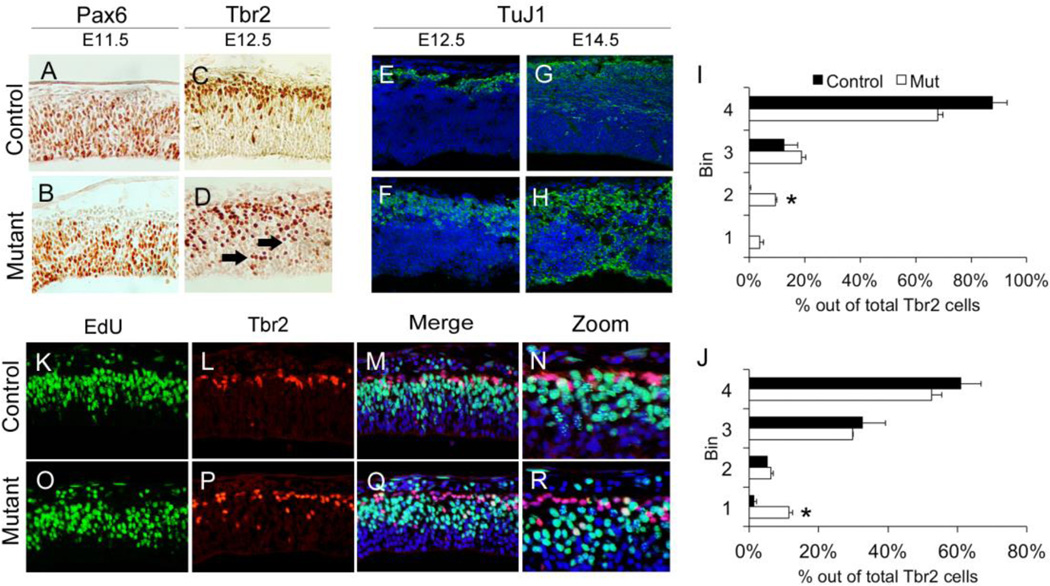
Because patterns of IKNM appeared abnormal in the Hsd17b7rud forebrain, we also determined the abundance of both apical and intermediate progenitors. IHC for Pax6-positive apical progenitors revealed a slight reduction in numbers in the mutant cortex at E11.5 as compared to control (Fig. 4A,B). Furthermore, IHC for Tbr2-positive intermediate progenitors at E12.5 showed a restricted area of Tbr2-positive cells in the SVZ of the control, whereas these cells were enriched in the Hsd17b7rud mutant (Fig. 4C,D, arrowheads). With regard to the number and distribution of Tbr2-positive cells at different developmental stages, while there were significantly more Tbr2 cells in the apical region of the mutant cortex compared to the control at E11.5 (Fig. 4I), by E12.5, there were significantly more Tbr2 cells present near the VZ in the mutant (Fig. 4J). These patterns of Pax6- and Tbr2-positive progenitors indicate dysfunctional mechanisms of migration and/or differentiation. We also analyzed neuronal differentiation using a marker, TuJ1, for post-mitotic neurons and noted an expansion of TuJ1-positive cells in the Hsd17b7rud mutant at E12.5, in contrast to the more restricted, dorsal location of TuJ1-positive cells in the control. This expansion continued at E14.5, when we occasionally saw a “double-cortex” characterized by neurons found at both the outer and inner layers of the mutant cortex (Fig. 4E–H). These results suggest defective migration or differentiation, consistent with our previous analysis of IKNM, and that this defect leads to reduced numbers and abnormal placement of progenitor populations. As a result, post-mitotic neurons appear throughout the cortex rather than being restricted to the extreme basal surface.
Figure 4. Neural progenitor and differentiated neuron populations are altered in the Hsd17b7rud cortex.
Pax6 apical progenitors at E11.5 (A,B), Tbr2 intermediated progenitors at E12.5 (C,D), and post-mitotic (TuJ1-positive) populations (E–H) at both E12.5 (E,F) and E14.5 (G,H) are shown in control and mutant. (I,J) Distribution and number of Tbr2 cells at E11.5 (I) and E12.5 (J; n=2 for each stage, error bars show mean ±S.E.M, *P<0.05). Double-immunostaining for EdU (K,O) and Tbr2 (L,P) at E11.5 shows increased double-positive cells in the control (M,N) compared to mutant (Q,R). (N and R are higher magnification or M and Q respectively).
To investigate further the possibility of abnormal differentiation, we performed a 1-hour EdU pulse followed by immunostaining for Tbr2 at E11.5. The EdU pulse labels apical progenitor cell bodies in S-phase that should migrate to the VZ for cell division to create either intermediate or apical progenitor cells but not immediately express the Tbr2 marker that indicates basal progenitor cell fate (Fig. 4K,O). If migrating progenitor nuclei were undergoing precocious differentiation, we would expect to see EdU/Tbr2 double-positive cells in the mid-cortical region. Indeed, Tbr2 staining showed a less restricted pattern of intermediate progenitor cells in the Hsd17b7rud cortex (Fig. 4L,P). In addition, dual-labeled Tbr2-postive/EdU-positive cells were detected in the Hsd17b7rud cortex but not in the control cortex (Fig. 4M,N,Q,R). Taken together, these results demonstrate that migrating nuclei in the Hsd17b7rud mutant cortex are precociously transitioning in vivo from apical progenitor fates to neurons.
Supplementation with cholesterol corrects cellular structural defects but not precocious differentiation of Hsd17b7rud neurons in vitro
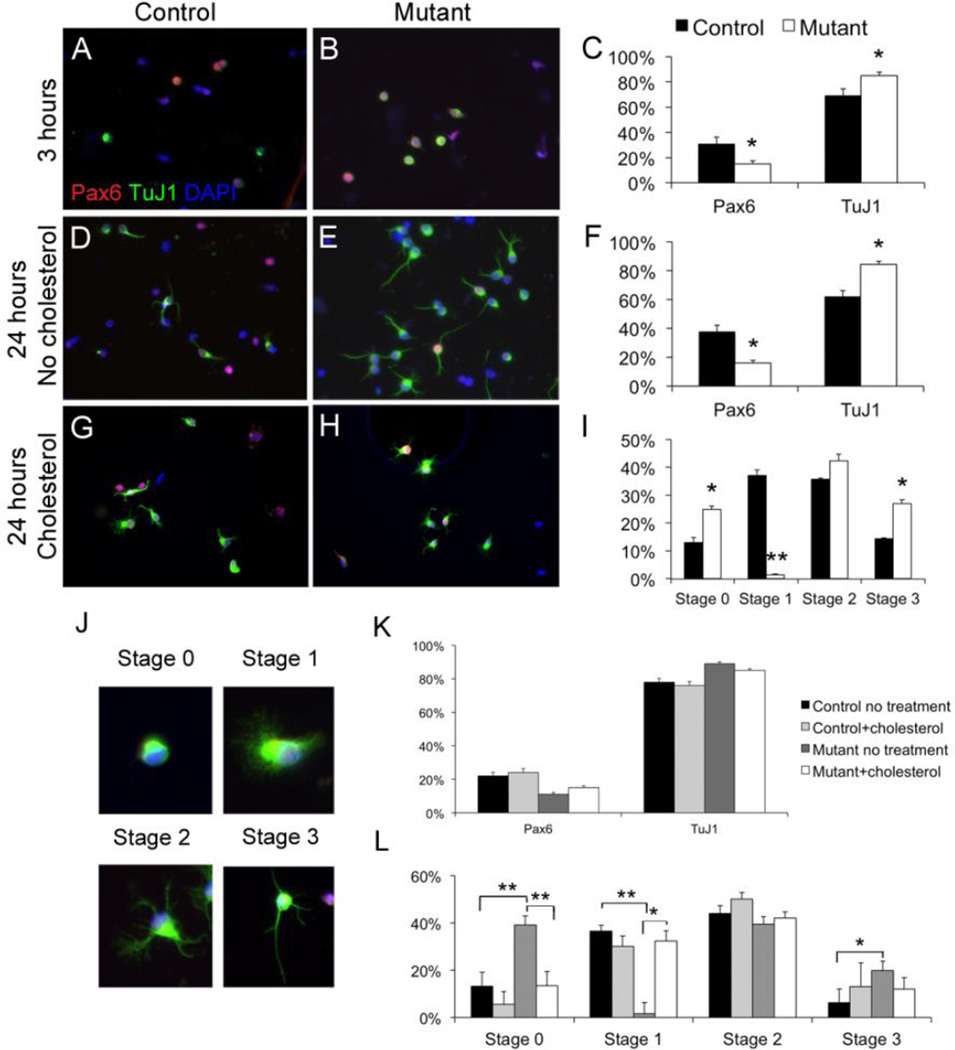
To determine if the precocious Hsd17b7rud neuronal differentiation is a cell-autonomous phenotype, we studied the differentiation of wild-type and Hsd17b7rud mutant neurons in vitro. For this, we isolated neural progenitor cells from E11.5 wild-type and Hsd17b7rud cortices and measured their rates of differentiation using immunohistochemical markers (Shen et al., 2002). Cells were quantified as being either progenitors (solely Pax6 positive) or differentiated (TuJ1-positive) neurons. Although TuJ1-positive cells could still show residual Pax6 immunoreactivity through perdurance of the protein, the product of TUBB3 protein, the TuJ1 antigen, definitively identifies a transition away from progenitor cell fate. As soon as 3-hours in vitro, there were significantly fewer Pax6-positive cells and increased numbers of TuJ1-positive cells in mutant compared to wild-type cultures (Fig. 5A–C). This pattern was again seen at 24-hours in vitro (Fig. 5D–F).
Figure 5. Hsd17b7rud primary neurons show differentiation and structural defects in vitro.
(A–H) Primary E11.5 neurons labeled for TuJ1 (green) and Pax-6 (red) at 3 hours in vitro (A–B), 24-hours (D–E), and 24 hours with cholesterol supplementation (G–H). (J) Neuronal staging is illustrated. Quantification of both Pax6/TuJI immunocytochemistry (C,F) and developmental stage (I) show significant differences between control and mutant. The effects of in vitro cholesterol supplementation are shown with respect to differentiation (K) and morphology (L). Graphs indicate mean ± S.E.M., *P<0.05, **P<0.01.
Neuronal TuJ1 immunoreactivity also allowed us to address neuron maturation in the 24-hour cultures. Early neuronal maturation involves a sequence of developmental stages culminating in neurite extension (Dotti et al., 1988). To better understand if mutant neurons are maturing at a faster rate compared to controls, we classified the neurons using an established grading system (Dotti et al., 1988; Gertz et al., 2010; Kwiatkowski et al., 2007). Neurons having no visible lamellipodia/filopodia structures (Fig. 5J) are defined as stage 0, those with dense lamellipodia/filopodia but no neurites (Fig. 5J) as stage 1, those with immature neurites but no defined axon as stage 2, and those with a defined, dominant axon (Fig. 5J) as stage 3. After 24-hours in culture, there were significantly more mutant cells in stage 0 compared to control (Fig. 5G–I). Interestingly, we found an absence of lamellipodia/filopodia with significantly fewer mutant cells in stage 1(Fig. 5I) and with significantly more mutant cells in stage 3 (with a primary axon) compared to control cells (Fig. 5I). These results indicate that Hsd17b7rud neurons have fewer lamellipodia and show a shift towards forming axons earlier than do control cells. Although we did not definitively test axonal/dendritic character in vitro, the patterns of process extension show clear differences in control vs. mutant neuron maturation.
Funfschilling et al. (2012) showed that in vitro cholesterol supplementation of neurons isolated from squalene synthase conditional (SQS/Nex-cre) mice improved neurite outgrowth and survival. Therefore, we supplemented neurons in culture with cholesterol to determine if cholesterol supplementation could ameliorate the observed defects in Hsd17b7rud neurons. Although Hsd17b7rud cultures continued to have an increased ratio of TuJ1:Pax6 with cholesterol supplementation (Fig. 5K), structural changes in the Hsd17b7rud neurons were nonetheless observed. For example, there were significantly more lamellipodia structures in mutant cells cultured with cholesterol compared to those cultured without (Fig. 5L). In addition, the percentage of mutant cells in stage 0 was closer to wild-type levels after cholesterol supplementation (Fig. 5L).
Overall, these data argue that the apical progenitor pool is prematurely depleted in the Hsd17b7rud cortex due to precocious differentiation of progenitors into intermediate progenitors and post-mitotic neurons. In addition, although cholesterol supplementation is able to improve structural integrity of the Hsd17b7rud cells, it does not correct the defects in cell fate.
Reduced Hsd17b7 activity affects cortical lamination and radial glia fiber integrity in vivo
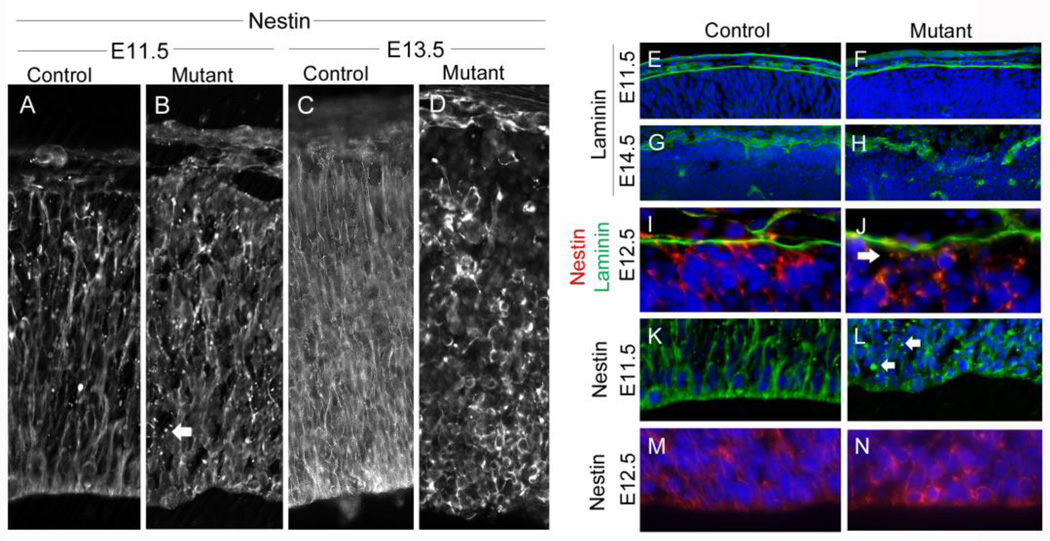
Structurally, apical progenitors connect to both the apical and basal surfaces of the cortex via glial endfeet, providing critical migratory tracts and signaling during early neurogenesis. Loss of apical endfeet has been implicated in the transition from apical to intermediate progenitors (Farkas and Huttner, 2008). In addition, Weimer et al. (2009) reported that disruption of radial glia resulted in abnormal patterns of progenitors with reduced Pax6/Tbr2 levels and that Tbr2-positive cells were shifted towards the ventricular surface, similar to what we observe in the Hsd17b7rud cortex. Therefore, we investigated if loss of radial glia structural integrity and/or endfeet could be contributing to the abnormal differentiation patterns in the Hsd17b7rud mutants. We first studied nestin, an intermediate filament protein that marks progenitor cells in the CNS, including the radial glia cell fibers (Dahlstrand et al., 1995), and found that nestin staining revealed disorganization of radial glial fibers occurring as early as E11.5 in the Hsd17b7rud mutants, leading to the appearance of small nodules along the length of the radial fibers (Fig. 6A,B, arrow). By E13.5, there is complete disruption of fibers in the Hsd17b7rud cortex, with clumping at the apical surface and lack of glial endfeet at the basal surface (Fig. 6C,D). By staining laminin to assess the integrity of the basement membrane, we found that their basal surfaces appeared intact at E11.5 in both the mutant and control cortex (Fig. 6E,F). By E14.5, however, laminin-staining shows a disrupted basement membrane in the mutant cortex, including some areas devoid of any staining (Fig. 6G,H). Using double-immunostaining for nestin and laminin to determine when the connection between the basal endfeet of the radial glia and laminin is lost, we noted a significant decrease in glial endfeet staining at E12.5 (Fig. 6I,J arrow). Although the basal endfoot connection was lost in the Hsd17b7rud mutants, (Haubst et al., 2006) showed that this connection is dispensable for IKNM and for differentiation. However, because loss of the apical endfoot appears to cause severe disruption in both IKNM and progenitor differentiation, we used nestin staining to assess apical integrity. At E11.5, the radial fibers at the apical surface of the mutant appear disrupted, again with the presence of small nodules (Fig. 6K,L, arrows). By E12.5, radial fibers appear densely packed in the control, whereas the staining in the mutant appears to surround individual cells, indicating complete disruption of the radial fiber (Fig. 6M,N). This suggests that the precocious differentiation and abnormal IKNM seen in the Hsd17b7rud mutants could be partly due to disrupted radial glia in the Hsd17b7rud cortex.
Figure 6. Radial glia and basement membrane defects in the Hsd17b7rud cortex.
(A–D) Nestin staining of radial glial fibers at E11.5 (A,B) and E13.5 (C,D) in control (A,C) and mutant (B,D). (E–J) Laminin marks the pial basement membrane at E11.5 (E,F) and E14.5 (G,H). Double-immunostaining for nestin (red) and laminin (green) at E12.5 show failed connection in the mutant (J, arrow) as compared to control (I). (K–N) Higher magnification of nestin at the apical surface (L, arrows indicate small nodules in mutants). Nestin staining at E12.5 surrounds mutant progenitor cells (N). n=3 for each comparison.
Transcriptional profiling of Hsd17b7rud embryos reveals changes in cholesterol biosynthesis and lipid signaling but not in canonical developmental pathways
To identify the molecular mechanism for these developmental defects, we first analyzed several candidate canonical signaling pathways, including sonic hedgehog, Wingless-Int, and Notch, using a combination of reporter assays and transcriptional profiling. We found no evidence that any of these pathways is involved in causing the Hsd17b7rud cortical phenotype (Fig. S2).
As would be predicted by the known roles for Hsd17b7 in cholesterol metabolism, RNA-Seq analysis revealed changes in a number of pathways involved in cholesterol biosynthesis and in oxysterol and lipid signaling (Table 1). Gαi signaling, LXR/FXR activation, and cAMP-mediated signaling were the most significantly affected pathways (P<0.01). However, there is no evidence that these signaling modules affect cortical development, and genetic perturbations of genes in these pathways do not phenocopy the Hsd17b7rud mutants (Dworkin et al., 2009). It appears that the Hsd17b7rud cortical phenotype could result from abnormal cholesterol biology and signaling proper, rather than an upstream effect on any of these canonical developmental pathways.
Table 1.
Pathways enriched between Hsd17b7rud and control E12.5 forebrain.
| Canonical Pathways | p value | # of molecules |
Molecules |
|---|---|---|---|
| Gαi Signaling | 0.006 | 4 | GRM3,CHRM4,PRKAR1B,GNG12 |
| LXR/RXR Activation | 0.006 | 4 | IL36G,NCOR2,GC,CYP51A1 |
| cAMP-mediated signaling | 0.040 | 4 | CAMK1D,GRM3,CHRM4,PRKAR1B |
| α-Adrenergic Signaling | 0.014 | 3 | PRKAR1B,PYGB,GNG12 |
| Nitric Oxide Signaling in the Cardiovascular System |
0.019 | 3 | CACNA1S,FLT1,PRKAR1B |
| Neuropathic Pain Signaling In Dorsal Horn Neurons |
0.021 | 3 | CAMK1D,GRM3,PRKAR1B |
| FXR/RXR Activation | 0.038 | 3 | IL36G,GC,SLCO1B3 |
| Cardiac β-adrenergic Signaling | 0.044 | 3 | CACNA1S,PRKAR1B,GNG12 |
| Prostanoid Biosynthesis | 0.001 | 2 | PTGES,PTGS1 |
| Glycogen Degradation III | 0.002 | 2 | PYGB,MGAM |
| D-myo-inositol (1,4,5)- trisphosphate Degradation |
0.005 | 2 | SEC16A,PMPCA |
| 1D–myo-inositol Hexakisphosphate Biosynthesis II (Mammalian) |
0.006 | 2 | SEC16A,PMPCA |
| D-myo-inositol (1,3,4)- trisphosphate Biosynthesis |
0.006 | 2 | SEC16A,PMPCA |
| Superpathway of D-myo-inositol (1,4,5)-trisphosphate Metabolism |
0.009 | 2 | SEC16A,PMPCA |
| Induction of Apoptosis by HIV1 | 0.048 | 2 | DIABLO,BID |
| Branched-chain α-keto acid Dehydrogenase Complex |
0.023 | 1 | DBT |
| Acetyl-CoA Biosynthesis I (Pyruvate Dehydrogenase Complex) |
0.035 | 1 | DBT |
| Zymosterol Biosynthesis | 0.035 | 1 | CYP51A1 |
| Glycoaminoglycan-protein Linkage Region Biosynthesis |
0.041 | 1 | XYLT1 |
P-value< 0.05
Alteration of cholesterol biosynthesis via simvastatin and dietary cholesterol supplementation results in partial restoration of the wild type cortical phenotype
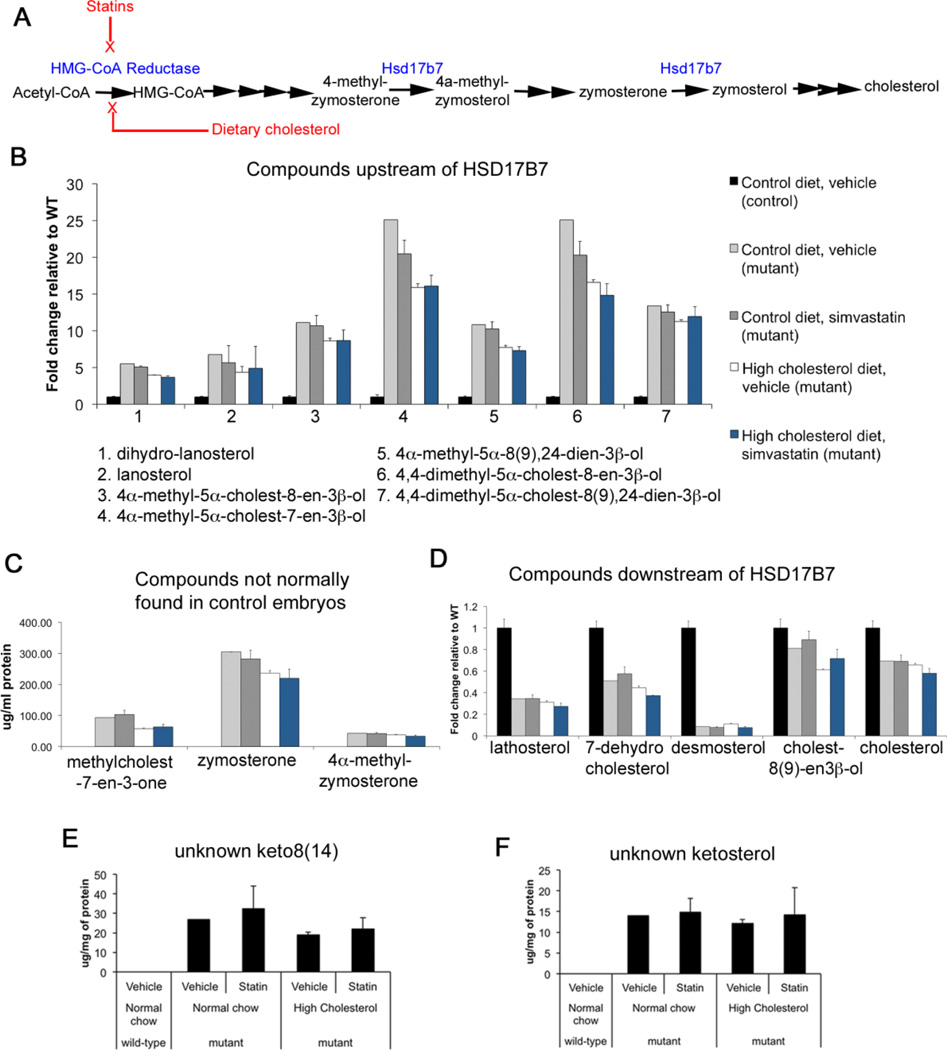
Hsd17b7rud mice show defects in cholesterol biosynthesis with a marked increase in forebrain levels of substrates of the Hsd17b7 enzyme (e.g., zymosterone and 4α-methylzymosterone), as well as intermediates further upstream from the enzymatic block (e.g., lanosterol) and several unidentified sterols in forebrain tissue (Stottmann et al., 2011). We hypothesized the precursor sterols present at increased levels in Hsd17b7rud mice contributed to abnormal forebrain neurogenesis, and that reducing these levels would improve cortical development. We pursued two modes of rescue: 1) treatment with simvastatin to reduce flux in the cholesterol biosynthesis pathway by inhibiting its rate liming step, HMG-CoA reductase, which would in turn cause SREBP2-mediated up-regulation of most enzymes involved in cholesterol biosynthesis, including residual activity of Hsd17b7; and 2) dietary supplementation with cholesterol (standard diet + 2% cholesterol by weight) to cause negative feedback on the pathway by providing its end product (Fig. 7A). We also administered a combined simvastatin + cholesterol treatment to try to enhance the effects of each individual treatment. We predicted that all treatments would lower the levels of possibly toxic intermediate compounds and at least partially rescue the wild-type phenotype, with the most effective treatment being the combination of statin and cholesterol.
Figure 7. Dietary cholesterol and statin administration alter the cholesterol biosynthesis profile in Hsd17b7rud embryonic brains.
(A) A schematic of treatment effects on cholesterol biosynthesis. (B) Levels of compounds upstream of the HSD17B7 enzyme after each treatment. (C) Substrates and compounds normally found at very low levels in the control. (D) Compounds downstream of the HSD17B7 enzyme. (E,F) Ketosterol levels. Color code legend in panel B applies to panels B–D. All graphs show values +/− S.E.M, n=3 for all treated animals, n=1 for untreated mutant and values are similar to other mutants we have analyzed.
Dietary cholesterol supplementation of female Hsd17brud mice was started prior to mating, whereas statin administration (simvastatin, 50mg/kg) started at E7.5, prior to development of the neural tube and forebrain. Embryo sterol levels were quantified at E14.5 by GC-MS (Table S1). The levels of compounds upstream of the Hsd17b7 enzyme were increased in mutants receiving control treatments but reduced in all experimental diets (Fig. 7B). In addition, two compounds (4-methylcholest-7-en-one and 4α-methylzymosterone) usually found at low levels or totally absent in controls but increased in the mutant were lower in the mutants treated with simvastatin and/or cholesterol compared to controls (Fig. 7C). For compounds downstream of Hsd17b7, statin treatment alone was associated with either no change or a slight increase in their levels (Fig. 7D). In contrast, treatment with cholesterol caused decreased levels of both 7-dehydrocholesterol and cholest-8(9)-en-3β-ol, metabolites distal to the Hsd17b7 enzymatic block (Fig. 7D). In the brains of all mutant groups, cholesterol levels were at least 30% lower than in wild-type brain tissue (Fig. 7D). Although we had hypothesized that the unidentified apparent ketosterol species detected in Hsd17b7rud brain would be reduced with treatment, their levels remained high in the mutants of all treatment groups (Fig. 7E,F). Overall, these results suggest that administration of simvastatin and/or cholesterol did effectively altered cholesterol biosynthesis in the Hsd17b7rud.
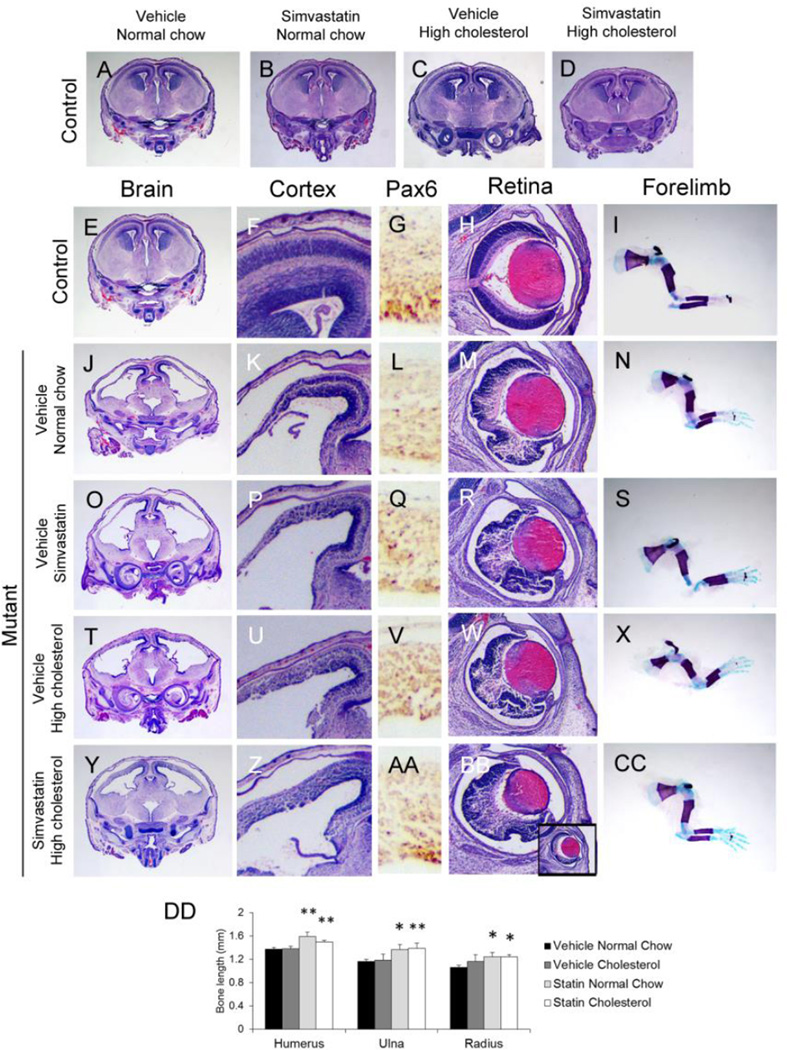
We next determined if manipulating cholesterol metabolism affected the phenotype. Among embryos collected at E16.5 from all treatment groups, none of the treatments affected forebrain development in wild-type embryos (Fig. 8A–D). Simvastatin treatment alone did not alter to any appreciable degree the phenotype of the Hsd17b7rud forebrain, which remained thin and disorganized (Fig. 8O,P), similar to the mutant given only vehicle (Fig. 8J,K). Although high cholesterol treatment alone appeared to have only a minimal effect on cortical differentiation (Fig. 8U,V), the combined treatment of high cholesterol and simvastatin led to substantial structural and histological improvement, yielding a more robust cortex and a partially restored cortical organization (Fig. 8Y,Z). Compared to mice given other treatments, the combined simvastatin and cholesterol treatment group was the only one that showed significant staining for Pax6-positive apical progenitors along the ventricular surface (Fig. 8L,Q,V,AA). We also noted improvement towards normal in Hsd17b7rud extra-cortical abnormalities, including retinal structure and long bone length. For example, the retinae of control animals had proper retinal lamination (Fig. 8H), whereas the vehicle-treated Hsd17b7rud mutant had severe disorganization of the retinal layers (Fig. 8M). Long bones (humerus, radius, and ulna), which were significantly shorter in the Hsd17b7rud embryos (Fig. 8I,N; Stottmann et al., 2011) were significantly less shortened in Hsd17b7rud mutants treated with simvastatin in both the normal chow and high cholesterol groups (Fig. 8I,S,CC,DD). Taken together, these results show that administration of cholesterol, with or without simvastatin, achieves recovery of normal cortical development and improved bone growth in a mouse mutant of a cholesterol biosynthesis enzyme. The most likely compounds exerting this teratogenic effect are the 4-methylsterols, which showed the most significant reduction in their levels with treatment.
Figure 8. Cholesterol and simvastatin treatment partially rescued Hsd17b7rud phenotypes.
(A–D) Control embryos in all treatments are unaffected. (E-CC) Representative whole brain (E,J,O,T,Y), cortical (F,K,P,U,Z), and retinal (H,M,R,W,BB) histology, immunohistochemistry for apical progenitors using Pax6 (G,L,Q,V,AA), and long bone skeletal preps (I,N,S,X,CC) are shown for control (E–I), and mutants treated with vehicle (J–N), simvastatin (O–S), high cholesterol (T–X) and combination of simvastatin and cholesterol (Y-CC). (DD) Quantification of long bone length.*P<0.05, **P<0.01.
Conclusions
We report here adverse effects on progenitor cell fate and function associated with impaired function of the cholesterol biosynthesis pathway enzyme, Hsd17b7. Specifically, we find that neuroprogenitors in the Hsd17b7rud cortex undergo improper IKNM and divide inappropriately leading to displacement of the apical progenitor stem cell pool. With the loss of apical progenitor cells from the VZ, normal polarity is disrupted by E13.5 in the Hsd17b7rud mutant. Further studies in vivo showed that apical progenitors shift into non-renewing intermediate progenitor cell and differentiated neural cell fates. In vitro culture of mutant neuroprogenitor cells also showed precocious differentiation and altered neurite development. In addition, we were able to partially recover normal cortical histology in the Hsd17b7rud mutant by treatment of pregnant dams with simvastatin and dietary cholesterol. These findings demonstrate that reduced activity of a cholesterol biosynthesis enzyme can cause abnormal neuroprogenitor fate determination, a phenomenon not previously documented.
Normal function of Hsd17b7 is required for proper maintenance of neural progenitor cell populations
Because of the unique ability of apical progenitor cells to self renew, their maintenance is critical to ensure that the developing brain has a constant source of stem cells. Cappello et al. (2006) showed that a loss of the Rho-GTPase, Cdc42, increased the production of basal progenitor cells and neurons at the expense of apical progenitors. The Hsd17b7rud progenitor cell resembles that of Cdc42-deficient cells in their abnormal migration and increased rates of mitosis distant from the VZ. However, the Hsd17b7rud mutant shows abnormal differentiation prior to the appearance of tissue polarity defects, suggesting that disrupted polarity is not the primary cause of abnormal neurogenesis in the Hsd17b7rud cortex.
For neural stem cells to self-renew, they must avoid premature cell cycle exit and differentiation. Our EdU pulse-chase analysis showed that migrating nuclei in the Hsd17b7rud cortex prematurely differentiate. In addition, our analysis of IKNM showed mutant cells accumulating in the VZ (with a lack of accumulating cells in the outer cortex) 8 hours after labeling, suggesting that the cells were able to migrate both apically and basally, only at a slower rate. Since IKNM is coordinated with the cell cycle, a lengthened cell cycle could explain the patterns we observed. Moreover, in view of the finding (Garcia-Garcia et al., 2012) that lengthening the cell cycle results in increased production of intermediate progenitor cells, a longer apical progenitor cell cycle in the Hsd17b7rud cortex could be stimulating differentiation into intermediate progenitors and neurons at a faster rate.
We found additional support for the precocious differentiation model in vitro. We saw decreased populations of Pax6-positive apical progenitor cells and correspondingly increased numbers of TuJ1-postive neurons in mutant cultures at both 3 and 24 hours in vitro. Evidence supporting the hypothesis that cholesterol pathway metabolites can affect differentiation in vitro comes from studies of Smith Lemli Opitz Syndrome models where redued DHCR7 activity leads to accumulation of the metabolite 3β,5α-dihydroxycholest-7-en-6-one (DHCEO). Treatment with DHCEO in vitro accelerates differentiation of primary cortical neurons (Xu et al., 2012). Of interest, cholesterol supplementation did not appear to rescue this phenotype but did improve the morphological defects. Namely, mutant cells more closely resembled wild-type cells in their increased number of lamellipodia. Although lamellipodia are actin-based structures, other work has suggested that lipid rafts, specific microdomains comprised of sphingolipids and cholesterol, mediate the reorganization of actin required for lamellipodial extension in growing neurites (Haglund et al., 2004). The abnormal sterol metabolism in the Hsd17b7rud mutant could be altering the composition of lipid rafts thereby affecting signaling necessary for lamellipodia formation. Abnormal membrane composition of Hsd17b7rud cells is further supported by the normalization of the Hsd17b7rud phenotype with cholesterol supplementation.
G-coupled signaling and oxysterol pathways involved in the etiology of the Hsd17b7rud CNS defects
Although the Shh, Notch, and Wnt signaling pathways have roles in early patterning and neural development and were logical candidates for analysis, they do not appear to be affected in the Hsd17b7rud forebrain. None of these pathways appeared significantly changed at the transcriptional level as determined using RNA-Seq and genetic analyses with reporter mice. However, transcriptional profiling identified several pathways involved in cellular and lipid signaling to be significantly altered in the Hsd17b7rud mutants, consistent with the known role of Hsd17b7 in cholesterol biosynthesis. Moreover, lipid second messenger systems have been shown to serve as mechanisms for differentiation for neural stem cells (Bieberich, 2012). In our studies, RNA-Seq analysis comparing Hsd17b7rud and wild-type E12.5 forebrains detected two lipid-activated pathways, Gαi and cAMP-mediated signaling, to be enriched in Hsd17b7rud vs. wild-type forebrains (Bieberich, 2012). These pathways regulate neurite outgrowth and differentiation in both the CNS and neural stem cell lines (Ma’ayan et al., 2009). CREB, a response-element protein in cAMP-signaling, has been shown to stimulate neurite outgrowth and to mediate the neurotrophin response (Finkbeiner et al., 1997; Schmid and Maness, 2008). These G-protein-coupled pathways also rely on lipid rafts for signaling. Abnormal lipid rafts have been described in a number of neurodegenerative conditions, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (Wang, 2014). It is possible, therefore, that impaired cholesterol biosynthesis in the Hsd17b7rud mutant increases the level of specific cholesterol precursors that activate differentiation pathways or impair signaling pathways dependent on normal lipid raft structure.
Our RNA-Seq studies also found enrichment of pathways involving nuclear receptors, such as LXR (liver X receptor), that can bind cholesterol and its derivatives (Ogundare et al., 2010). LXR, a sensor of tissue levels of cholesterol, aids in regulating cholesterol metabolism and controlling the plasma membrane cholesterol transporter, ABCA1. Both FXR and RXR are hypothesized to have roles in neural stem cell maintenance and differentiation (Eendebak et al., 2011). For example, mouse embryonic stem cells can be induced to follow a neural fate through binding of retinoic acid to its nuclear receptor, RXR, after which retinoic acid response elements activate downstream transcription factors to induce in vitro differentiation into neuronal lineages (Guan et al., 2001). The action of these receptors in determining neural fate support a role for disturbed lipid and cholesterol homeostasis in determining the Hsd17b7rud forebrain phenotype.
Manipulating cholesterol biosynthesis can improve forebrain development in Hsd17b7rud embryos
As we have shown, simultaneous administration of simvastatin and dietary cholesterol to pregnant dams causes partial amelioration of numerous defects in the Hsd17b7rud embryos, including restoration of retinal organization, long bone length, and forebrain cortical thickness. The greatest improvement in forebrain cortical thickness was noted in the simvastatin plus high cholesterol treatment group. Of note, although we achieved partial recovery of normal brain structures, total plasma cholesterol levels were not increased in any of the mutant treatment groups as compared to vehicle control diet. In animal models of other defects in cholesterol biosynthesis, there is also evidence that specific cholesterol precursors and their by-products compromise embryonic development (Gaoua et al., 1999). For example, Korade et al. (2010) demonstrated in vitro that Dhcr7-deficient Neuro2a cells produce increased levels of abnormal oxysterols that induce abnormal proliferation and differentiation. Further work by (McLarren et al., 2010; Xu et al., 2012) reported that exposure of neurons to a DHCR7-mutant oxysterol mixture resulted in accelerated differentiation of neurons. In addition, increased levels of 4-methyl- and 4,4’-dimethylsterols have been reported in granule cell precursors from Nsdhl mouse mutants (Cunningham et al., 2015). Our initial GC-MS analysis identified a number of sterols at significantly increased levels in mutant vs. wild-type, not all of which could be identified (Stottmann et al., 2011), but we were able to show decreases in methylsterol levels and an overall reduction in cholesterol biosynthesis coincident with our phenotypic rescue. This was likely due to a global activation of cholesterol biosynthesis enzymes via sterol response element binding protein 2 (SREBP-2), a critical regulator of cholesterol biosynthesis. It has been shown up-regulation of SREBP-2 leads to increased expression of cholesterol biosynthesis enzymes throughout the pathway (Sakakura et al., 2001), both in low cellular cholesterol conditions and during statin exposure (Wong et al., 2006). Therefore, it is likely that our treatments activated SREBP-2 resulting in augmentation of residual Hsd17b7 activity and other cholesterol biosynthetic enzymes, thereby alleviating the accumulation of sterol intermediate compounds.
Increased methyl sterol levels have been reported in both humans and mice with NSDHL mutations, which can lead to conditions such as CHILD and CK syndromes (McLarren et al., 2010). In both mouse models and human patients with disorders of cholesterol biosynthesis, there are a number of neural deficits reported. In the case of the Bare patches (Bpa) mouse, male embryos undergo lethality by E10.5 with severe cortical thinning due to abnormal apoptosis and proliferation (McLarren et al., 2010). In surviving male patients with CK syndrome, there are a variety of CNS abnormalities including intellectual disability, cortical malformations, and seizures (McLarren et al., 2010). In a recent study by Cunningham et al. (2015), methylsterols were implicated as the cause of abnormal development of granule cell precursors in a mouse mutant of Nsdhl, the enzyme of cholesterol biosynthesis immediately upstream of Hsd17b7. Reducing the levels of methylsterols, including 4α-methylcholest-7-en-3β-ol and 4α-methylcholest-8(9),24-dien-3β-ol, led to increased mutant cell viability and morphological improvement (Cunningham et al., 2015). Interestingly, the levels of the methylsterols, including 4α-methylcholest-8(9),24-dien-3β-ol and 4α-methy-5α-cholest-7-en-3β-ol, that were increased in the Nsdhl mutants decreased toward normal in our Hsd17b7rud mutants under all treatment conditions (Fig. 6B). Therefore, methylsterols could also be contributing to the abnormal neural development in the Hsd17b7rud mutants similar to what is being reported in Nsdhl mutants. In addition, a number of the 4,4’-dimethysterol compounds accumulated upstream of Hsd17b7 have known roles as meiosis activating sterols (MAS). Detected in both follicular fluid (FF-MAS) and testicular tissue (T-MAS), these sterols have a role in resumption of meiosis (Byskov et al., 1999). In addition, research has shown that FF-MAS promotes both nuclear and cytoplasmic maturation of mouse oocytes in vitro (Marin Bivens et al., 2004). An increased level of T-MAS in vitro also alters the morphology of cerebellar granule cell precursors and reduced their viability (Cunningham et al., 2015). Therefore, this evidence suggests that these sterols with steroid-like actions could be contributing to the cortical phenotype in the Hsd17b7rud forebrain.
Our studies show that inducing changes in cholesterol metabolism and biosynthesis can improve but not normalize neural development in the Hsd17b7rud mutant. Based on their studies of maternal-fetal cholesterol economy in mice, Yoshida and Wada (2005) concluded that very little of the maternal cholesterol transported to the embryo reaches the brain, which appears to synthesize de novo almost all needed cholesterol. Moreover, others have shown that, postnatally, dietary cholesterol is unable to cross the blood-brain-barrier (BBB) (Jurevics et al., 1997; Pardridge and Mietus, 1980). Therefore, our cholesterol treatment might have been effective prior to formation of the BBB, but was less effective thereafter. Because simvastatin can cross the BBB, however, simvastatin in combination with cholesterol could have suppressed the accumulation of cholesterol precursors by up-regulation of residual Hsd17b7 activity in mutants long enough after early BBB formation to allow better neural development in the combined cholesterol + simvastatin group compared to cholesterol-only treatment. Regardless, our study has shown that in vivo manipulation of cholesterol metabolism can improve neural development in the embryonic forebrain and retina of the Hsd17b7rud mutant.
A model for forebrain development in the Hsd17b7rud mutant
Based on these results, we present the following model of abnormal cortical development in the Hsd17b7rud mutant: the telencephalon patterns normally by E10.5, showing proper polarity and proliferation kinetics. Between E9.5 and E12.5, neuroepithelial cells begin transitioning to radial glia (Anthony et al., 2004). As the apical progenitors in the Hsd17b7rud mutant undergo IKNM, the radial structures become compromised, and IKNM patterns are therefore altered. In contrast to the normal renewal of apical progenitors at the VZ, the Hsd17b7rud apical progenitor cells precociously undergo neurogenesis and thereby expand the intermediate progenitor cell population. This continual loss of stem cell-preserving divisions results in progressive loss of the stem cell pool in the Hsd17b7rud cortex. In addition, radial glia fibers fail to span the cortex due to a loss of structural integrity. Apical endfeet are lost, and adherens junctions no longer form, leading to a reduced anchoring base for the remaining apical cells. As a result, the apical progenitors organize into neural rosettes. With progressive loss of the progenitor pool, the brain is unable to grow, resulting in an extremely thinned forebrain. In addition, the precocious differentiation causes post-mitotic neurons to locate throughout the cortex. The result is a forebrain with a limited and progressively absent stem cell pool, which prevents proper expansion and development of the embryonic telencephalon.
Materials and Methods
Animals
All animals were housed under an approved protocol and standard conditions. For embryo collections, noon of the day of vaginal plug detection was designated as E0.5.
Immunohistochemistry
Immunohistochemistry (IHC) samples were fixed for 2 hours in 4% paraformaldehyde at 4°C and cryo-embedded. Sections were treated with an antigen retrieval solution and blocked in 5% normal goat serum prior to overnight incubation at 4°C in primary antibody. Primary antibodies used were: phalloidin (Sigma, 1:100), β-catenin (Abcam, 1:100), ZO-1 (Abcam, 1:100), Ki67 (Abcam, 1:1000), TuJ1 (Abcam, 1:500), anti-pHH3 (Sigma, 1:500), anti-Nestin (Nakafuku and Nakamura, 1995), Laminin (Sigma, 1:1:250), Pax6 (1:1000, MBL, International Corp), and anti-Tbr2/eomes (Abcam, 1:200). Alexa-Fluor® 488 or 594 goat secondary antibodies (Life Technologies) were used (1 hour at room temperature), followed by DAPI (1:1000) counter staining. All paired images are at the same magnification.
For EdU labeling, pregnant dams were injected with a 20mg/kg EdU solution i.p. at E11.5. Visualization was performed using the Click-iT® EdU Alexa Fluor® 488 Imaging Kit (1:500, Life Technologies).
Quantification of Immunohistochemistry
Quantification used either Imaris Imaging (Bitplane) or NIS Elements (Nikon). To quantify mitotic cells, the VZ (defined at the ventricular surface plus two cell diameters) was compared to the non-VZ (remainder to the pia) and pHH3-positive cells were measured per µm2. For IKNM, the cortical width was divided into four equal-sized bins and cells in each were graphed as a percentage of total. All statistical analyses were done a student’s t-test (P<0.05).
Cortical neuron culture and immunostaining
Cortical neurons were harvested from E11.5 Hsd17b7rudolph and wild-type embryos using standard methods (neuron media of DMEM/F12, 1% Pen-Strep, N2, and B27) and cultured on Poly-d-lysine coated coverslips. Cells were supplemented with EGF (20ng/uL, R&D Systems) and bFGF (10ng/uL, R&D Systems). Neurons were double-immunostained for TuJ1 and Pax6 to and neurite measurements obtained with NeuronJ (Meijering et al., 2004). Data comparing differentiation at 3 and 24 hours in vitro between control and mutant were collected from three independent experiments and multiple embryos (cell number ≥ 467; Fig. 5C,F). Data on the direct comparison of differentiation at 24H in vitro between control and mutant, with and without cholesterol, were collected from two independent experiments and multiple embryos (cell number ≥ 216; Fig. 5K).
RNA Sequencing
RNA was prepared from three E12.5 dorsal forebrains and RNASeq performed on the Illumina HiSeq2500. Analysis was with DESeq normalization and Avadis-NGS at a statistical threshold of P<0.05.
Simvastatin and cholesterol administration
Pregnant dams (n=3 per treatment group) were given either 50 mg/kg of simvastatin (10mg/mL; Sigma) or vehicle (DMSO/1% carboxymethycellulose) via daily oral gavage from E7.5–15.5. Cholesterol supplementation was through supplemented chow starting one week prior to breeding (standard diet + 2% cholesterol, Harland/Teklan Diets, Diet#TD.01383). In vitro cholesterol (Sigma) culture media supplementation was at 15µg/ml (15mg/ml stock in 95% ethanol; (Cunningham et al., 2015).
Sterol analysis via gas chromatography-mass spectrometry (GC-MS)
Embryonic brain and liver samples from each experimental group were collected at E14.5 (n=3 for all treated animals and compared to a single control mutant for which all values were consistent with previous experiments) and snap frozen for GC-MS analysis as previously described (Kelley, 1995; Stottmann et al., 2011). Serum was collected from dams for measuring sterol levels. Sterol concentrations are reported as µg/mg of protein for tissues and µg/mL for plasma.
Phenotype analysis for statin/cholesterol experiments
Embryos were collected at E16.5, and prepared for histology (formalin fixation) or snap frozen for skeletal preparations. Histology and bone/cartilage stains used standard protocols. All images were taken using a Zeiss V8 Dissection Scope and Axiocam MRc5. Measurements were taken using Zeiss Axiovision software. All paired images were taken at the same magnification.
Supplementary Material
(A) Lentiviral knockdown using siRNA resulted in ∼90% reduction of Hsd17b7 mRNA. Spheroid culture and staining for phalloidin (green) shows proper formation in siRNA-treated (C) compared to control cultures (B).
(A,B) Given that Hsd17b7rud mouse embryonic fibroblasts (MEFs) have reduced intracellular signaling downstream of Shh ligand binding (Stottmann et al., 2011), we assayed Shh signaling in the E12.5 mouse forebrain using a Gli-LacZ mouse reporter. At E12.5 there was little to no Shh activity in the dorsal cortex where we noted defects. (C-F) Wnt signaling assays with either Bat-gal (C,D) or Axin-LacZ show no significant alterations between mutant (D,F) and control (C,E) embryos at E11.5. (G) Levels of the Notch intracellular domain (NICD) protein were measured in forebrain lysates from E11.5 Hsd17b7rud and wild-type embryos. If reduced Notch signaling were contributing to the Hsd17b7rud phenotype, there would be lower levels of NICD in the mutant brain. However, there were similar levels between control and mutant cortices (G). A GAPDH loading control is shown below the NICD bands.
Highlights.
-
-
A recessive mutation in Hsd17b7rud causes precocious neural progenitor differentiation.
-
-
Proliferation, polarity, and IKNM are disrupted in the Hsd17b7rud mutant forebrain.
-
-
Hsd17b7rud mutant phenotypes are primarily due to altered cholesterol biosynthesis.
-
-
In utero administration of statins and dietary cholesterol results in partial rescue.
Acknowledgments
Funding
This work was supported by funding to RWS from the Cincinnati Children’s Research Foundation and the National Institutes of Health [NS085023].
We would like to thank K. Campbell and R. Waclaw for discussions on the project and reading this manuscript prior to publication. M. Nakafuku provided the nestin antibody. N. DasGupta helped with the RNA-Seq analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
A.M.D and R.W.S conceived the experiments. A.M.D. and L.E.K. carried out the experiments. A.M.D., L.E.K., R.I.K., and R.W.S. analyzed and interpreted all data. A.M.D., R.I.K, and R.W.S. wrote the manuscript.
References
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Bieberich E. It’s a lipid’s world: bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochemical research. 2012;37:1208–1229. doi: 10.1007/s11064-011-0698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2’-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2’-deoxyuridine antibodies. BioTechniques. 2008;44:927–929. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- Byskov AG, Andersen CY, Leonardsen L, Baltsen M. Meiosis activating sterols (MAS) and fertility in mammals and man. The Journal of experimental zoology. 1999;285:237–242. [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nature neuroscience. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Cunningham D, DeBarber AE, Bir N, Binkley L, Merkens LS, Steiner RD, Herman GE. Analysis of hedgehog signaling in cerebellar granule cell precursors in a conditional Nsdhl allele demonstrates an essential role for cholesterol in postnatal CNS development. Human molecular genetics. 2015;24:2808–2825. doi: 10.1093/hmg/ddv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain research. Developmental brain research. 1995;84:109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin S, Malaterre J, Hollande F, Darcy PK, Ramsay RG, Mantamadiotis T. cAMP response element binding protein is required for mouse neural progenitor cell survival and expansion. Stem cells. 2009;27:1347–1357. doi: 10.1002/stem.56. [DOI] [PubMed] [Google Scholar]
- Eendebak RJ, Lucassen PJ, Fitzsimons CP. Nuclear receptors and microRNAs: Who regulates the regulators in neural stem cells? FEBS letters. 2011;585:717–722. doi: 10.1016/j.febslet.2011.01.039. [DOI] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Current opinion in cell biology. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S, Tavazoie SF, Maloratsky A, Jacobs KM, Harris KM, Greenberg ME. CREB: a major mediator of neuronal neurotrophin responses. Neuron. 1997;19:1031–1047. doi: 10.1016/s0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
- Fitzky BU, Moebius FF, Asaoka H, Waage-Baudet H, Xu L, Xu G, Maeda N, Kluckman K, Hiller S, Yu H, et al. 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith-Lemli-Opitz/RSH syndrome. The Journal of clinical investigation. 2001;108:905–915. doi: 10.1172/JCI12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funfschilling U, Jockusch WJ, Sivakumar N, Mobius W, Corthals K, Li S, Quintes S, Kim Y, Schaap IA, Rhee JS, et al. Critical time window of neuronal cholesterol synthesis during neurite outgrowth. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:7632–7645. doi: 10.1523/JNEUROSCI.1352-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaoua W, Chevy F, Roux C, Wolf C. Oxidized derivatives of 7-dehydrocholesterol induce growth retardation in cultured rat embryos: a model for antenatal growth retardation in the Smith-Lemli-Opitz syndrome. Journal of lipid research. 1999;40:456–463. [PubMed] [Google Scholar]
- Garcia-Garcia E, Pino-Barrio MJ, Lopez-Medina L, Martinez-Serrano A. Intermediate progenitors are increased by lengthening of the cell cycle through calcium signaling and p53 expression in human neural progenitors. Molecular biology of the cell. 2012;23:1167–1180. doi: 10.1091/mbc.E11-06-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz CC, Leach MK, Birrell LK, Martin DC, Feldman EL, Corey JM. Accelerated neuritogenesis and maturation of primary spinal motor neurons in response to nanofibers. Developmental neurobiology. 2010;70:589–603. doi: 10.1002/dneu.20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Chang H, Rolletschek A, Wobus AM. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell and tissue research. 2001;305:171–176. doi: 10.1007/s004410100416. [DOI] [PubMed] [Google Scholar]
- Haglund K, Ivankovic-Dikic I, Shimokawa N, Kruh GD, Dikic I. Recruitment of Pyk2 and Cbl to lipid rafts mediates signals important for actin reorganization in growing neurites. Journal of cell science. 2004;117:2557–2568. doi: 10.1242/jcs.01148. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, Gotz M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development. 2006;133:3245–3254. doi: 10.1242/dev.02486. [DOI] [PubMed] [Google Scholar]
- Jokela H, Rantakari P, Lamminen T, Strauss L, Ola R, Mutka AL, Gylling H, Miettinen T, Pakarinen P, Sainio K, et al. Hydroxysteroid (17beta) dehydrogenase 7 activity is essential for fetal de novo cholesterol synthesis and for neuroectodermal survival and cardiovascular differentiation in early mouse embryos. Endocrinology. 2010;151:1884–1892. doi: 10.1210/en.2009-0928. [DOI] [PubMed] [Google Scholar]
- Jurevics HA, Kidwai FZ, Morell P. Sources of cholesterol during development of the rat fetus and fetal organs. Journal of lipid research. 1997;38:723–733. [PubMed] [Google Scholar]
- Kamei Y, Inagaki N, Nishizawa M, Tsutsumi O, Taketani Y, Inagaki M. Visualization of mitotic radial glial lineage cells in the developing rat brain by Cdc2 kinase-phosphorylated vimentin. Glia. 1998;23:191–199. doi: 10.1002/(sici)1098-1136(199807)23:3<191::aid-glia2>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Kelley RI. Diagnosis of Smith-Lemli-Opitz syndrome by gas chromatography/mass spectrometry of 7-dehydrocholesterol in plasma, amniotic fluid and cultured skin fibroblasts. Clinica chimica acta; international journal of clinical chemistry. 1995;236:45–58. doi: 10.1016/0009-8981(95)06038-4. [DOI] [PubMed] [Google Scholar]
- Korade Z, Xu L, Shelton R, Porter NA. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. Journal of lipid research. 2010;51:3259–3269. doi: 10.1194/jlr.M009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, Kimura A, Matsuzaki F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. The EMBO journal. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, et al. Ena/VASP Is Required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Ma’ayan A, Jenkins SL, Barash A, Iyengar R. Neuro2A differentiation by Galphai/o pathway. Science signaling. 2009;2:cm1. doi: 10.1126/scisignal.254cm1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marijanovic Z, Laubner D, Moller G, Gege C, Husen B, Adamski J, Breitling R. Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Molecular endocrinology. 2003;17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- Marin Bivens CL, Grondahl C, Murray A, Blume T, Su YQ, Eppig JJ. Meiosis-activating sterol promotes the metaphase I to metaphase II transition and preimplantation developmental competence of mouse oocytes maturing in vitro. Biology of reproduction. 2004;70:1458–1464. doi: 10.1095/biolreprod.103.026351. [DOI] [PubMed] [Google Scholar]
- McLarren KW, Severson TM, du Souich C, Stockton DW, Kratz LE, Cunningham D, Hendson G, Morin RD, Wu D, Paul JE, et al. Hypomorphic temperature-sensitive alleles of NSDHL cause CK syndrome. American journal of human genetics. 2010;87:905–914. doi: 10.1016/j.ajhg.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Nakafuku M, Nakamura S. Establishment and characterization of a multipotential neural cell line that can conditionally generate neurons, astrocytes, and oligodendrocytes in vitro. Journal of neuroscience research. 1995;41:153–168. doi: 10.1002/jnr.490410203. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ogundare M, Theofilopoulos S, Lockhart A, Hall LJ, Arenas E, Sjovall J, Brenton AG, Wang Y, Griffiths WJ. Cerebrospinal fluid steroidomics: are bioactive bile acids present in brain? The Journal of biological chemistry. 2010;285:4666–4679. doi: 10.1074/jbc.M109.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth M, Bellosta S. Cholesterol: its regulation and role in central nervous system disorders. Cholesterol. 2012;2012:292598. doi: 10.1155/2012/292598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E, Azzarelli R, Guillemot F. Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nature communications. 2013;4:1635. doi: 10.1038/ncomms2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge WM, Mietus LJ. Palmitate and cholesterol transport through the blood-brain barrier. Journal of neurochemistry. 1980;34:463–466. doi: 10.1111/j.1471-4159.1980.tb06621.x. [DOI] [PubMed] [Google Scholar]
- Porter FD, Herman GE. Malformation syndromes caused by disorders of cholesterol synthesis. Journal of lipid research. 2011;52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakura Y, Shimano H, Sone H, Takahashi A, Inoue N, Toyoshima H, Suzuki S, Yamada N. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis. Biochemical and biophysical research communications. 2001;286:176–183. doi: 10.1006/bbrc.2001.5375. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Current opinion in neurobiology. 2008;18:245–250. doi: 10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehu A, Mao J, Gibori GB, Halperin J, Le J, Devi YS, Merrill B, Kiyokawa H, Gibori G. Prolactin receptor-associated protein/17beta-hydroxysteroid dehydrogenase type 7 gene (Hsd17b7) plays a crucial role in embryonic development and fetal survival. Molecular endocrinology. 2008;22:2268–2277. doi: 10.1210/me.2008-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. The Journal of clinical investigation. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Development, growth & differentiation. 2012;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Turbe-Doan A, Tran P, Kratz LE, Moran JL, Kelley RI, Beier DR. Cholesterol metabolism is required for intracellular hedgehog signal transduction in vivo. PLoS genetics. 2011;7:e1002224. doi: 10.1371/journal.pgen.1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tint GS, Yu H, Shang Q, Xu G, Patel SB. The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. Journal of lipid research. 2006;47:1535–1541. doi: 10.1194/jlr.M600141-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Lipid rafts: a signaling platform linking cholesterol metabolism to synaptic deficits in autism spectrum disorders. Frontiers in behavioral neuroscience. 2014;8:104. doi: 10.3389/fnbeh.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer JM, Yokota Y, Stanco A, Stumpo DJ, Blackshear PJ, Anton ES. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development. 2009;136:2965–2975. doi: 10.1242/dev.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Quinn CM, Brown AJ. SREBP-2 positively regulates transcription of the cholesterol efflux gene, ABCA1, by generating oxysterol ligands for LXR. The Biochemical journal. 2006;400:485–491. doi: 10.1042/BJ20060914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Mirnics K, Bowman AB, Liu W, Da J, Porter NA, Korade Z. DHCEO accumulation is a critical mediator of pathophysiology in a Smith-Lemli-Opitz syndrome model. Neurobiology of disease. 2012;45:923–929. doi: 10.1016/j.nbd.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Wada Y. Transfer of maternal cholesterol to embryo and fetus in pregnant mice. Journal of lipid research. 2005;46:2168–2174. doi: 10.1194/jlr.M500096-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Lentiviral knockdown using siRNA resulted in ∼90% reduction of Hsd17b7 mRNA. Spheroid culture and staining for phalloidin (green) shows proper formation in siRNA-treated (C) compared to control cultures (B).
(A,B) Given that Hsd17b7rud mouse embryonic fibroblasts (MEFs) have reduced intracellular signaling downstream of Shh ligand binding (Stottmann et al., 2011), we assayed Shh signaling in the E12.5 mouse forebrain using a Gli-LacZ mouse reporter. At E12.5 there was little to no Shh activity in the dorsal cortex where we noted defects. (C-F) Wnt signaling assays with either Bat-gal (C,D) or Axin-LacZ show no significant alterations between mutant (D,F) and control (C,E) embryos at E11.5. (G) Levels of the Notch intracellular domain (NICD) protein were measured in forebrain lysates from E11.5 Hsd17b7rud and wild-type embryos. If reduced Notch signaling were contributing to the Hsd17b7rud phenotype, there would be lower levels of NICD in the mutant brain. However, there were similar levels between control and mutant cortices (G). A GAPDH loading control is shown below the NICD bands.