Abstract
Small RNAs can be used to target and eliminate expression of virtually any disease causing gene or infectious virus, resulting in their pre-clinical and clinical development for treating disease1. To ensure success of RNAi therapeutics, small hairpin RNAs (shRNAs) must co-opt sufficient quantities of endogenous microRNA machinery to elicit efficient gene knockdown without impeding normal cellular function. We previously observed liver toxicity including hepatocyte turnover, loss of gene repression and lethality2 in mice receiving high doses of a recombinant adeno-associated virus (rAAV) vector expressing shRNAs; however the mechanism by which toxicity ensues has not been elucidated. Using rAAV-shRNAs, we have now determined that hepatotoxicity arises when exogenous shRNAs exceed 12% of liver microRNAs. Once this threshold was surpassed, shRNAs specifically reduced the initial synthesized 22-nucleotide isoform of miR-122-5p without substantially affecting other microRNAs resulting in functional de-repression of miR-122 target mRNAs. Delivery of an rAAV-shRNA vector expressing miR-122 could circumvent toxicity despite accounting for 70% of microRNAs. Toxicity was also not observed in miR-122 knockout mice regardless of the level or sequence of shRNA. Our study establishes limits to the microRNA machinery available for therapeutic siRNAs and suggests new paradigms for the role of miR-122 in liver homeostasis in mice.
RNAi-mediated cleavage of cognate mRNAs can be induced by delivery of siRNAs or DNA vectors expressing small hairpin RNAs (shRNAs). Actionable mutant genes have been successfully targeted and degraded in clinical trials, particularly in tissues where high rates of delivery can be achieved3-5, such as for the liver, where >95% of hepatocytes can be transduced by recombinant adeno-associated virus (rAAV) vectors6. However, overt overexpression of rAAV-shRNAs has the potential to overwhelm existing RNAi machinery utilized by both microRNAs and shRNAs in the liver leading to hepatotoxicity.2,7 This is surprising considering that the liver is tolerant to perturbations of key microRNA components, resulting in relatively mild phenotypes in mice with Dicer deficiency that are devoid of microRNAs8,9 or mice with germline ablation of the predominant liver microRNA, miR-12210,11. As a result, the mechanism of shRNA induced liver toxicity is not clear.
To evaluate the consequence of delivering shRNAs on existing microRNA pools, we sequenced liver small RNAs 7 days after tail-vein injection of 5×1011 vector genomes of AAV8-shRNAs of various stem lengths (19nt, 21nt and 25nt) that are dependent on Dicer processing (Supplementary Fig. 1a) and target human alpha-anti-trypsin (hAAT). AAV-hAAT-shRNAs were delivered driven by high-expressing (U6) or low-expressing (H1) pol-III promoters in hAAT transgenic animals yielding robust initial in vivo knockdown of hAAT (Fig. 1a,b and Supplementary Fig. 1b). Substantial but non-lethal toxicity ensued as measured by increased serum alanine transaminase (ALT) levels (Fig. 1c and Supplementary Fig. 1c).
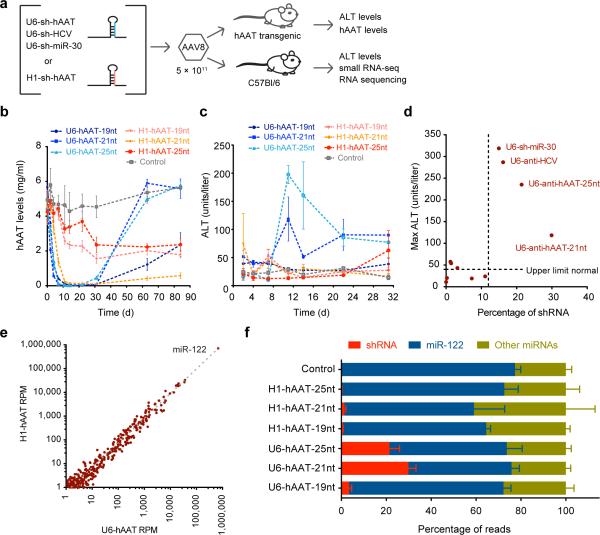
Figure 1.
Evaluation of toxicity relative to efficacy of delivered shRNAs. (a) rAAV injection schematic. (b) Human α-1-antitrypsin (hAAT) levels decrease following AAV8-U6- and H1-shRNA delivery in mice transgenic for human AAT, n = 3; data are mean ± s.e.m. (c) Serum alanine aminotransferase (ALT) levels, indicative of shRNA toxicity, start to rise after approximately a week, but only in U6-driven constructs, n = 3; data are mean ± s.e.m. (d) Comparison of maximum mean ALT levels over 1 month relative to percent shRNA relative to microRNAs at livers extracted at day 7, n = 3 mice per condition. U6-shRNA AAV species with greater than 12% expression coming from shRNAs are noted. (e) Mean microRNA expression from H1-delivered vectors versus U6-delivered vectors reveals no differentially expressed microRNAs, n = 9; RPM = reads per million mapped microRNAs. (f) Percentage of reads that map to shRNAs, miR-122 and all other microRNAs, n = 3; data are mean ± s.e.m.
Sequence reads from rAAV-H1-shRNA constructs contributed to a mean of 0.78% of the total microRNA pool, equivalent to the ~15th most expressed microRNA (Supplementary Table 1). In contrast, shRNA reads constituted 29.9% (U6-hAAT-21nt) and 21.4% (U6-hAAT-25nt) of total reads, higher than all microRNAs in the liver except for miR-122 in both total small RNA pools and those immunoprecipitated with Ago2 (Supplementary Table 1). The U6-sh-hAAT-19nt vector constituted 2.2% of total reads without elevated ALT levels, likely from its poor processing (Supplementary Fig. 1a). To identify the level of shRNA expression that can lead to toxicity, we tested additional U6-shRNA constructs including those against HCV and expressing miR-3012 and H1-shRNA-hAAT-21nt at increased doses of 1×1012 and 2.5×1012 vector genomes. We found that the shRNA expression levels much reach >12% of total microRNA reads to be toxic (Fig. 1d). This provides an upper limit for the spare endogenous cellular machinery available for safe exogenous RNAi activity.
Despite the high accumulation of shRNA reads, we detected no microRNAs with overt relative expression differences between mice receiving toxic U6-shRNAs and non-toxic H1-shRNAs including miR-122 which accounts for ~70% of all human and mouse liver microRNAs13-15 and ~65% in toxic rAAV-U6-shRNA treated mice (Fig. 1e and Supplementary Tables 2,3). However, when we combined reads mapping to microRNAs and shRNAs, we found a ~22% drop in miR-122 expression for toxic U6-shRNAs which actually exceeds the number of reads mapping to the second and third highest expressed microRNAs, miR-21 and let-7a combined (Supplementary Table 1). The miR-122 reads displaced by exogenous shRNAs was far greater than the total non-miR-122 microRNAs, which remained at ~30% of read counts (Fig. 1f and Supplementary Fig. 1d,e). This was not specific to high-level expression of one shRNA because we confirmed these findings using six additional AAV-U6-shRNA constructs targeting HCV or expressing miR-30. Thus, high shRNA levels selectively displaced miR-122.
Since our small RNA sequencing suggests that miR-122 reads are outcompeted by shRNAs, we evaluated the small RNA sequencing reads that map to miR-122 to determine if their composition is altered in any way upon U6-shRNA delivery. miR-122-5p is present in human and mouse livers in four primary isoforms of 21-23nt (Fig. 2a and Supplementary Fig. 2a,b) including a 23nt isoform terminating in a non-templated adenosine16. Surprisingly, delivery of U6-shRNAs specifically reduced the 22-nucleotide isoform (22mer) of miR-122-5p while relative levels of the 21mer isoform increased and 23nt species remained constant (Fig. 2b). This is in marked contrast to the combined length distribution of all other microRNAs including the distribution of miR-122-3p species (20–22nt) which remained unchanged regardless of the delivered AAV-shRNA vector (Fig. 2c and Supplementary Fig. 2c). The 22mer to 21mer ratio of miR-122-5p isoforms was reduced in all U6 samples relative to H1 samples and controls both in the Ago2-IP and total small RNA fractions (Fig. 2d–f) but was not a common feature of other microRNAs (Supplementary Fig. 2d,e). In total, 17 shRNAs targeting six different genes had the same disruption in miR-122 isoform profiles (Supplementary Fig. 2f,g). Together, this suggests that microRNA isoform changes are of consequence only for miR-122-5p. Thus, the exogenously delivered shRNAs in the liver specifically inhibit generation of, compete with, or displace the miR-122-5p 22mer isoform.
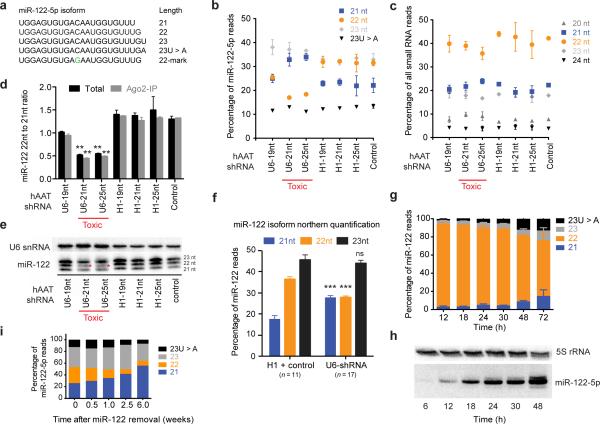
Figure 2.
shRNAs specifically outcompete the 22-nucleotide isoform of miR-122-5p. (a) Isoforms generated by mmu-miR-122-5p along with an artificially marked sequence (22-mark). (b) Only the 22mer of miR-122 is reduced in samples with high shRNA expression, n = 3 mice per condition; data are mean ± s.d. (c) The combined length distribution of all other microRNAs is unchanged, n = 3 mice; data are mean ± s.d. (d) The ratio between 22nt and 21nt isoforms is altered in toxic liver samples both for total and Ago2-immunoprecipitated miR-122-5p, n = 3 mice per condition except control Ago2-IP where n = 1; data are mean ± s.d; ** P < 0.01 relative to all H1-shRNA conditions by one-way ANOVA with Tukey post-hoc test. (e) Northern blots validate the loss of the miR-122-5p 22nt isoform (asterisks). (f) Quantification of northern blot bands shows a consistently lower 22nt isoform in toxic (U6) liver samples (n = 17) relative to H1 and control samples (n = 11; mean ± s.e.m) *** P < 0.001 by paired Student's t test comparing H1 + control samples relative to U6 samples. (g) CMV-driven time course of miR-122-5p biogenesis, transfected in HEK293 cells, n = 2–5 separate sequences per time point, (see Supplementary Fig. 3a,b); data are mean ± s.e.m. (h) The 22nt isoform of miR-122 is the first species to accumulate followed by 21nt and 23nt species. (i) Cre-mediated removal of miR-122 in miR-122-floxed mice10 at various time points following Cre delivery, n = 1 mice per time point.
The differential effect of shRNAs on miR-122 isoforms prompted us to evaluate how the different isoforms arise with the intention of determining if shRNAs affect miR-122 biogenesis or turnover. Consequently, we expressed the miR-122 precursor in HEK293 cells with a nucleotide modification in miR-122-5p to allow us to distinguish it from endogenous miR-122 (Fig. 2a). Time course analysis of miR-122 expression revealed that the 22mer initially contributes ~90% of miR-122-5p reads 12 hours post-transfection, dropping to ~60% by 72 hours (Fig. 2g,h and Supplementary Fig. 3a, 3b). Further support that the miR-122-5p 22mer is the first to be synthesized came when we performed a targeted excision of the miR-122 locus in vivo and found that this isomer was predominantly lost (Fig. 2i). Together, these results suggest that the 22mer isoform of miR-122 is the initial product of Dicer cleavage, and then is subsequently trimmed or tailed to 21 or 23nt species. Therefore our data is consistent with the idea that the loss of the 22nt isoform we observe in vivo in liver tissues receiving toxic levels of AAV-shRNAs represents the loss of newly synthesized miR-122.
If miR-122 loss is indeed the primary consequence of high shRNA expression, we anticipate increased expression of mRNAs bearing miR-122 target matches. To test this, we performed RNA sequencing on control, H1-shRNA and U6-shRNA livers (Supplementary Fig. 4a and Supplementary Table 4), confirmed by RNA-seq of mouse livers receiving one of five additional U6-shRNAs that target HCV or express miR-30a (Supplementary Fig. 4b and Supplementary Table 5). The overlap of differentially expressed transcripts in both RNAseq experiments was considerable (P < 1e−173 by hypergeometric distribution; Fig. 3a). Gene ontology processes such as oxidative phosphorylation, steroid metabolism, and cholesterol biosynthesis were enriched in genes significantly decreased in U6-shRNA livers. Conversely, innate immune response, viral response and programmed cell death pathways were enriched in significantly upregulated genes (Fig. 3b,c and Supplementary Fig. 4c,d).
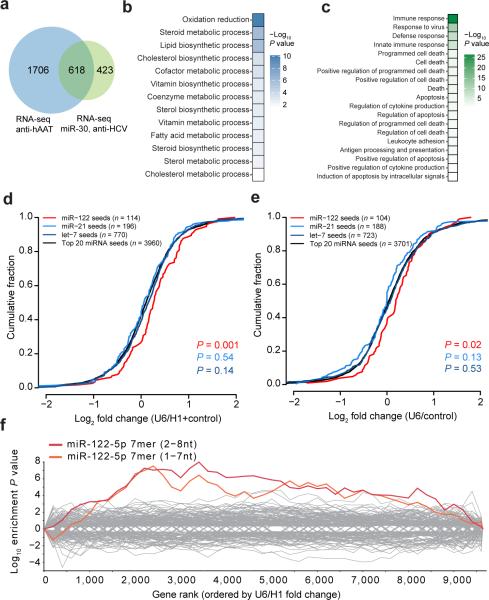
Figure 3.
miR-122 targets are specifically de-repressed in livers receiving toxic shRNAs. (a) Overlap of differentially expressed genes (P < 0.05) from RNA sequencing of U6-hAAT shRNAs compared with U6-sh-HCV and U6-miR-30 shRNAs. (b) Gene ontology (GO) term enrichments (Benjamini P < 0.05) from overlapping genes down-regulated in U6-shRNA toxic livers. (c) GO term enrichments (Benjamini P < 0.05) from up-regulated in U6-shRNA toxic livers. (d) RNAseq analysis indicates that targets of miR-122 are significantly de-repressed when comparing two U6-driven shRNAs versus four H1-driven and control liver samples P values are from a Wilcoxon rank-sum test. (e) RNAseq evaluation of five additional U6-shRNA constructs compared to one control. The number of genes in each condition is listed in parentheses. P values are from a Wilcoxon rank-sum test. (f) Seed enrichment analysis reveals that target mRNAs bearing miR-122 7mer seed matches are de-repressed; genes are ranked in order of differential expression.
Several lines of evidence from our RNAseq analysis pointed to specific de-repression of miR-122 targets. First, 3′UTRs that harbor predicted miR-122 targets had enriched expression in U6-shRNA livers (Fig. 3d,e), in contrast to predicted gene targets for miR-21 or let-7. Second, 73 validated targets of miR-12211 were also significantly upregulated in U6-shRNA livers (Supplementary Fig. 4e). Finally, only the 7mer seed of miR-122 was enriched compared with all microRNA seeds (Fig. 3f). Thus, high U6-shRNA expression appears to primarily affect hepatic miR-122 gene targets.
We examined liver pathology after delivery of shRNAs inducing toxicity that revealed features reminiscent of hereditary fructose intolerance, caused by mutations in Aldolase B (ALDOB)17. We found that decreased Aldob levels significantly correlated with the degree of toxicity in mice and similar to patients with ALDOB mutations, the phenotype was exacerbated by a fructose-rich diet (Supplementary Fig. 5 and Supplementary Table 6). We also found de-repression of Aldoa, a common feature of miR-122 inhibition10,11,18-23. Of note, Aldoa often has opposing expression patterns with miR-122 and Aldob in the developing and mature liver14,24,25. These findings indicate that in situations where Aldob is also suppressed – such as in conjunction with AAV-shRNA delivery – functional metabolic consequences in the liver can occur.
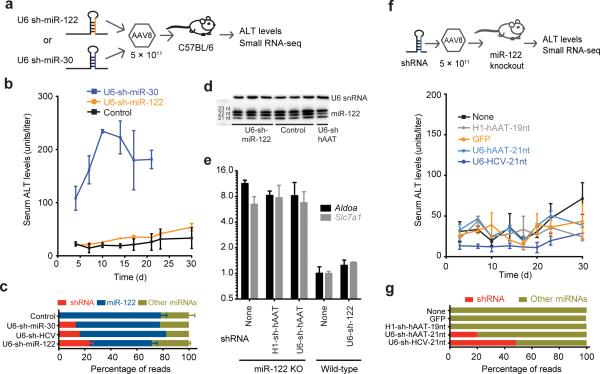
We reasoned that if toxicity was related to competition with miR-122, then delivery of a U6-shRNA construct expressing mature miR-122-5p should be tolerated in mice. Mice receiving this U6-shRNA-miR-122 construct incorporating a marked internal nucleotide (Fig. 2a) maintained normal ALT levels in contrast to mice receiving U6-shRNA-miR-30 (Fig. 4a,b). We detected a displacement of 23.4% of reads mapping to the total microRNA pool for U6-shmiR-122 recipients one-week post-AAV administration, well higher than the minimum proportion (12%) of reads necessary to induce toxicity (Fig. 4c). miR-122 isoform ratios also were unchanged (Fig. 4d). Remarkably, the exogenous miR-122 levels rose to 72% of reads by 8 weeks, almost completely replacing endogenous miR-122 (Supplementary Fig. 6a), all while maintaining normal levels of miR-122 mRNA targets including Aldoa and Slc7a126 (Fig. 4e). This indicates that miR-122 is specifically displaced by any delivered non-miR-122 U6-shRNA resulting in toxicity; however, because exogenous U6-sh-miR-122 functionally replaces endogenous miR-122 toxicity is abrogated.
Figure 4.
Toxicity is abrogated by delivering a U6 construct expressing miR-122-5p or a U6-shRNA in miR-122 knockout mice. (a) Schematic of experimental design. (b) ALT levels measured when mice receive U6-driven shRNAs expressing mature miR-122-5p in control (n = 4), U6-sh-miR-122 (n = 9) and U6-sh-miR-30 (n = 3) mice. Error bars are mean ± s.e.m. (c) The proportion of reads from U6-shRNAs relative to microRNAs including toxic shRNAs compared to U6-sh-miR-122 sequences. Controls are from Fig. 1f; n = 3 mice for U6-sh-miR-122 and controls, n = 1 for sh-miR-30 and sh-HCV. Error bars are mean ± s.e.m. (d) Small RNA northern blot reveals sustained expression of the 22nt isoform from AAV-sh-miR-122 delivered samples after one week. (e) qPCR levels show that miR-122 knockout mice have elevated levels of Aldoa that do not change when additional shRNAs are delivered, n = 3 mice per condition; error bars are mean ± s.e.m. (f) ALT levels remain low in miR-122 knockout (KO) mice receiving U6-shRNAs against hAAT, U6-shRNAs against HCV and H1-shRNAs against hAAT along with injected and non-injected controls, n = 3 mice per condition; error bars are mean ± s.e.m. (g) Proportion of reads that map to shRNAs and microRNAs in miR-122 knockout mice (one mouse per condition).
Based on our studies, we tested the prediction that toxicity would not be induced by AAV-shRNAs in the miR-122 germline knockout mouse10 since no copies of miR-122 are available to be out-competed. We indeed found that delivery of AAV8-U6-shRNAs shown to be toxic in normal mice had normal ALT levels in miR-122−/− mice (Fig. 4f). This occurred despite the robust accumulation of small RNAs, accounting for 22% of the total microRNA pool for U6-hAAT-25nt shRNAs and ~50% for U6-HCV-21nt shRNAs (Fig. 4g and Supplementary Fig. 6b-e). miR-122+/− mice exhibited slightly elevated toxicity upon U6-shRNA delivery but nonetheless had a similar reduction in miR-122-5p 22:21nt isoform ratios and loss of shRNA expression 1-month post-transduction (Supplementary Fig. 6d,f). Aldoa and Slc7a1 levels are already high in miR-122−/− mice, with no further changes incurred by U6-shRNA constructs (Fig. 4e). miR-122−/− mice are generally more fragile than their wildtype counterparts to several physiological perturbations making it particularly notable that that they tolerate AAV-U6-shRNAs more readily than controls.
Our data taken in total provides strong evidence that shRNA-induced toxicity is solely related to competition with and a reduction in the first produced mature isoform of miR-122 (Supplementary Fig. 7). This might explain why a small reduction in the whole miR-122 pool is sufficient for de-repression of target mRNAs. This result is also consistent with the effects of HCV infection where despite a 103–105 fold excess of miR-122 compared to HCV RNA18,27 miR-122 targets are nonetheless efficiently de-repressed18. Blocking miR-122 expression has been pursued for therapeutic purposes19,20,22,23,28. However, competition of the 22mer by shRNAs is different than what is observed through antisense inhibition approaches to miR-122 that differentially target the various isoforms23 (Supplementary Fig. 8a). It is possible that newly synthesized miR-122 products have a unique function or that miR-122 needs to be continually synthesized to sequester targets in a timely manner21. Notably, the seed sequence of miR-122-5p is unchanged regardless of the length of the isoform that is generated; however, in certain situations regions other than the seed are important for target repression29-31.
Despite the overwhelming abundance of miR-122 in the liver, miR-122 germline knockout mice are viable but over a long period develop liver pathology including hepatosteatosis, hepatitis and ultimately hepatocellular carcinoma10,11. Perhaps the lack of severe acute toxicity in mice with a congenital absence of miR-122 is a consequence of their livers that never develop a fully differentiated hepatocyte expression pattern and retain expression of genes typically restricted to development (Supplementary Fig. 8b).
Complications and toxicity arising from high doses of shRNA delivery needs to be recognized and avoided to ensure safe and sustained gene knockdown in particular in situations where rAAV-shRNA constructs are being considered in clinical trials1. The similarities between human, non-human primate and mouse miR-122 concentrations and isoforms in healthy and disease states, as well as the change in liver gene regulation after the infusion of anti-miR-122 oligonucleotides20,22,28, point to the potential translation of these findings to human studies. Our study provides important guidelines for in vivo shRNA expression and suggests that RNAi-induced silencing complexes have limits to which they are safely accessible for piggybacking therapeutic siRNAs. Moreover, our work provides new insights into the functional role of miR-122, the interplay between miR-122 and exogenously expressed shRNAs in the liver and the level to which cellular miRNA levels may be altered by shRNAs without affecting metabolic homeostasis in the liver.
Online Methods
Mouse breeding and rAAV transduction
The Stanford Institute of Medicine Animal Care and Use Committee approved all protocols. The AAV8 serotype was used for all in vivo samples. To test for shRNA efficacy, we generated AAVs bearing shRNAs against the human Alpha-1-antitrypsin (AAT) gene (SERPINA1). 5×1011 vector genomes (in final volume of 200 μl of PBS) of these shRNAs were injected via the tail vein in mice transgenic for human AAT that have been described previously32. For small RNA sequencing samples, we used 6–8 week old female C57Bl/6 mice, purchased from Jackson Laboratory (Bar Harbor, ME). Three mice in each condition received 5×1011 vector genomes shRNAs that were designed to generate a small RNA product against hAAT of 19nt, 21nt or 25nt stem lengths driven either by the high-expressing U6 promoter or the low-expressing H1 promoter. Group sizes for animal studies were selected based on numbers historically suitable for detecting effects in ALT levels. Animals were randomly selected to receive various rAAV vectors with respect to control littermates. No animals were excluded from analysis.
Additional shRNAs that target HCV, that mimic miR-30 and that mimic miR-122-5p were delivered under the same parameters. For shRNAs targeting HCV, three stem-lengths were used, 21nt, 29nt and 29nt with an internal bulge (29-B). Those expressing miR-30 contained stem lengths of 21nt, 24nt and 24nt with an internal bulge (24-B). miR-122 knockout mice were obtained from The Ohio State University10. Female miR-122−/− mice 7–9 weeks of age were used for infections. The Stanford Department of Comparative Medicine Diagnostic Lab generated metabolic panel values (Supplementary Table 6).
miR-122 knockout mice and miR-122 floxed mice with loxP sites surrounding miR-122 have been described previously10. To remove the miR-122 precursor, an rAAV8 vector was generated that expresses Cre under the Ef1a promoter (Addgene plasmid #55636) and was delivered at 1×1012 vector copies per mouse by tail vein injection. Mice heterozygous for miR-122 were generated by breeding miR-122 knockout male mice with C57Bl/6 female mice.
Plasmid design for expressing shRNAs and miR-122
Primers for generating various shRNAs (Supplementary Table 7) have been previously reported2,12. Primers were chemically synthesized (Integrated DNA Technologies), T4-PNK kinased and annealed. shRNA constructs bearing miR-122 and anti-hAAT sequences were ligated into a BbsI restriction site downstream of a U6 or H1 promoter in a scAAV construct. shRNAs expressing a miR-30 sequence or anti-HCV were first ligated into a pBluescript II KS(+) plasmid (Stratagene) containing a U6-promoter using an introduced BglII site and an existing KpnI restriction site. This fragment was then sub-cloned into a scAAV construct using restriction sites NdeI and SalI. AAVs were generated by triple-transfection of pAd5, pAAV8 and various AAV-shRNA constructs as described previously2.
The miR-122 sequence along with ~500 bp of surrounding mouse genomic sequence was PCR amplified using the primers 5′-AATTGTTGCAAACAGAGTTCCTG and 5 ′-TGCAATCTTTTTATTGACTTGGGAA with additional NotI and SalI restriction enzymes that enabled cloning into a CMV-expressing construct.
Small RNA deep sequencing
Small RNAs were purified from mouse liver samples snap frozen and ground into powder in liquid nitrogen using the miRvana kit (Ambion). 1 μg of small RNAs were ligated using T4 RNA ligase in a buffer lacking ATP with 3′ linker-1 (Integrated DNA Technologies) and run on a 15% polyacrylamide gel. A 17–28 nucleotide fragment was excised. Samples were 5′ barcoded (4 nucleotides) for multiple 36 bp sequencing performed on an Illumina GAII machine at the Stanford Functional Genomics Facility (SFGF). Barcodes and linkers were trimmed using custom Perl scripts and then aligned to mouse microRNAs from miR-base (release 15)33 using Bowtie version 0.12.734 allowing for two mismatches. 5 μg of Trizol extracted RNA was used for additional shRNAs to confirm findings. These were sequenced using the same protocol on a MiSeq machine (Illumina) at the SFGF. The Deseq R package35 was utilized to evaluate microRNA expression between samples. A Benjamini-Hochberg adjusted P value of < 0.05 considered to be significant. To detect microRNA isoform ratios (Fig. 2d), a minimum mean 20 reads per million mapped microRNA hits was required for both 22nt and 21nt isoforms, leaving 66 microRNAs.
For Ago2-immunoprecipitated fractions, a Wako antibody (catalog #015-22031) was used against Ago2. Liver powder samples were incubated with Anti-Ago2 overnight at 4° C, washed three times with cold IP buffer and eluted in 1 ml of Trizol.
Small RNA deep sequencing data of miR-122-5p isoforms from human liver samples was obtained from GEO submission GSE5738136 and data from “tough decoys” against miR-122 was obtained from GEO record GSE2597123.
RNA sequencing and analysis
RNA samples were extracted using Trizol (Invitrogen) from mouse liver samples, snap frozen and ground into powder in liquid nitrogen. RNAs were subjected to poly-A purification prior to sequencing using a poly-A spin kit (NEB). In total 6 samples were prepared using a TruSeq kit (Illumina), barcoded and pooled. Sequencing was performed on an Illumina HiSeq 2500 machine at the Stanford Center for Genomics and Personalized Medicine, generating one lane of paired end 101-bp reads. We obtained a moderate depth of sequencing (~15 million 100bp paired end reads per sample). Reads were aligned to the mouse mm9 genome using TopHat (v.2.0.14)37,38 to generate BAM files. Sequences were aligned to mouse mRNAs to generate FPKM values using Cufflinks v.2.2.137,38 with the following parameters: “--library-norm-method quartile” and removing ribosomal, snoRNA and mitochondrial sequences. Differentially expressed transcripts (Jensen-Shannon divergence false discovery rate < 0.05) were obtained via Cuffdiff as part of the Cufflinks package. Replicate RNAseq libraries were generated using the same protocol described above except for the use of the ScriptSeq v2 strand-specific RNA extraction kit (Epicenter), and the option “--library-type frsecondstrand” in cufflinks transcript mapping. Also, differentially expressed transcripts with a P value <0.05 for replicate RNAseq samples when comparing the overlap of differentially expressed genes. The overlap of differentially expressed genes between the two sets of samples was evaluated by the DAVID v6.7 Gene Ontology program using a Benjamini P value of < 0.05 for significance39,40. Transcripts significantly differentially expressed (by Cuffdiff analysis) between all control + H1 liver samples relative to all AAV-U6-shRNAs (false discovery rate < 0.001) were analyzed by the Ingenuity Pathway Analysis program using default settings. Seed matches of genes ranked in order (U6 to H1 + control) were evaluated for significantly enriched 7nt seed sequences by the Sylamer algorithm41. Cluster analysis was performed with the DeSeq R package35. miR-122 and miR-21 3′UTR targets were obtained from DIANA microT-CDS v542. Sequence data has been deposited in the NCBI GEO repository with the accession number GSE69186.
ALT and hAAT measurements
Mouse serum was collected by centrifugation of blood obtained by retro-orbital bleeding. Alanine aminotransferase (ALT) levels were calculated using an ALT kinetic measurement kit (Teco Diagnostics). hAAT levels were determined by enzyme-linked immunosorbent assay (ELISA) as described previously43. The investigators were blinded to samples for hAAT and ALT measurements.
Cell culture
Human Embryonic Kidney (HEK) 293 cells were grown in Dulbecco's modified Eagle medium (Gibco-BRL) supplemented with non-essential amino acids, pen-strep antibiotics, sodium pyruvate, L-glutamine and 10% heat-inactivated fetal bovine serum. HEK293 cells were obtained from ATCC, were routinely tested for mycoplasma contamination and were used solely as a vehicle for determining small RNA read counts from transfected plasmids. Cells were transfected with 1 μg of CMV-miR-122 in 12-well plates using Lipofectamine 2000 and RNA was extracted by Trizol at various time points post-transfection. Dicer knockout cell lines were obtained from the Cullen laboratory44, transfected with various H1 and U6 sh-hAAT plasmids in 12-well plates and extracted 24 hours post-transfection.
Quantitative RT-PCR
Probes for Aldoa (Mm00833172_g1), Aldob (Mm00523293_m1), Slc7a1 (Mm01219060_m1), Dlk1 (Mm00494477_m1), Meg3 (Mm00522599_m1), Igf2 (Mm00439564_m1) and Alb (Mm00802090_m1) along with an actin internal control were ordered from Life Technologies for quantitative RT-PCR analysis. 2 μg of total RNA was reverse-transcribed using a superscript II RT kit (Invitrogen). 30 ng of cDNA was used for quantitative RT-PCR analysis, run on a CFX384 Real-Time system machine (Biorad) with mouse beta-actin probes used for normalization purposes. Fold change was calculated using ΔΔ-cT calculations.
Northern blot studies
500 ng of miR-Vana or 5 μg of Trizol extracted RNA was loaded on an 18% (w/v) acrylamide/7 M urea gel to analyze miR-122 isoform differences at nucleotide resolution and transferred to a Hybond-N+ membrane (Amersham). miR-122 was detected using a 32P-labelled miRCURY LNA detection probe (Exiqon). Isoforms were quantified using ImageJ software. A similar LNA probe (Exiqon) was used to detect mmu-miR-107. Human liver RNA was purchased from Zyagen.
Luciferase analysis
The ALDOA 3′UTR fragment was cloned into the psi-CHECK2 dual-luciferase reporter system between XhoI and SpeI restriction sites using the primers found in Supplementary Table 7. A miRIDIAN microRNA mimic for miR-122-5p was obtained for the 22nt isoform or custom synthesized for the 21nt isoform (GE Healthcare). For transfection, 250 ng of psi-CHECK2 reporter plasmids were co-transfected with a final concentration of 25 nM of synthetic miRNA mimics in 24-well plates of HEK293 cells using the Tranist-TKO transfection reagent (Mirus). Cells were lysed in 1× Passive Lysis Buffer and evaluated using a Dual Luciferase Reporter Assay (Promega) 24 hours after transfection, detected on a Modulas Microplate Luminometer (Turner Biosystems).
Fructose and glucose rich diet delivery
A 60% fructose (product # TD.89247) and corresponding 60% glucose (product # TD.05256) chow was obtained from Teklad Diets. Five female C57Bl/6 mice in each group were provided each diet ad libitum one week prior to either saline or AAV8-shRNA delivery.
Statistical analyses
Data are presented as means ± s.d. or s.e.m. Statistical significance was tested using GraphPad Prism (version 5.0b). Statistical analysis was performed using Student's t-test (two-tailed), one-way ANOVA with Tukey's post-hoc test and Wilcoxon rank-sum tests for cumulative frequency plots. P values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by grant NIH R01DK078424 and R01AI071068 (M.A.K.) and CA193244 (K.G.). P.N.V. was supported by a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research. We would like to thank all members of the Kay laboratory for input and suggestions and H. Vogel for EM image analysis. Dicer knockout cells were kindly provided by B. Cullen (Duke University). We would like to thank Stanford Functional Genomics Facility and Stanford Center for Genomics and Personalized Medicine for high throughput sequencing services.
Footnotes
Author Contributions
P.N.V., S.G., L.L., and M.A.K. conceived the project and designed the experiments. P.N.V., S.G., K.C., L.J., F.Z., Y.H. and L.L. performed the experiments. Y.Z. provided bioinformatics support. H.K. and K.G. provided the miR-122 knockout mice. P.N.V., S.G., E.M.M. and L.L. analyzed the data. P.N.V. and M.A.K. wrote the manuscript with critical review from S.G., E.M.M., K.G., and L.L. and input from all other co-authors.
Accession Codes
RNA sequencing and small RNA sequencing data has been deposited in the NCBI GEO repository www.ncbi.nlm.nih.gov/geo with the accession number GSE69186.
Competing Financial Interest Statement
The authors declare no competing financial interests.
References
- 1.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 2.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 3.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maguire AM, et al. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009;374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodino-Klapac LR, Chicoine LG, Kaspar BK, Mendell JR. Gene therapy for duchenne muscular dystrophy: expectations and challenges. Arch Neurol. 2007;64:1236–1241. doi: 10.1001/archneur.64.9.1236. [DOI] [PubMed] [Google Scholar]
- 6.Nakai H, et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol. 2005;79:3606–3614. doi: 10.1128/JVI.79.6.3606-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimm D, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine S, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315. e2301–2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu SH, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai WC, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu S, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 15.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh T, et al. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross NC, Tolan DR, Cox TM. Catalytic deficiency of human aldolase B in hereditary fructose intolerance caused by a common missense mutation. Cell. 1988;53:881–885. doi: 10.1016/s0092-8674(88)90349-2. [DOI] [PubMed] [Google Scholar]
- 18.Luna JM, et al. Hepatitis C Virus RNA Functionally Sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castoldi M, et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krutzfeldt J, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nature methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hommes FA, Draisma MI. The development of L- and M-type aldolases in rat liver. Biochim Biophys Acta. 1970;222:251–252. doi: 10.1016/0304-4165(70)90380-6. [DOI] [PubMed] [Google Scholar]
- 25.Numazaki M, Tsutsumi K, Tsutsumi R, Ishikawa K. Expression of aldolase isozyme mRNAs in fetal rat liver. European journal of biochemistry / FEBS. 1984;142:165–170. doi: 10.1111/j.1432-1033.1984.tb08265.x. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Kandathil AJ, et al. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology. 2013;145:1404–1413. e1401–1410. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb GB, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 32.Jenkins DD, et al. Donor-derived, liver-specific protein expression after bone marrow transplantation. Transplantation. 2004;78:530–536. doi: 10.1097/01.tp.0000130180.42573.b1. [DOI] [PubMed] [Google Scholar]
- 33.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selitsky SR, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Scientific reports. 2015;5:7675. doi: 10.1038/srep07675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartonicek N, Enright AJ. SylArray: a web server for automated detection of miRNA effects from expression data. Bioinformatics. 2010;26:2900–2901. doi: 10.1093/bioinformatics/btq545. [DOI] [PubMed] [Google Scholar]
- 42.Paraskevopoulou MD, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Zhang F, Kay MA. A mini-intronic plasmid (MIP): a novel robust transgene expression vector in vivo and in vitro. Mol Ther. 2013;21:954–963. doi: 10.1038/mt.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.