Abstract
Long term treatment with therapies aimed at blocking the estrogen- (ER) or androgen receptor (AR) action often leads to the development of resistance to selective modulators of the estrogen receptor (SERMs) in ERα-positive breast cancer, or of the androgen receptor (SARMs) in AR-positive prostate cancer. Many underlying molecular events that confer resistance are known, but a unifying theme is yet to be revealed. Receptor tyrosine kinases (RTKs) such EGFR, ERBB2 and IGF1R are major mediators that can directly alter cellular response to the SERM, tamoxifen, but the mechanisms underlying increased expression of RTKs are not clear. A number of HOX genes and microRNAs and non-coding RNAs residing in the HOX cluster, have been identified as important independent predictors of endocrine resistant breast cancer. Recently, convincing evidence has accumulated that several members belonging to the four different HOX clusters contribute to endocrine therapy resistant breast cancer, but the mechanisms remain obscure. In this article, we have reviewed recent progress in understanding of the functioning of HOX genes and regulation of their expression by hormones. We also discuss, in particular, the contributions of several members of the HOX gene family to endocrine resistant breast cancer.
1. Introduction
Breast cancer is the second most common cancer among women in the United States, affecting one out of eight females during their lifetime. In 2015, an estimated 231,840 new cases of invasive breast cancer were diagnosed, and during this time, almost 40,290 died from this disease [1]. At least 70% of breast cancers are identified as ER-positive in that their growth is dependent on the continued presence of estrogen. Despite the relative safety and significant therapeutic effects of endocrine therapy such as tamoxifen and aromatase inhibitors, about thirty percent of breast tumors develop resistance and eventually recur as distant metastasis. For decades, extensive studies have identified multiple mechanisms including activation of EGFR, HER2, ERα and IGF1R signaling pathways by which breast cancers mediate endocrine resistance [2]. However, which critical pathway should be targeted in order to overcome endocrine resistance in breast cancer is not clear.
The HOX proteins contain a homeodomain that is a highly conserved 61 amino acid helix-turn-helix motif. The homeodomain recognizes and binds to the consensus core DNA sequence 5’-TAAT-3’ in the proximal promoter or enhancer regions of its target genes, along with cofactors such PBX1, PBX2, MEIS1, PBC and TALE, to promote transcription [3–5]. First found in the fruit fly, Drosophila melanogaster, in two clusters consisting of the bithorax and antennapedia complexes, HOX genes are intimately involved in spatial patterning of the body during development [6]. In human, there are 39 HOX genes distributed in 4 paralogous clusters named A, B, C, and D which are located on chromosomes 7p15, 17q21.2, 12q13, and 2q31 respectively, and are named according to their 3’ to 5’ order of alignment. The number of HOX genes in each cluster is not uniform and varies from 9 to 11 genes in mammals [7].
HOX genes are transcription factors involved in the specification of segmental pathways during embryonic development [8]. Belonging to one of the largest superfamily of homeotic genes, members of the HOX gene family are primarily responsible for maintaining cell pluripotency, determining cell fate and promoting differentiation in multicellular organisms [9, 10]. Continued expression of some of the members of the HOX clusters into adulthood suggests a functional requirement of HOX proteins in maintaining cellular homeostasis [11]. Aberrant changes in HOX clusters including DNA hypermethylation and altered gene expression are frequently associated with cancer development [12]. In cancer, HOX genes have been reported to regulate cell cycle and apoptosis [13]. A number of strong pieces of evidence suggest that HOX genes play a critical role in the development of resistance against therapy in cancers. In this review, we will discuss studies on the HOX clusters including HOX genes, long non-coding RNA and microRNAs along with their functional characterization, clinical implications of their dysregulation, and regulation of gene expression in endocrine resistant breast cancer.
2. Germline mutations in HOX genes
Germline mutations in HOX genes manifest themselves as abnormal human phenotypes at birth or during sexual development (Table 1) [14–31]. Germline missense mutations are found in HOXB13 (G84E) in men with early-onset or with a positive history of prostate cancer (up to 3.1%) and is linked less frequently with familial breast cancer (0.7%) [25]. In sporadic prostate [32] and breast cancer [33], HOXB13 overexpression predicts poor prognosis [34]. In breast cancer cells, HOXB13 expression is regulated by estrogen, and its overexpression is an important marker of tamoxifen-resistant ER+ breast cancer [35] . In prostate cancer, HOXB13 interacts directly with AR, thus regulating AR-target gene expression [36]. These results imply that germ line mutations of HOXB13 are also likely associated with endocrine regulation. In addition, HOX genes play an important role in homeostasis in adult tissues. Several HOX genes are responsible for endometrial receptivity, pregnancy and hematopoiesis [37]. As transcription factors, HOX genes function to regulate crucial processes in embryonic development as well as cell differentiation, proliferation and motility in adult [10].
Table 1.
Disorders associated with mutations in HOX genes
| HOX gene | Condition | References |
|---|---|---|
| HOXA1 | Bosley–Salih–Alorainy syndrome | 14 |
| HOXA1 | Athabascan brainstem dysgenesis syndrome | 15 |
| HOXA2 | Microtia, hearing impairment and cleft palate | 16 |
| HOXA11 | Radioulnar synostosis with amegakaryocytic thrombocytopenia |
17 |
| HOXA13 | Hand–foot–genital syndrome | 18–20 |
| HOXA13 | Guttmacher syndrome | 21 |
| HOXB1 | Hereditary congenital facial paresis-3 | 22 |
| HOXB13 | Prostate, Colon and Breast | 23–25 |
| HOXC13 | Ectodermal dysplasia 9, hair/nail type | 26 |
| HOXD4 | Lymphoid malignancy and skeletal malformations | 27 |
| HOXD10 | Congenital vertical talus and Charcot–Marie–Tooth disease | 28 |
| HOXD13 | Synpolydactyly type II | 29–31 |
| HOXD13 | Brachydactyly types D and E | |
| HOXD13 | Syndactyly type V | |
| HOXD13 | Brachydactyly–syndactyly |
3. Regulation of HOX gene expression
3.1. Retinoic acid
Retinoic acid (RA) plays a critical role in embryogenesis. When RA binds to RA receptors (RARs), a member of the ligand activated nuclear receptor family, the RAR homo-, or heterodimeric complex with rexinoid receptors (RXR), is recruited to the proximal promoter regions at HOXA1, HOXB1 and HOXD4 genes to enhance their transcription [38–42]. The RA signaling pathways are involved in various types of cancer and thus the RARs and their downstream genes are potential therapeutic targets due to their functions in differentiation, proliferation, apoptosis, and oxidation [43]. In 2007, Chen et al. reported that RA can induce HOXA5 expression in RARβ-positive breast cancer cells [44]. RARβ binds directly to the RA response element (RARE) present in the 3' region of HOXA5 gene. In addition, we observed a decrease in both HOXA5 and RARβ expression during neoplastic transformation and progression. Knockdown of HOXA5 partially abrogates retinoid-induced apoptosis and promotes cell survival upon RA treatment, suggesting that HOXA5 acts directly downstream of RARβ and may contribute to its differentiation functions [44].
3.2. Vitamin D, Testosterone, and Progesterone
Vitamin D and its receptor, VDR (Vitamin D receptor) regulate HOXA10 and HOXA11 expression in both hematopoietic and endometrial cells [45]. Also, testosterone binds to the androgen receptor causing a decrease in HOXA10 and HOXA11 mRNA expression in an endometrial adenocarcinoma cell line [46]. The progesterone receptor (PR), by binding to its cognate ligand, progesterone, co-ordinates morphogenesis of the mammary gland Progesterone upregulates HOXA10 and HOXA11 expression in myocytes, in endometrium and in primary endometrial stromal cells [47–49]. In breast cells, on the other hand, HOXA5 directly activates PR gene expression and function through binding to a single HOX-binding site in its promoter region; this is not the case for HOXB4, HOXB5, or HOXB7 [50].
3.3. Estrogen
Estrogens are required for embryonic development of the female reproductive tract as well as functional differentiation of the adult endometrium. Several papers have shown that the HOXA genes are regulated by estrogen. In the endometrium, HOXA10 expression is significantly increased in a menstrual cycle stage–dependent manner. HOXA10 mRNA levels is not altered upon pretreatment with cycloheximide, which supported the notion that estrogen bound ER (estrogen receptor) binds to ERE (ER binding element) at the HOXA10 promoter to directly enhance HOXA10 transcription [51, 52]. In addition, the level of HOXA10 expression is dramatically increased in MCF-7 cells treated with estradiol and tamoxifen as determined by semi-quantitative RT-PCR and northern blot analysis [53]. In endometrial cells, HOXA10 expression is regulated by estrogen and tamoxifen [54]. HOXA1 is also an estrogen-responsive gene [55]. The authors show that ACK1, a ubiquitously expressed non-receptor tyrosine kinase, interacts with ER and the ER co-activator, KDM3A, a H3K9 demethylase, by phosphorylation in presence of tamoxifen. The complex of activated ACK1, ER and KDM3A binds to the HOXA1 promoter regions directly and regulates HOXA1 transcriptional activity. These results suggest that the ACK1-KDM3A-HOXA1 signaling axis might be a new target in acquired tamoxifen-resistant breast cancer [55]. Another HOXA protein, HOXA7, influences ER expression in MCF-7 cells. Knockdown of HOXA7 decreases cell proliferation and ER downregulation [56]. In ER+ MCF-7 cells, in contrast to ER-negative cells, the expression level of HOXB7 and HOXB13 shows a dramatic decrease upon treatment of the cells with estrogen. However, this effect is abrogated when MCF-7 cells are treated with tamoxifen [57, 58]. HOXC10 is upregulated by estrogen, through interaction with several ERE elements in its promoter region, but is downregulated by promoter methylation when the breast cancer cells acquire aromatase inhibitor resistance [59]. In osteoblast-like cell lines, the expression of HOXD11 is increased by exemestane, an aromatase inhibitor (AI) [60]. Since the expression of many HOX genes is altered by estrogen and SERMs, HOX genes have been implicated as significant players in endocrine-resistant breast cancer.
4. HOX genes in endocrine resistant breast cancer
4.1 HOX genes
It is well established that, at diagnosis, overexpression of HOXB13 is a strong marker of tamoxifen resistant breast cancer. By analysis of gene expression profiles in ER+ primary breast cancer in patients treated with adjuvant tamoxifen monotherapy, the ratio of HOXB13 to interleukin-17B receptor (IL17BR) was identified as an important independent predictor of outcome [35]. A high ratio of HOXB13/IL17BR (H:I) is associated with increased relapse, tumor aggressiveness, tamoxifen treatment failure, and death in breast cancer patients [61]. Breast cancers in tamoxifen resistant patients is also found to exhibit high HOXB13 expression, where HOXB13 overexpression is shown to be repressed by estrogen and this repression is lifted by tamoxifen. Refining this signature further, they developed the Breast Cancer Index or BCI by combining the H:I ratio with molecular grade index or MGI. MGI utilizes five other tamoxifen resistance biomarkers [62], shown to predict long-term recurrence risk after 5 years to not only tamoxifen, but also to aromatase inhibitor treatment, making this biomarker relevant to the clinical management of breast cancer [63]. For nearly a decade, HOXB13 has been recognized as an important biomarker of tamoxifen resistance, but its mode of action remained unclear. Our recent studies provides some insights into the mechanism whereby HOXB13 mediates tamoxifen resistance and metastasis. We showed that HOXB13 confers tamoxifen resistance via repression of ER expression, as well as induction of IL6 expression [34]. Using anti-IL6-antibodies [34] or Tocilizumab (an anti-IL6R antibody) (our unpublished data) combined with tamoxifen, we found that the inhibition of IL-6 signaling pathway reduces tumor growth of high HOXB13 expressing breast cancer cells. Thus, the IL-6 signaling pathway may be a novel therapeutic target in HOXB13-overexpressing tamoxifen resistant breast cancer.
Among the HOX genes, HOXB7 has emerged as a new master regulator of tamoxifen resistance. In preclinical models, HOXB7 expression promotes the growth of ER+ breast tumors. Interestingly, even in the absence of exogenous estrogen supplementation, HOXB7 overexpressing tumors grow rapidly in athymic nude mice. We showed that HOXB7 overexpression renders breast cancer cells resistant to tamoxifen through activation of several receptor tyrosine kinase pathways, including the EGFR pathway [58]. Recently, we investigated other pathways that may be involved, and the interplay between them. HOXB7 enhances ER target gene expression via the ER–HOXB7 complex directly bound to the promoter regions of key ER target genes such as CA12, MYC and HER2. What is the consequence of overexpression of HER2 in tamoxifen-resistant breast cancer? Studying the upstream regulation of HOXB7, we discovered the HOXB7-regulatory function of miR-196a, which, in turn, is transcriptionally regulated by c-MYC. High c-MYC, stabilized by activated HER2, downregulates miR-196a, which results in upregulation of HOXB7 in tamoxifen-resistant breast cancer cells. The identification of the cooperative role of HOXB7 and ER in controlling the expression of a myriad of genes, including HER2, the phosphorylation mediated activation of c-MYC by HER2 leading to downregulation of miR-196a, and its effects on HOXB7 expression revealed the existence of the HER2-MYC-miR-196a axis and its function. With the uncovering of this axis also came the identification of multiple, yet unrecognized ER/HOXB7 targets that are capable of attenuating tamoxifen resistance in breast cancer [64].
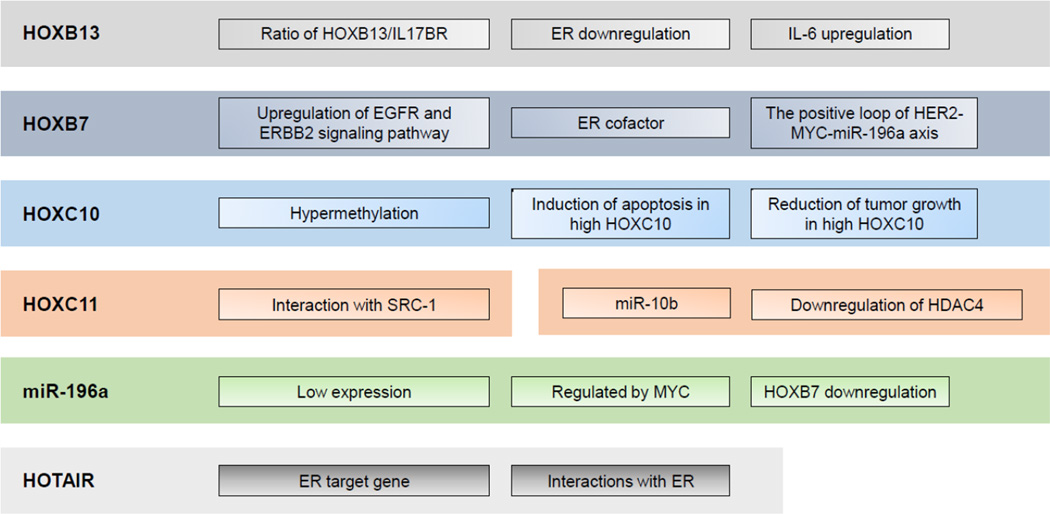
The role for epigenetic silencing of HOX genes was long suspected due to the frequent presence of dense CpG islands in their promoter regions. Using a genome-wide methylation screen it was reported that HOXC10 expression is repressed by promoter hypermethylation in two independent long-term estrogen-deprived cell lines, C4-12 and LTED [65]. Downregulation of HOXC10 by estrogen supplementation enhances breast tumor growth. On the contrary, upregulation of HOXC10 expression by aromatase inhibitors that suppressed estrogen production initiates apoptosis and subsequently reduces tumor growth. Thus, epigenetic silencing of HOXC10 in tumor cells results in continued growth and eventually to acquired aromatase inhibitor resistance [65]. In addition, it was demonstrated that HOXC11 is involved in endocrine resistance by interacting with SRC-1 as ER coactivator [66]. While the frequent presence of CpG island and promoter methylation may be one mechanism of epigenetic control of expression of HOX genes, multiple other factors play a role in the regulation of their expression (Figure 1).
Figure 1.
Overview of known HOX gene functions in endocrine resistant breast cancer.
4.2. HOX cluster non-coding RNA (ncRNA)
The HOX gene clusters harbor five different microRNAs: miR-10a, miR-10b, miR-196a1, miR-196a2 and miR-196b. The miR-10a gene is located between HOXB4 and HOXB5 [67]. Using a global microRNA screen (1,105 human miRNAs) of primary tumors of patients with ER-positive breast cancer treated with tamoxifen, who were either recurrence-free or had suffered a recurrence, miR-10a was found as one of top 20 deregulated miRNAs. It is also a major predictor of ER-positive tumor relapse in early postmenopausal breast cancer patients who are treated with tamoxifen [68]. In addition, miR-10b gene, located upstream of HOXD4, is a well-known enhancer of breast cancer invasion and metastasis [69]. Recently it was shown that in MCF-7 and T47D cells, overexpression of miR-10b renders cells tamoxifen resistant by reduction of HDAC4 expression, while depletion of miR-10b enhances sensitivity to tamoxifen in MCF-7 tamoxifen-resistant cells [70]. Thus, the miR-10b-HDAC4 axis is a potential therapeutic target in tamoxifen resistant breast cancer [70].
The HOXB7 downregulator, miR-196a, is located upstream of HOXB7, and is repressed by MYC protein, which is stabilized by phosphorylation by the highly activated HER2 signaling pathway in tamoxifen resistant MCF-7 cells. Predictably, overexpression of miR-196a sensitizes MCF-7-TMR cells to tamoxifen by downregulation of HOXB7. In order to overcome tamoxifen resistance induced by miR-196a, both HER2 and MYC signaling pathway could be potentially exploited through novel targeted therapeutic approaches [64].
The long non-coding RNA, HOTAIR, which is located upstream of HOXC11, is well known to promote breast tumorigenesis and metastasis by silencing, in trans, the expression of genes in the HOXD cluster. Recently, it was reported that HOTAIR is overexpressed in tamoxifen-resistant breast cancers [71]. In addition, the expression HOTAIR is directly inhibited by estrogen. The interaction between HOTAIR and ER promotes ER target gene transcription in estrogen-depleted conditions. Thus, HOTAIR also promotes growth and tamoxifen resistance in breast cancer cells. HOTAIR could serve as an additional potential therapeutic target and a crucial biomarker (Figure 1) [71].
5. Future clinical directions
Over the past few decades, many HOX genes and noncoding RNAs in the HOX clusters have been found to be deregulated in different tumor types and to actively drive diverse hallmarks of cancer. Despite their obvious clinical significance, it has been difficult to therapeutically target them with high specificity because of their extreme shared homology. Recent advances in gene therapy and computational biology might help in overcoming this limitation: by using siRNAs/shRNAs or RNAs, stable, selective and efficient knockdown or overexpression of genes can be achieved. Brock et al. [72] took it one step further. Gene network inference allowed them to identify HOXA1 to be causally involved in cancer development. They then designed HOXA1 siRNA nanoparticles and injected the particles intraductally through the nipple of transgenic mice with early-stage breast disease. This led to a decrease of proliferation and mammary tumor incidence. This approach, taking advantage of novel therapeutics and a unique delivery modality offers a novel noninvasive strategy to identify and specifically target oncogenes such as the HOX genes [72].
Other RNA targeting tools (like ribozymes, aptamers, ASOs and microRNAs) might be as effective and even have unique features that provide them advantages over siRNAs, especially for targeting the HOX lncRNAs. For example, at least 30 of the 39 HOX gene 3’UTR harbor conserved matches to miRNAs, many of these have been validated experimentally, and thus represent new opportunities for clinical intervention. Interestingly, the interaction of microRNAs with lncRNAs- for example, miR-141 with the oncogenic lncRNA, HOTAIR- may serve not only to decrease transcript levels but also to interfere with binding to their factors, and thus blunt their function [73].
Recently, the high levels of expression of the lncRNA H19 in cancer cells was exploited by BioCancell Therapeutics, who have developed a plasmid (BC-819) composed of the H19 promoter driving the expression of diphtheria toxin. BC-819 is being tested in phase I/II clinical trials to target invasive bladder, ovarian, lung and pancreatic cancers [74]. Given that HOTAIR is highly expressed in many aggressive and chemo-resistant cancers, using such a gene therapy strategy might provide promising clinical benefits.
Pharmaceutically, though targeting individual HOX proteins may still be not feasible, designing drugs that interfere with their interaction with downstream factors is a promising approach to inhibit their function. HXR9 is a successful example: HXR9, a small, cell-permeable peptide, acts by antagonizing HOX/PBX interactions (which influence HOX DNA binding specificity). HXR9 inhibits growth of prostate, melanoma, breast and ovarian cancer in cell culture and in preclinical models [75]. With further discoveries of novel HOX partners and the importance of these interactions in oncogenesis and drug resistance, and with advances in computational structural biology, it might be possible to design new drugs in future that specifically interfere with HOX functions.
Finally, the expression, methylation, translocation or mutation of many of the HOX genes and ncRNAs is increasingly found to function as novel biomarkers for diagnosis, prognosis, metastasis, and predictors of responsiveness to therapy. So far, only HOXB13 testing has entered clinical practice where high HOXB13/IL-17BR expression ratio is associated with increased relapse and death in ER-positive breast cancer patients treated with tamoxifen [76]. Strong enrichment of HOX genes is present within the 100 methylated markers that correlate with recurrence markers in breast cancer [77]. In fact, methylated HOXB4 in circulating tumor DNA is one among the 10-gene panel of biomarkers that can detect more than 90% of cases with metastatic breast cancer, help monitor response to treatment, and possibly prognosticate long term outcome [78]. The potential of HOX genes and lncRNAs as markers in human fluids is still unexplored and may provide new specific and facile diagnostic biomarkers, and minimize unnecessary biopsies.
In summary, the exploration of the HOX clusters has taken an interesting twist in the last few decades as we begin to unveil their complex interactions and discover their roles in diverse cellular processes, far beyond just the control of embryonic development. The investigation of HOX genes for understanding, discovering and treating endocrine resistant breast cancer might still be at its early stages of development, but is full of future promise.
Highlights.
HOX genes are often overexpressed in breast cancer.
Hormones regulate expression of many HOX genes.
HOX genes and miRNAs/ncRNA in the HOX cluster can contribute to endocrine resistance.
Targeting specific HOX gene can overcome endocrine-resistance in breast cancer.
Acknowledgments
We thank Susan G. Komen fellowship KG101506 (to KJ), the Commonwealth Foundation Grant and the Core Grant P30 CA006973 (to SS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: SS and KJ are named co-inventors in the U.S. Provisional Application 61/448,009, filed Mar. 01, 2012, named P11436-02 on the subject of “HOXB7 and tamoxifen resistance”.
References
- 1.A.C. Society. Cancer Facts & Figures 2015. Atlanta: American Cancer Society; 2015. [Google Scholar]
- 2.Garcia-Becerra R, Santos N, Diaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. International journal of molecular sciences. 2012;14:108–145. doi: 10.3390/ijms14010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. Journal of molecular medicine. 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 4.Moens CB, Selleri L. Hox cofactors in vertebrate development. Developmental biology. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 5.Shanmugam K, Green NC, Rambaldi I, Saragovi HU, Featherstone MS. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol Cell Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 7.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature reviews. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi Y. Hox transcription factors: modulators of cell-cell and cell-extracellular matrix adhesion. BioMed research international. 2014;2014:591374. doi: 10.1155/2014/591374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippmann ES, Williams CE, Ruhl DA, Estevez-Silva MC, Chapman ER, Coon JJ, Ashton RS. Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem cell reports. 2015;4:632–644. doi: 10.1016/j.stemcr.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of Hox genes in stem cell differentiation. World journal of stem cells. 2015;7:583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller DF, Holtzman SL, Kalkbrenner A, Kaufman TC. Homeotic Complex (Hox) gene regulation and homeosis in the mesoderm of the Drosophila melanogaster embryo: the roles of signal transduction and cell autonomous regulation. Mechanisms of development. 2001;102:17–32. doi: 10.1016/s0925-4773(01)00300-8. [DOI] [PubMed] [Google Scholar]
- 12.Pilato B, Pinto R, De Summa S, Lambo R, Paradiso A, Tommasi S. HOX gene methylation status analysis in patients with hereditary breast cancer. Journal of human genetics. 2013;58:51–53. doi: 10.1038/jhg.2012.118. [DOI] [PubMed] [Google Scholar]
- 13.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 14.Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nature genetics. 2005;37:1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- 15.Bosley TM, Alorainy IA, Salih MA, Aldhalaan HM, Abu-Amero KK, Oystreck DT, Tischfield MA, Engle EC, Erickson RP. The clinical spectrum of homozygous HOXA1 mutations. American journal of medical genetics. Part A. 2008;146A:1235–1240. doi: 10.1002/ajmg.a.32262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alasti F, Sadeghi A, Sanati MH, Farhadi M, Stollar E, Somers T, Van Camp G. A mutation in HOXA2 is responsible for autosomal-recessive microtia in an Iranian family. American journal of human genetics. 2008;82:982–991. doi: 10.1016/j.ajhg.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson AA, Nguyen LT. Amegakaryocytic thrombocytopenia and radio-ulnar synostosis are associated with HOXA11 mutation. Nature genetics. 2000;26:397–398. doi: 10.1038/82511. [DOI] [PubMed] [Google Scholar]
- 18.Mortlock DP, Innis JW. Mutation of HOXA13 in hand-foot-genital syndrome. Nature genetics. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- 19.Goodman FR, Bacchelli C, Brady AF, Brueton LA, Fryns JP, Mortlock DP, Innis JW, Holmes LB, Donnenfeld AE, Feingold M, Beemer FA, Hennekam RC, Scambler PJ. Novel HOXA13 mutations and the phenotypic spectrum of hand-foot-genital syndrome. American journal of human genetics. 2000;67:197–202. doi: 10.1086/302961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innis JW, Goodman FR, Bacchelli C, Williams TM, Mortlock DP, Sateesh P, Scambler PJ, McKinnon W, Guttmacher AE. A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Human mutation. 2002;19:573–574. doi: 10.1002/humu.9036. [DOI] [PubMed] [Google Scholar]
- 21.Guttmacher AE. Autosomal dominant preaxial deficiency, postaxial polydactyly, and hypospadias. American journal of medical genetics. 1993;46:219–222. doi: 10.1002/ajmg.1320460223. [DOI] [PubMed] [Google Scholar]
- 22.Webb BD, Shaaban S, Gaspar H, Cunha LF, Schubert CR, Hao K, Robson CD, Chan WM, Andrews C, MacKinnon S, Oystreck DT, Hunter DG, Iacovelli AJ, Ye X, Camminady A, Engle EC, Jabs EW. HOXB1 founder mutation in humans recapitulates the phenotype of Hoxb1−/− mice. American journal of human genetics. 2012;91:171–179. doi: 10.1016/j.ajhg.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. The New England journal of medicine. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbari MR, Anderson LN, Buchanan DD, Clendenning M, Jenkins MA, Win AK, Hopper JL, Giles GG, Nam R, Narod S, Gallinger S, Cleary SP. Germline HOXB13 p.Gly84Glu mutation and risk of colorectal cancer. Cancer Epidemiol. 2013;37:424–427. doi: 10.1016/j.canep.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alanee S, Couch F, Offit K. Association of a HOXB13 variant with breast cancer. The New England journal of medicine. 2012;367:480–481. doi: 10.1056/NEJMc1205138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Z, Chen Q, Shi L, Lee M, Giehl KA, Tang Z, Wang H, Zhang J, Yin J, Wu L, Xiao R, Liu X, Dai L, Zhu X, Li R, Betz RC, Zhang X, Yang Y. Loss-of-function mutations in HOXC13 cause pure hair and nail ectodermal dysplasia. American journal of human genetics. 2012;91:906–911. doi: 10.1016/j.ajhg.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Scherpenzeel Thim V, Remacle S, Picard J, Cornu G, Gofflot F, Rezsohazy R, Verellen-Dumoulin C. Mutation analysis of the HOX paralogous 4–13 genes in children with acute lymphoid malignancies: identification of a novel germline mutation of HOXD4 leading to a partial loss-of-function. Human mutation. 2005;25:384–395. doi: 10.1002/humu.20155. [DOI] [PubMed] [Google Scholar]
- 28.Shrimpton AE, Levinsohn EM, Yozawitz JM, Packard DS, Jr, Cady RB, Middleton FA, Persico AM, Hootnick DR. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. American journal of human genetics. 2004;75:92–96. doi: 10.1086/422015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akarsu AN, Stoilov I, Yilmaz E, Sayli BS, Sarfarazi M. Genomic structure of HOXD13 gene: a nine polyalanine duplication causes synpolydactyly in two unrelated families. Human molecular genetics. 1996;5:945–952. doi: 10.1093/hmg/5.7.945. [DOI] [PubMed] [Google Scholar]
- 30.Muragaki Y, Mundlos S, Upton J, Olsen BR. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science. 1996;272:548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 31.Goodman FR. Limb malformations and the human HOX genes. American journal of medical genetics. 2002;112:256–265. doi: 10.1002/ajmg.10776. [DOI] [PubMed] [Google Scholar]
- 32.Zabalza CV, Adam M, Burdelski C, Wilczak W, Wittmer C, Kraft S, Krech T, Steurer S, Koop C, Hube-Magg C, Graefen M, Heinzer H, Minner S, Simon R, Sauter G, Schlomm T, Tsourlakis MC. HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy. Oncotarget. 2015;6:12822–12834. doi: 10.18632/oncotarget.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Shah N, Jin K, Cruz LA, Park S, Sadik H, Cho S, Goswami CP, Nakshatri H, Gupta R, Chang HY, Zhang Z, Cimino-Mathews A, Cope L, Umbricht C, Sukumar S. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERalpha and inducing IL-6 expression. Cancer Res. 2013;73:5449–5458. doi: 10.1158/0008-5472.CAN-13-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, Joseph JD, McDonnell DP. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daftary GS, Taylor HS. Endocrine regulation of HOX genes. Endocrine reviews. 2006;27:331–355. doi: 10.1210/er.2005-0018. [DOI] [PubMed] [Google Scholar]
- 38.Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 39.Popperl H, Featherstone MS. Identification of a retinoic acid response element upstream of the murine Hox-4.2 gene. Mol Cell Biol. 1993;13:257–265. doi: 10.1128/mcb.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogura T, Evans RM. Evidence for two distinct retinoic acid response pathways for HOXB1 gene regulation. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:392–396. doi: 10.1073/pnas.92.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura T, Evans RM. A retinoic acid-triggered cascade of HOXB1 gene activation. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:387–391. doi: 10.1073/pnas.92.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3' of the murine homeobox gene Hox-1.6. Mechanisms of development. 1992;38:217–227. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 43.di Masi A, Leboffe L, De Marinis E, Pagano F, Cicconi L, Rochette-Egly C, Lo-Coco F, Ascenzi P, Nervi C. Retinoic acid receptors: from molecular mechanisms to cancer therapy. Molecular aspects of medicine. 2015;41:1–115. doi: 10.1016/j.mam.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Zhang H, Lee J, Liang X, Wu X, Zhu T, Lo PK, Zhang X, Sukumar S. HOXA5 acts directly downstream of retinoic acid receptor beta and contributes to retinoic acid-induced apoptosis and growth inhibition. Cancer Res. 2007;67:8007–8013. doi: 10.1158/0008-5472.CAN-07-1405. [DOI] [PubMed] [Google Scholar]
- 45.Du H, Daftary GS, Lalwani SI, Taylor HS. Direct regulation of HOXA10 by 1,25-(OH)2D3 in human myelomonocytic cells and human endometrial stromal cells. Molecular endocrinology. 2005;19:2222–2233. doi: 10.1210/me.2004-0336. [DOI] [PubMed] [Google Scholar]
- 46.Cermik D, Selam B, Taylor HS. Regulation of HOXA-10 expression by testosterone in vitro and in the endometrium of patients with polycystic ovary syndrome. The Journal of clinical endocrinology and metabolism. 2003;88:238–243. doi: 10.1210/jc.2002-021072. [DOI] [PubMed] [Google Scholar]
- 47.Cermik D, Karaca M, Taylor HS. HOXA10 expression is repressed by progesterone in the myometrium: differential tissue-specific regulation of HOX gene expression in the reproductive tract. The Journal of clinical endocrinology and metabolism. 2001;86:3387–3392. doi: 10.1210/jcem.86.7.7675. [DOI] [PubMed] [Google Scholar]
- 48.Mesiano S, Chan EC, Fitter JT, Kwek K, Yeo G, Smith R. Progesterone withdrawal and estrogen activation in human parturition are coordinated by progesterone receptor A expression in the myometrium. The Journal of clinical endocrinology and metabolism. 2002;87:2924–2930. doi: 10.1210/jcem.87.6.8609. [DOI] [PubMed] [Google Scholar]
- 49.Chau YM, Pando S, Taylor HS. HOXA11 silencing and endogenous HOXA11 antisense ribonucleic acid in the uterine endometrium. The Journal of clinical endocrinology and metabolism. 2002;87:2674–2680. doi: 10.1210/jcem.87.6.8527. [DOI] [PubMed] [Google Scholar]
- 50.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 51.Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. The Journal of clinical investigation. 1998;101:1379–1384. doi: 10.1172/JCI1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akbas GE, Song J, Taylor HS. A HOXA10 estrogen response element (ERE) is differentially regulated by 17 beta-estradiol and diethylstilbestrol (DES) J Mol Biol. 2004;340:1013–1023. doi: 10.1016/j.jmb.2004.05.052. [DOI] [PubMed] [Google Scholar]
- 53.Chu MC, Selam FB, Taylor HS. HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol Ther. 2004;3:568–572. doi: 10.4161/cbt.3.6.848. [DOI] [PubMed] [Google Scholar]
- 54.Kulak J, Jr, Ferriani RA, Komm BS, Taylor HS. Tissue selective estrogen complexes (TSECs) differentially modulate markers of proliferation and differentiation in endometrial cells. Reproductive sciences. 2013;20:129–137. doi: 10.1177/1933719112463251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan K, Lawrence HR, Lawrence NJ, Mahajan NP. ACK1 tyrosine kinase interacts with histone demethylase KDM3A to regulate the mammary tumor oncogene HOXA1. J Biol Chem. 2014;289:28179–28191. doi: 10.1074/jbc.M114.584425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Cheng JC, Huang HF, Leung PC. Homeobox A7 stimulates breast cancer cell proliferation by up-regulating estrogen receptor-alpha. Biochemical and biophysical research communications. 2013;440:652–657. doi: 10.1016/j.bbrc.2013.09.121. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, Shioda T, Ma XJ, Sgroi DC. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 58.Jin K, Kong X, Shah T, Penet MF, Wildes F, Sgroi DC, Ma XJ, Huang Y, Kallioniemi A, Landberg G, Bieche I, Wu X, Lobie PE, Davidson NE, Bhujwalla ZM, Zhu T, Sukumar S. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2736–2741. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 60.Miki Y, Suzuki T, Hatori M, Igarashi K, Aisaki KI, Kanno J, Nakamura Y, Uzuki M, Sawai T, Sasano H. Effects of aromatase inhibitors on human osteoblast and osteoblast-like cells: a possible androgenic bone protective effects induced by exemestane. Bone. 2007;40:876–887. doi: 10.1016/j.bone.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Dahiya S, Provencher H, Muir B, Carney E, Coser K, Shioda T, Ma XJ, Sgroi DC. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 62.Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, Durbecq V, Harris A, Goss P, Sotiriou C, Erlander M, Sgroi D. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 63.Sgroi DC, Carney E, Zarrella E, Steffel L, Binns SN, Finkelstein DM, Szymonifka J, Bhan AK, Shepherd LE, Zhang Y, Schnabel CA, Erlander MG, Ingle JN, Porter P, Muss HB, Pritchard KI, Tu D, Rimm DL, Goss PE. Prediction of late disease recurrence and extended adjuvant letrozole benefit by the HOXB13/IL17BR biomarker. J Natl Cancer Inst. 2013;105:1036–1042. doi: 10.1093/jnci/djt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin K, Park S, Teo WW, Korangath P, Cho SS, Yoshida T, Gyorffy B, Goswami CP, Nakshatri H, Cruz LA, Zhou W, Ji H, Su Y, Ekram M, Wu Z, Zhu T, Polyak K, Sukumar S. HOXB7 Is an ERalpha Cofactor in the Activation of HER2 and Multiple ER Target Genes Leading to Endocrine Resistance. Cancer discovery. 2015;5:944–959. doi: 10.1158/2159-8290.CD-15-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, Lee AV, Chen J, Shea MJ, Santen RJ, Gannon F, Kangaspeska S, Jelinek J, Issa JP, Richer JK, Elias A, McIlroy M, Young LS, Davidson NE, Schiff R, Li W, Oesterreich S. Epigenetic Reprogramming of HOXC10 in Endocrine-Resistant Breast Cancer. Sci Transl Med. 2014;6:229ra241. doi: 10.1126/scitranslmed.3008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McIlroy M, McCartan D, Early S, P OG, Pennington S, Hill AD, Young LS. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC-1 mediates resistance to endocrine therapy in breast cancer [corrected] Cancer Res. 2010;70:1585–1594. doi: 10.1158/0008-5472.CAN-09-3713. [DOI] [PubMed] [Google Scholar]
- 67.Woltering JM, Durston AJ. The zebrafish hoxDb cluster has been reduced to a single microRNA. Nature genetics. 2006;38:601–602. doi: 10.1038/ng0606-601. [DOI] [PubMed] [Google Scholar]
- 68.Hoppe R, Achinger-Kawecka J, Winter S, Fritz P, Lo WY, Schroth W, Brauch H. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. European journal of cancer. 2013;49:3598–3608. doi: 10.1016/j.ejca.2013.07.145. [DOI] [PubMed] [Google Scholar]
- 69.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 70.Ahmad A, Ginnebaugh KR, Yin S, Bollig-Fischer A, Reddy KB, Sarkar FH. Functional role of miR-10b in tamoxifen resistance of ER-positive breast cancer cells through downregulation of HDAC4. BMC Cancer. 2015;15:540. doi: 10.1186/s12885-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2015 doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brock A, Krause S, Li H, Kowalski M, Goldberg MS, Collins JJ, Ingber DE. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Sci Transl Med. 2014;6:217ra212. doi: 10.1126/scitranslmed.3007048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The international journal of biochemistry & cell biology. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 74.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- 75.Morgan R, Boxall A, Harrington KJ, Simpson GR, Michael A, Pandha HS. Targeting HOX transcription factors in prostate cancer. BMC Urol. 2014;14:17. doi: 10.1186/1471-2490-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goetz MP, Suman VJ, Ingle JN, Nibbe AM, Visscher DW, Reynolds CA, Lingle WL, Erlander M, Ma XJ, Sgroi DC, Perez EA, Couch FJ. A two-gene expression ratio of homeobox 13 and interleukin-17B receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin Cancer Res. 2006;12:2080–2087. doi: 10.1158/1078-0432.CCR-05-1263. [DOI] [PubMed] [Google Scholar]
- 77.Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvananthan K, Marks J, Ethier S, Gray JW, Wolff AC, Cope LM, Sukumar S. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71:6195–6207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fackler MJ, Lopez Bujanda Z, Umbricht C, Teo WW, Cho S, Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C, Lyman JP, de Brot M, Ingle JN, Boughey J, McGuire K, King TA, Carey LA, Cope L, Wolff AC, Sukumar S. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Res. 2014;74:2160–2170. doi: 10.1158/0008-5472.CAN-13-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]