Abstract
Aging is a predominant risk factor for developing cardiovascular disease. Therefore, the cellular processes that contributes to aging are attractive targets for therapeutic interventions that can delay or prevent the development of age-related diseases. Our understanding of the underlying mechanisms that contribute to the decline in cell and tissue functions with age has greatly advanced over the past decade. Classical hallmarks of aging cells include increased levels of reactive oxygen species, DNA damage, accumulation of dysfunctional organelles, oxidized proteins and lipids. These all contribute to a progressive decline in the normal physiological function of the cell and to the onset of age-related conditions. A major cause of the aging process is progressive loss of cellular quality control. Autophagy is an important quality control pathway and is necessary to maintain cardiac homeostasis and to adapt to stress. A reduction in autophagy has been observed in a number of aging models and there is compelling evidence that enhanced autophagy delays aging and extents life span. Enhancing autophagy counteracts age-associated accumulation of protein aggregates and damaged organelles in cells. In this review, we discuss the functional role of autophagy in maintaining homeostasis in the heart, and how a decline is associated with accelerated cardiac aging. We will also evaluate therapeutic approaches being researched in an effort to maintain a healthy young heart.
Keywords: aging, autophagy, heart, mitochondria
1. Introduction
Cardiovascular disease is the leading cause of death in developed countries and represents a major financial burden to the health care systems. Aging is a major risk factor for developing cardiovascular disease and 4 out of 5 people who succumb to coronary heart disease are over 65 years old [1]. Furthermore, the prevalence of cardiovascular disease is expected to increase over the next several years in the baby boomer population. Therefore, it is of great interest and importance to gain increased understanding into the mechanisms underlying aging and how these cellular alterations contribute to age-related pathologies. A hallmark of aging is the accumulation of dysfunctional organelles, DNA mutations, oxidized proteins and lipids which all contribute to a progressive decline in the normal physiological function of the cell and to the onset of age-related conditions. Post-mitotic cells, such as cardiac myocytes, are particularly vulnerable to age-related changes and cellular damage since they are not easily replaced. Instead, cardiac myocytes rely on cellular quality control mechanisms to stay young [2].
Macroautophagy (hereafter referred to as autophagy), first observed and coined by Christian de Duve in 1963, is derived from the Greek language and means to “eat itself”. It constitutes a mechanism through which cargo is sequestered in a double membrane vesicle which subsequently delivers the content to a lysosome for degradation [3]. Autophagy is a major cellular quality control pathway and responsible for removing long lived proteins, dysfunctional organelles and protein aggregates in cells. Autophagy also maintains cellular energy levels during nutrient limiting conditions by catabolic recycling. Studies have reported that autophagy declines with age and it is believed that this defect in autophagy is a major contributor to aging. Pharmacologic or genetic enhancements of autophagy extend life span [4–9], whereas interfering with autophagy shortens life span [4, 10–12]. Therefore, a better understanding of the underlying molecular mechanisms that contribute to impaired autophagy in aging myocytes has significant scientific and clinical importance. In this review, we discuss the functional role of autophagy in maintaining cellular homeostasis and how a decline is associated accelerated cardiac aging. We also evaluate therapeutic approaches currently under investigation in an effort to maintain a healthy young heart.
2. Overview of Autophagy
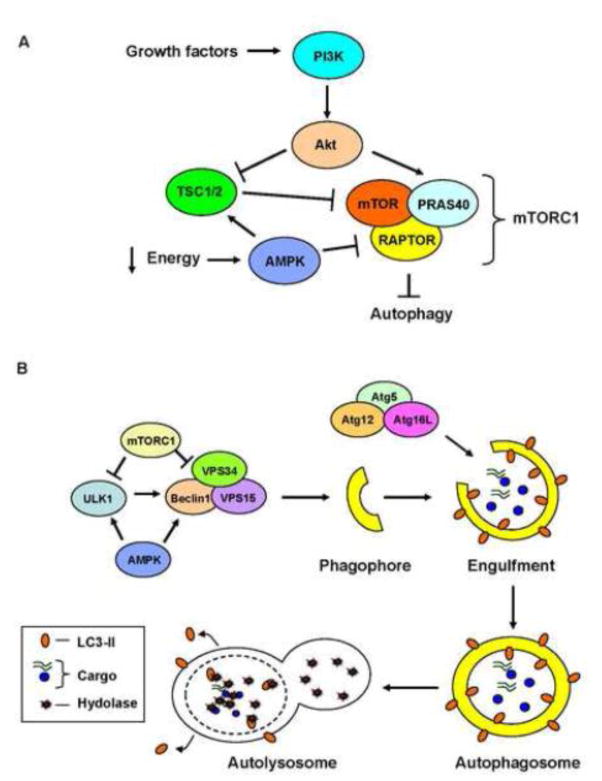
Autophagy is a tightly regulated degradation pathway and its activation involves the coordination of multiple protein complexes at several different steps (Fig. 1). The autophagy process consists of at least four steps: activation, formation and elongation of the phagophore, engulfment of cargo and delivery to the lysosome for degradation. Autophagic activity is regulated by both extra- and intracellular signals and the mammalian target of rapamycin (mTOR) is a central regulator of autophagy. It is part of the mTORC1 complex which integrates signals from various sources including growth factors, amino acids and energy levels. When activated, this complex inhibits autophagy (Fig. 1). For instance, during nutrient rich conditions, the PI3K/Akt signaling pathway inhibits autophagy through positive regulation of mTORC1 activity. Specifically, Akt phosphorylates and inhibits the mTORC1 repressor TSC1/2, resulting in activation of mTORC1 [13–15]. Akt can also regulate mTORC1 by directly phosphorylating and inactivating PRAS40, an endogenous inhibitor of the complex. Phosphorylation of PRAS40 leads to activation of mTORC1 and inhibition of autophagy [16, 17]. The activated mTORC1 complex hinders activation of autophagy by inhibiting the Unc-51 like kinase 1 (ULK1) [18] and by inhibiting the Beclin1-VPS34-Vps15, the Class III PI3 kinase complex involved in initiating formation of the phagophore [19]. In contrast, during nutrient limiting conditions, autophagy is activated by the energy sensor AMPK. Studies have reported that AMPK activates autophagy via several different mechanisms. It activates autophagy by inhibiting mTORC1 through phosphorylation and inhibition of RAPTOR, a critical subunit in the complex [20] or by activating the repressor TSC1/2 [21] (Fig 1A). AMPK also directly promotes autophagy by activating ULK1 [18] and the downstream Beclin1 complex [22] (Fig. 1B).
Figure 1.
Overview of autophagy. A) The PI3K/Akt signaling pathway regulates autophagy through actions on PRAS40 in the mTORC1 complex. Akt can also indirectly promote mTORC1 activity through direct inhibition of the mTORC1 repressor TSC1/2. During nutrient limiting conditions, the energy sensor AMPK acts on both TSC1/2 and mTORC1 to inhibit autophagy. B) Upon activation of autophagy, the ULK1 complex activates the Class III PI3K complex (Beclin1-VPS34-VPS15) which is responsible for initiating formation of the phagophore. Subsequent elongation of the phagophore is regulated by two ubiquitin-like conjugation systems, Atg5-Atg12-Atg16L and LC3. After engulfing cargo, the autophagosome fuses with a lysosome to form the autolysosome. The sequestered cargo and the inner membrane of the autophagosome are degraded by lysosomal hydrolases. The LC3-II on the outer membrane of the autophagosome is detached after fusion with the lysosome.
The second step involves formation of a small vesicular sac known as the phagophore (Fig. 1B). Although it is known that initiation of autophagy begins with formation of the phagophore through lipid acquisition, this is still a poorly understood process. Studies have reported that the plasma membrane, mitochondria, ER and Golgi can serve as lipid sources for autophagosomes [23]. However, it is possible that autophagosomes are composed of membranes from multiple sources to avoid affecting the homeostasis of a specific subcellular compartment. It is also possible that different types of autophagy induction lead to the formation of different types of autophagosomes. For instance, removal of mitochondria might involve deriving lipids from both mitochondria and ER, whereas removal of soluble cargo might involve deriving lipids from the plasma membrane. Additional studies on the formation of the phagophore in response to various stressors are needed to shed more light on these questions.
The biogenesis of the autophagosome is regulated by a number of conserved autophagy-related (ATG) proteins. The ULK1 complex activates the Class III PI3K (PI3K) complex which consists of core proteins Beclin1 (Atg6), Vps34 and Vps15. This complex is responsible for promoting nucleation of the phagophore [24]. The elongation of the phagophore into an autophagosome involves two ubiquitin-like conjugation systems, Atg5-Atg12-Atg16L and microtubule-associated protein 1 light chain 3 (LC3) (Reviewed in [25]). Soluble LC3 is cleaved by Atg4 [26] and then conjugated to phosphatidylethanolamine (PE). The resulting LC3II is integrated into the membrane and is important for phagophore elongation and cargo recognition [27]. The growing phagophore elongates until it closes on itself, engulfing the cargo to be degraded. Once the cargo-loaded autophagosome has been formed, it fuses with a lysosome to form the autolysosome. The cargo and the inner membrane of the autophagosome are subsequently degraded by the lysosomal hydrolases.
3. Selective Autophagy
It was initially believed that sequestration of cargo by autophagosomes was a non-selective process. However, studies have now demonstrated that autophagy can selectively target bacteria, protein aggregates, and organelles including peroxisomes, ER and mitochondria for degradation [27–31]. A majority of studies have focused on the selective degradation of mitochondria by autophagosomes, a process also termed mitophagy. These studies have contributed to the discovery of signaling pathways and proteins involved in selective autophagy.
The PINK1/Parkin pathway is involved in marking depolarized mitochondria for autophagic degradation [29]. Upon loss of mitochondrial membrane potential, the serine/threonine kinase PINK1 accumulates on the outer mitochondrial membrane (OMM) [32]. PINK1-mediated phosphorylation of Mfn2 and ubiquitin turns them into mitochondrial receptors for Parkin [33–35]. After recruitment to mitochondria, Parkin proceeds to ubiquitinate several different OMM proteins and the ubiquitination of these substrates serves as a signal for autophagic degradation. Autophagy adaptor proteins, such as p62/Sqstm1 and NBR1, bind to the ubiquitinated proteins via their ubiquitin-associated (UBA) domains and to LC3 via their LIR motif [27, 36], bringing the ubiquitinated cargo to the autophagosome. p62/Sqstm1 plays a role in clearing dysfunctional mitochondria, misfolded proteins and protein aggregates [27, 37]. However, studies have reported that p62/Sqstm1 is dispensable for PINK1/Parkin-mediated mitophagy [38, 39], suggesting that a redundancy in adaptor proteins exists. Recent studies have identified optineurin and NDP51 as important autophagy adaptor proteins that facilitate mitophagy [38, 40].
In addition, mitophagy can occur independent of ubiquitination and adaptor proteins through the use of proteins and lipids on the mitochondria functioning as autophagy receptors. The two most well characterized autophagy receptors are BNIP3 and NIX/BNIP3L which facilitate mitophagy by directly binding to LC3 or gamma-aminobutyric acid receptor-associated protein on the autophagosome via their LIR motifs [28, 41, 42]. Cardiolipin and FUNDC1 also function as autophagy receptors on mitochondria [43, 44]. More recently, Murakawa et al. identified Bcl-2-like protein 13 (Bcl2-L-13) to be a mitophagy receptor [45]. Similar to BNIP3 and NIX, overexpression of Bcl2-L-13 induces mitophagy in the absence of any other stress. Although studies have clearly demonstrated that selective autophagy exists in cells, there are still a number of outstanding questions that need to be answered. For instance, are there distinct autophagy receptors on other organelles such as peroxisomes and ER/SR? Are there age related changes in the autophagy receptors with age?
4. Autophagy and Aging
Conceptually, enhancing autophagy should counteract age-associated accumulations of protein aggregates and damaged organelles in cells. In agreement with this, there is extensive evidence that enhancing autophagy in diverse organisms delays aging and extents life-span (Table 1). For instance, pharmacological activation of autophagy using the mTOR inhibitor rapamycin or sperimidine promotes longevity in Drosophila Melanogaster [6, 8]. Rapamycin administration to aging mice also significantly extends the life span of both female and male mice [46, 47]. Similarly, mice with reduced mTOR expression also have longer life spans [48]. Transgenic mice with systemic overexpression of the autophagy protein Atg5 have enhanced autophagic flux in all tissues examined including heart, lungs, skeletal muscle and brain [9]. Consistent with a beneficial effect of enhancing autophagic activity, these mice have extended survival and exhibit anti-aging phenotypes such as increased insulin sensitivity, leanness, and improved motor function [9]. In contrast, impaired autophagy is associated with disease development and a reduced life span (Table 1). Loss-of-function mutations in critical autophagy genes lead to decreased life-span in C. elegans [10] and in D. Melanogaster [4]. In mice, disruption of autophagy in neurons leads to progressive neurodegeneration [49, 50], and mice lacking the autophagy adaptor p62/Sqstm1 have a premature aging phenotype and reduced life span [11]. Overall, these studies suggest that autophagy plays an important role in preventing premature aging.
Table 1.
Autophagy and aging studies.
| Activation of autophagy | Species | Effect on aging | References |
|---|---|---|---|
| Overexpression of Atg8a | D. Melanogaster | Increased autophagy and enhanced lifespan | [4] |
| Spermidine treatment | D. Melanogaster | Increased autophagy and enhanced lifespan | [8] |
| Resveratrol treatment | C. Elegance | Increased autophagy and enhanced lifespan | [5] |
| Rapamycin treatment | D. Melanogaster | Increased autophagy and enhanced lifespan | [6] |
| Rapamycin treatment | Mice | Enhanced lifespan; “young” cardiac mitochondrial proteome in aged mice; preserved cardiac function with age | [46, 47, 54, 55, 108] |
| Reduced mTOR expression | Mice | Enhanced lifespan | [48] |
| Resveratrol treatment | Mice | Improved health, increased lifespan | [102] |
| Caloric restriction | Mice, Rats | Elevated autophagy and preserved contractile function in aged mice | [7, 52, 53] |
| Systemic overexpression of Atg5 | Mice | Elevated autophagy, anti-aging phenotype, extended lifespan | [9] |
| Cardiac specific Atg7 overexpression | Mice | Elevated cardiac autophagy and increased clearance of protein aggregates | [56] |
| Cardiac specific Parkin overexpression | Mice | Improved mitochondrial turnover and delayed cardiac aging | [74] |
| Inhibition of autophagy | |||
| Loss-of-function mutations in autophagy genes | D. Melanogaster | Impaired autophagy, reduced lifespan | [4] |
| Loss-of-function mutations in autophagy genes | C. Elegance | Impaired autophagy, reduced lifespan | [10] |
| p62/Sqstm1−/− | Mice | Premature aging, reduced lifespan | [11] |
| Cardiac specific Atg5−/− | Mice | Accelerated cardiac dysfunction, reduced lifespan | [12] |
Manipulation of autophagy also impacts cardiovascular function and health. Long term excessive fat intake inhibits autophagic flux in the heart and is associated with myocyte apoptosis and cardiac dysfunction in rats [51]. In contrast, caloric restriction (CR) is a potent inducer of autophagy in the heart [52] and long-term CR treatment in aged mice leads to AMPK activation, increased autophagy, and preserved contractile function [7]. Similarly, an increase in cardiac autophagic activity correlates with improved left ventricular diastolic function in aged rats subjected to lifelong CR [53]. Administration of the mTOR inhibitor rapamycin improves the mitochondrial proteome in aging hearts [54] and reverses the age-related decline in cardiac function in mice [55]. While it is unknown exactly how CR and rapamycin improve cardiac health and delay aging, it has been hypothesized that increased autophagy is one of the beneficial effects of these two interventions. However, it is important to bear in mind that many other cellular processes are affected by CR and rapamycin that might also contribute to their anti-aging effects.
Moreover, selectively enhancing autophagy in the heart via cardiac specific overexpression of Atg7 leads to increased autophagic flux and protection against cardiac proteinopathy by increasing clearance of protein aggregates. [56]. It has not been determined yet whether increased Atg7-mediated autophagic flux in myocytes will delay cardiac aging. Other studies have demonstrated that abrogating autophagy has negative consequences for the heart. Cardiac-specific Atg5 knockout mice have an accelerated aging phenotype, including development of left ventricular hypertrophy, decreased fractional shortening, and premature death [12]. These autophagy deficient hearts accumulate ubiquitinated proteins and p62, confirming that clearance of damaged proteins is an important cellular quality control mechanism that prevents premature aging. In addition, p62/Sqstm1-deficiency leads to accumulation of abnormal mitochondria in cardiac tissue. Unfortunately, this study did not investigate how the absence of this important autophagy adaptor protein impacts autophagic activity in cells. Thus, there are still many outstanding questions that need to be answered such as 1) which forms of selective autophagy are abrogated in p62/Sqstm1-deficient mice, 2) how is autophagic flux affected, 3) can other autophagy adaptors compensate for lack of p62/Sqstm1 in the young mice, and 4) can overexpression of p62 enhance selective autophagy in myocytes?
5. Aging is Associated with Insufficient Autophagy
Although few studies have directly assessed autophagic activity in aged tissues, it is generally accepted that autophagy is reduced with age [57–63]. For instance, Russ et al. reported an age-related reduction in basal autophagy in skeletal muscle in rats [62] and Uddin et al. observed decreased LC3 protein levels and reduced number of autophagosomes in thymus and liver of aged mice [59]. However, studies on autophagy in the aging heart are inconsistent and have reported that autophagy is increased, decreased, or unchanged in the heart with age. It has been observed that expression of autophagy-related genes are unchanged in the aged mouse heart [64] or that aging mouse hearts have higher levels of Beclin1 and LC3II/I [65]. Another study found an increase in LC3II levels with age but no change in Beclin1 and p62 levels [66], whereas Taneike et al. reported that LC3II levels are reduced with age in the mouse heart [12]. Wohlgemuth et al. observed an increase in Beclin1 and LC3I with age in rat hearts, whereas Atg9 and LAMP1 are reduced [52]. It is unclear why the results from these studies varies so much, but experimental variables such as differences in experimental design, species and genetic backgrounds are possible factors. Also, it is also important to note that none of these studies examined autophagic flux in the aged hearts. Therefore, it is difficult to interpret the data from the gene and protein analyses and how they relate to cardiac autophagic flux. Although an increase in LC3II levels indicate an increase in the number of autophagosomes, it is unknown if this is due to increased autophagic activity or an accumulation of autophagosomes due to reduced flux. Similarly, a decrease in LC3II could reflect increased degradation of autophagosomes due to enhanced flux or a decrease in the number of autophagosomes formed. Clearly, additional studies are needed to firmly establish how aging affects autophagic flux in the heart.
6. Mechanisms Underlying Reduced Autophagy
Although there are many potential causes underlying the alterations in cardiovascular function with age, it is very likely that a major determinant of the aging process is the progressive loss of quality control due to reduced autophagy. Thus, the accumulation of cytotoxic proteins and dysfunctional organelles as autophagy function is reduced contributes to the development of age related pathologies, such as skeletal muscle atrophy, neurodegeneration and cardiac dysfunction. The underlying mechanisms for the reduction in autophagy in aging tissues are not well understood. Given the intricate regulation of autophagy at multiple steps by numerous proteins (Fig. 1), it is conceivable that disruptions in one or even several of the regulatory steps contribute to the decline in autophagy with age. For instance, a study comparing expression of autophagy genes in young versus old human brain samples found that expression of several key autophagy genes, such as Atg5 and Atg7, are down-regulated in aged brains [63]. This suggests that the normal aging process might be associated with transcriptional down-regulation of autophagy which could contribute to the observed age-dependent development of neurodegenerative diseases. In addition, changes in metabolism and hormonal responses with age might also be involved in altering autophagic activity. Aging is associated with hyperactivation of mTOR [67] which has been linked to accelerated aging [66, 68]. The mechanisms leading to reduced expression of autophagy genes and hyperactivation of mTOR, and their relationship to the aging heart still need to be investigated.
Cardiac myocytes contain a lot of mitochondria to meet their high energy demand. It has been proposed that accumulation of dysfunctional mitochondria in myocytes play a major role in the aging process and development of age-related cardiomyopathy [69]. Reactive oxygen species (ROS) are generated in the cell as a byproduct of mitochondrial respiration. Under normal conditions, low levels of ROS have important signaling functions including regulation of autophagy at homeostatic levels [70–72]. However, when mitochondria become dysfunctional, they can become a major source of ROS. Excessive ROS negatively affects cellular processes by modifying proteins, lipids and inducing DNA damage. It is possible that excessive ROS production in aging cells contributes to impaired autophagy via the modification of one or several of the proteins involved in regulating the autophagy process. In the young healthy heart, aberrant mitochondria are rapidly removed by autophagosomes [73]. However, if autophagic activity is reduced with age, then the removal of dysfunctional mitochondria will also be decreased. This will lead to accumulation of dysfunctional mitochondria in aging myocytes. Moreover, it was recently reported that the PINK1/Parkin mitochondrial quality control pathway is impaired with age in mouse hearts. Hoshino et al. found that although the expression of PINK1 and Parkin are unaltered in the aged heart, the recruitment of Parkin to dysfunctional mitochondria is significantly attenuated in the aged myocardium [74]. This suggests that a decrease in mitochondrial clearance also contributes to aging. Their findings indicate that upregulation of p53 in senescent cells contributes to the defect in Parkin-mediated mitochondrial clearance, where p53 interacts with Parkin and sequesters it in the cytosol. Interestingly, overexpression of Parkin in the heart improves mitochondrial turnover and ameliorates cardiac aging, suggesting that Parkin is a potential therapeutic target to delay or prevent aging.
Mitochondria contain their own genome which encodes for several subunits involved mitochondrial respiration. Mitochondrial DNA (mtDNA) mutations accumulate with age in various tissues in humans and rodents, which leads to impaired mitochondrial function [75–79]. Homoplasmic mtDNA mutations and deletions have been observed in aged human cardiac myocytes [80]. The contribution of mtDNA mutations to the aging process has been confirmed by studies in mice expressing a proofreading-deficient mitochondrial DNA polymerase γ (POLGm/m) [81, 82]. These mice accumulate mtDNA mutations in cells at a faster rate than wild type mice, which leads to accelerated aging and reduced lifespan. Interestingly, the development of age-related cardiomyopathy in the POLGm/m mice is rescued by overexpression of a mitochondrial targeted catalase, which reduces levels of mitochondrial ROS [83]. Even though this study clearly indicates an important connection between mitochondrial ROS and aging, the effect on autophagy was not assessed. However, Li-Harms et al. recently reported that accumulation of mtDNA mutations leads to hyperactivation of mTOR and subsequent suppression of autophagy in erythroid cells and MEFs from the POLGm/m mice [84]. The reduced autophagy leads to decreased mitochondrial clearance and subsequent accumulation of dysfunctional mitochondria. Exactly how mtDNA mutations contribute to aberrant increase in mTOR signaling is currently unclear. Future studies also need to focus on how accumulation of mtDNA mutations in aging mice affects autophagic activity in the heart.
Lipofuscins are structures primarily composed of non-degradable cross-linked proteins, lipids, and small amounts of carbohydrates and metals [85–87]. They are a hallmark of aging and post-mitotic cells such as cardiac myocytes and neurons accumulate large amounts of lipofuscin with age. Autophagic degradation of mitochondria also contributes to lipofuscin formation. Lipofuscins have been proposed to contribute to reduced autophagic flux in aging cells by impairing lysosomal function. Lipofuscins are resistant to hydrolytic enzyme degradation and, as a result, accumulate inside lysosomes [87, 88]. With age, accumulation of large lipofuscin aggregates inside lysosomes affects their function by filling most of the bulk volume of lysosomes [89]. In addition, even though lipofuscin cannot be degraded, the lysosomal hydrolases are still directed towards these structures resulting in a futile accumulation of lytic hydrolases to lipofuscin filled lysosomes leaving a meager amount of hydrolases for effective autophagy [2].
7. Targeting Autophagy to Delay Cardiac Aging
Many life-style modifications that can delay the aging process may be effective, in part, by enhancing autophagy. For instance, exercise is known to increase autophagy in the heart [90] and have beneficial effects against cardiovascular diseases and aging [91, 92]. Mice with cardiac-specific overexpression of CryABR120G accumulate misfolded proteins and aggregates which lead to the development of heart failure [93]. Although baseline autophagic flux is reduced in these hearts, inducing autophagy by exercising the mice promotes increased clearance of the protein aggregates and significantly extends survival [56, 93]. Another study using a mouse model of chronic graft-versus-host disease found that chronic exercise increases autophagy in the heart and positively impacts cardiovascular function and extends survival [94]. Moreover, caloric restriction (CR) is a potent inducer of autophagy and increases autophagy in both young and aged hearts [52]. This intervention has consistently shown a positive anti-aging effect and an ability to increase lifespan in different species [52, 95–97].
However, these interventions are not practical for all people, especially patients with severely compromised cardiovascular function. Therefore, pharmacologic induction of autophagy is a more desirable option and is being pursued extensively to combat aging and various age-related diseases. CR is known to activate autophagy via the deacetylate SIRT1 [98]. Therefore, the plant polyphenol resveratrol, an activator of SIRT1, is being explored as a potential therapeutic strategy. Studies have reported that resveratrol extends life span in lower organism [5, 99, 100] and in mice fed a high calorie diet [101, 102]. Resveratrol is also cardioprotective and slows the progression of heart failure after myocardial infarction [103], pressure overload [104] and chemotherapy-induced cardiotoxicity [105]. However, the beneficial effects of resveratrol on cardiac cells is dose-dependent. At lower doses, resveratrol enhances autophagy and has anti-apoptotic properties, whereas high doses attenuate autophagy and promote apoptosis [106, 107]. More comprehensive pharmacodynamic studies are needed to determine better understand the cardioprotective versus cardiotoxic effects of resveratrol. It will also be important to investigate whether resveratrol treatment also leads to long-term cardiac damage that doesn’t become apparent until much later, as observed with the anthracyclines.
The mTOR inhibitor Rapamycin has also shown promise as an anti-aging therapeutic agent, and is known to confer functional benefits to the aging heart [54, 55, 108]. As discussed in previous sections, long-term administration of rapamycin enhances longevity in mice [46, 47, 109, 110]. Also, Wilkinson et al. observed that rapamycin-treated mice have delayed development of various age-related pathologies, including degenerative changes in the heart [108]. There is a significant increase in abnormalities of nuclear size and chromatin conformation in the aged myocardium, and rapamycin-treated mice have a lower incidence of myocardial nuclear atypia [108]. Unfortunately, chronic rapamycin treatment can lead to some undesirable side effects such as testicular degeneration and cataracts [108], insulin resistance [111] glucose intolerance and hyperlipidemia [112].
8. Concluding Remarks
Over the past decade, our understanding of the molecular and cellular mechanisms regulating the decline in cell and tissue function with age has been greatly advanced. Interventions that target the underlying aging process can potentially delay or interrupt the development of age-related diseases. It is clear that autophagy has effective anti-aging properties by clearing cytotoxic protein aggregates and dysfunctional organelles. Autophagy is a very complex and highly regulated pathway and there are a number of proteins that could potentially be targeted to modulate this pathway therapeutically. Currently, there are no pharmacological interventions known to selectively increase autophagy without also targeting other processes. Effective autophagy inducers, such as exercise, CR or rapapmycin treatment, affect multiple cellular processes besides autophagy. In addition, recent studies have revealed that several of the Atg proteins in the autophagy machinery have non-autophagic functions in cells [113]. Therefore, future research needs to focus on identifying which proteins in the pathway are specific regulators of autophagy before therapeutic targets can be selected. In addition, another important objective in future research will be to better understand the mechanisms underlying the improved cardiac function following induction of autophagy. Greater insights into the regulation of autophagy and how this process delays myocardial aging will allow for the development of novel autophagy targeted therapies to attenuate cardiac aging and disease.
Highlights.
Autophagy is a major cellular quality control pathway in the heart
Autophagy is involved in degrading protein aggregates and dysfunctional organelles
Autophagy is reduced with age and contributes to the aging process
Evidence exists that enhanced autophagy delays aging and extents life span
Acknowledgments
Å.B. Gustafsson is supported by an AHA Established Investigator Award, and by NIH R01HL087023 and P01HL085577. L.J. Leon is supported by a Supplement from NIH/NHLBI (R01HL087023-S1).
Footnotes
Disclosure Statement
The authors have no conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terman A, Brunk UT. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc Res. 2005;68:355–65. doi: 10.1016/j.cardiores.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 5.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X, Turdi S, Hu N, Guo R, Zhang Y, Ren J. Influence of long-term caloric restriction on myocardial and cardiomyocyte contractile function and autophagy in mice. The Journal of nutritional biochemistry. 2012;23:1592–9. doi: 10.1016/j.jnutbio.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 9.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–8. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 11.Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, et al. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13:150–6. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6:600–6. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 13.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 14.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Molecular cell. 2002;10:151–62. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 15.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nature cell biology. 2002;4:658–65. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 16.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nature cell biology. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. The Journal of biological chemistry. 2007;282:20036–44. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–95. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SN, Tang BL. Location and membrane sources for autophagosome formation - from ER-mitochondria contact sites to Golgi-endosome-derived carriers. Molecular membrane biology. 2013;30:394–402. doi: 10.3109/09687688.2013.850178. [DOI] [PubMed] [Google Scholar]
- 24.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 26.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 27.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 28.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Interacts with Bnip3 Protein to Selectively Remove Endoplasmic Reticulum and Mitochondria via Autophagy. J Biol Chem. 2012;287:19094–104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–41. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 31.Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–28. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–5. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 35.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 38.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–48. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–8. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 43.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature cell biology. 2013;15:1197–205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 45.Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nature communications. 2015;6:7527. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10:4230–6. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- 48.Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4:913–20. doi: 10.1016/j.celrep.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu HC, Chen CY, Lee BC, Chen MF. High-fat diet induces cardiomyocyte apoptosis via the inhibition of autophagy. European journal of nutrition. 2015 doi: 10.1007/s00394-015-1034-7. [DOI] [PubMed] [Google Scholar]
- 52.Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, et al. Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res. 2007;10:281–92. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- 53.Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, et al. Impact of long-term caloric restriction on cardiac senescence: caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. J Mol Cell Cardiol. 2011;50:117–27. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 54.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–39. doi: 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynn JM, O’Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12:851–62. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, et al. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–97. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi O, Otsu K. Role of autophagy in aging. Journal of cardiovascular pharmacology. 2012;60:242–7. doi: 10.1097/FJC.0b013e31824cc31c. [DOI] [PubMed] [Google Scholar]
- 58.Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38:519–27. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 59.Uddin MN, Nishio N, Ito S, Suzuki H, Isobe K. Autophagic activity in thymus and liver during aging. Age (Dordr) 2012;34:75–85. doi: 10.1007/s11357-011-9221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 61.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Russ DW, Boyd IM, McCoy KM, McCorkle KW. Muscle-specificity of age-related changes in markers of autophagy and sphingolipid metabolism. Biogerontology. 2015 doi: 10.1007/s10522-015-9598-4. [DOI] [PubMed] [Google Scholar]
- 63.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–9. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inuzuka Y, Okuda J, Kawashima T, Kato T, Niizuma S, Tamaki Y, et al. Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation. 2009;120:1695–703. doi: 10.1161/CIRCULATIONAHA.109.871137. [DOI] [PubMed] [Google Scholar]
- 65.Boyle AJ, Shih H, Hwang J, Ye J, Lee B, Zhang Y, et al. Cardiomyopathy of aging in the mammalian heart is characterized by myocardial hypertrophy, fibrosis and a predisposition towards cardiomyocyte apoptosis and autophagy. Exp Gerontol. 2011;46:549–59. doi: 10.1016/j.exger.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou J, Freeman TA, Ahmad F, Shang X, Mangano E, Gao E, et al. GSK-3alpha is a central regulator of age-related pathologies in mice. J Clin Invest. 2013;123:1821–32. doi: 10.1172/JCI64398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–45. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–46. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terman A, Brunk UT. The aging myocardium: roles of mitochondrial damage and lysosomal degradation. Heart Lung Circ. 2005;14:107–14. doi: 10.1016/j.hlc.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 70.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song M, Chen Y, Gong G, Murphy E, Rabinovitch PS, Dorn GW., 2nd Super-suppression of mitochondrial reactive oxygen species signaling impairs compensatory autophagy in primary mitophagic cardiomyopathy. Circ Res. 2014;115:348–53. doi: 10.1161/CIRCRESAHA.115.304384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sciarretta S, Zhai P, Shao D, Zablocki D, Nagarajan N, Terada LS, et al. Activation of NADPH oxidase 4 in the endoplasmic reticulum promotes cardiomyocyte autophagy and survival during energy stress through the protein kinase RNA-activated-like endoplasmic reticulum kinase/eukaryotic initiation factor 2alpha/activating transcription factor 4 pathway. Circulation research. 2013;113:1253–64. doi: 10.1161/CIRCRESAHA.113.301787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J. 2007;405:407–15. doi: 10.1042/BJ20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 75.Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–97. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–9. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 77.Wanagat J, Wolff MR, Aiken JM. Age-associated changes in function, structure and mitochondrial genetic and enzymatic abnormalities in the Fischer 344 x Brown Norway F(1) hybrid rat heart. J Mol Cell Cardiol. 2002;34:17–28. doi: 10.1006/jmcc.2001.1483. [DOI] [PubMed] [Google Scholar]
- 78.Khaidakov M, Heflich RH, Manjanatha MG, Myers MB, Aidoo A. Accumulation of point mutations in mitochondrial DNA of aging mice. Mutat Res. 2003;526:1–7. doi: 10.1016/s0027-5107(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 79.Bratic A, Larsson NG. The role of mitochondria in aging. The Journal of clinical investigation. 2013;123:951–7. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khrapko K, Bodyak N, Thilly WG, van Orsouw NJ, Zhang X, Coller HA, et al. Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic acids research. 1999;27:2434–41. doi: 10.1093/nar/27.11.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–4. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 82.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 83.Dai DF, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, et al. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–44. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li-Harms X, Milasta S, Lynch J, Wright C, Joshi A, Iyengar R, et al. Mito-protective autophagy is impaired in erythroid cells of aged mtDNA-mutator mice. Blood. 2015;125:162–74. doi: 10.1182/blood-2014-07-586396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Porta EA. Advances in age pigment research. Archives of gerontology and geriatrics. 1991;12:303–20. doi: 10.1016/0167-4943(91)90036-p. [DOI] [PubMed] [Google Scholar]
- 86.Jolly RD, Douglas BV, Davey PM, Roiri JE. Lipofuscin in bovine muscle and brain: a model for studying age pigment. Gerontology. 1995;41(Suppl 2):283–95. doi: 10.1159/000213750. [DOI] [PubMed] [Google Scholar]
- 87.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free radical biology & medicine. 2002;33:611–9. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 88.Brunk UT, Jones CB, Sohal RS. A novel hypothesis of lipofuscinogenesis and cellular aging based on interactions between oxidative stress and autophagocytosis. Mutation research. 1992;275:395–403. doi: 10.1016/0921-8734(92)90042-n. [DOI] [PubMed] [Google Scholar]
- 89.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. European journal of biochemistry / FEBS. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 90.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–5. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson MG, Ellison GM, Cable NT. Basic science behind the cardiovascular benefits of exercise. Heart. 2015;101:758–65. doi: 10.1136/heartjnl-2014-306596. [DOI] [PubMed] [Google Scholar]
- 92.Giannuzzi P, Temporelli PL, Corra U, Tavazzi L, Group E-CS. Antiremodeling effect of long-term exercise training in patients with stable chronic heart failure: results of the Exercise in Left Ventricular Dysfunction and Chronic Heart Failure (ELVD-CHF) Trial. Circulation. 2003;108:554–9. doi: 10.1161/01.CIR.0000081780.38477.FA. [DOI] [PubMed] [Google Scholar]
- 93.Maloyan A, Gulick J, Glabe CG, Kayed R, Robbins J. Exercise reverses preamyloid oligomer and prolongs survival in alphaB-crystallin-based desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2007;104:5995–6000. doi: 10.1073/pnas.0609202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fiuza-Luces C, Delmiro A, Soares-Miranda L, Gonzalez-Murillo A, Martinez-Palacios J, Ramirez M, et al. Exercise training can induce cardiac autophagy at end-stage chronic conditions: insights from a graft-versus-host-disease mouse model. Brain, behavior, and immunity. 2014;39:56–60. doi: 10.1016/j.bbi.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 95.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010;9:92–5. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–61. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–55. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 100.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 101.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–68. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gu XS, Wang ZB, Ye Z, Lei JP, Li L, Su DF, et al. Resveratrol, an activator of SIRT1, upregulates AMPK and improves cardiac function in heart failure. Genetics and molecular research : GMR. 2014;13:323–35. doi: 10.4238/2014.January.17.17. [DOI] [PubMed] [Google Scholar]
- 104.Sung MM, Das SK, Levasseur J, Byrne NJ, Fung D, Kim TT, et al. Resveratrol treatment of mice with pressure-overload-induced heart failure improves diastolic function and cardiac energy metabolism. Circ Heart Fail. 2015;8:128–37. doi: 10.1161/CIRCHEARTFAILURE.114.001677. [DOI] [PubMed] [Google Scholar]
- 105.Danz ED, Skramsted J, Henry N, Bennett JA, Keller RS. Resveratrol prevents doxorubicin cardiotoxicity through mitochondrial stabilization and the Sirt1 pathway. Free Radic Biol Med. 2009;46:1589–97. doi: 10.1016/j.freeradbiomed.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 106.Gurusamy N, Lekli I, Mukherjee S, Ray D, Ahsan MK, Gherghiceanu M, et al. Cardioprotection by resveratrol: a novel mechanism via autophagy involving the mTORC2 pathway. Cardiovascular research. 2010;86:103–12. doi: 10.1093/cvr/cvp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baarine M, Thandapilly SJ, Louis XL, Mazue F, Yu L, Delmas D, et al. Pro-apoptotic versus anti-apoptotic properties of dietary resveratrol on tumoral and normal cardiac cells. Genes & nutrition. 2011;6:161–9. doi: 10.1007/s12263-011-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, et al. Rapamycin slows aging in mice. Aging Cell. 2012;11:675–82. doi: 10.1111/j.1474-9726.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, et al. Rapamycin extends maximal lifespan in cancer-prone mice. The American journal of pathology. 2010;176:2092–7. doi: 10.2353/ajpath.2010.091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang GR, Wu YY, Chiu YS, Chen WY, Liao JW, Hsu HM, et al. Long-term administration of rapamycin reduces adiposity, but impairs glucose tolerance in high-fat diet-fed KK/HlJ mice. Basic & clinical pharmacology & toxicology. 2009;105:188–98. doi: 10.1111/j.1742-7843.2009.00427.x. [DOI] [PubMed] [Google Scholar]
- 112.Houde VP, Brule S, Festuccia WT, Blanchard PG, Bellmann K, Deshaies Y, et al. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–48. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–51. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]