Abstract
Purpose
Utilization of sequencing to screen the general population for preventable monogenic conditions is receiving substantial attention due to its potential to decrease morbidity and mortality. However, the selection of which variants to return is a serious implementation challenge. Procedures must be investigated to ensure optimal test characteristics and avoidance of harm from false positive test results.
Methods
We scanned exome sequences from 478 well-phenotyped individuals for potentially pathogenic variants in 17 genes representing 11 conditions that are among the most medically actionable Mendelian disorders in adults. We developed 5 variant selection algorithms with increasing sensitivity and measured their specificity in these 17 genes.
Results
Variant selection algorithms with increasing sensitivity exhibited decreased specificity, and performance was highly dependent on the genes analyzed. The most sensitive algorithm ranged from 88.8% to 99.6% specificity among the 17 genes.
Conclusion
For very low prevalence conditions, small reductions in specificity greatly increase false positives. This inescapable test characteristic governs the predictive value of genomic sequencing in the general population. To address this issue, test performance must be evaluated systematically for each condition so that the false negatives and false positives can be tailored for optimal outcomes, depending on the downstream clinical consequences.
INTRODUCTION
Screening programs can be valuable public health tools. Universal newborn screening has been highly successful in detecting severe but preventable genetic disorders. Such programs utilize defined mechanisms to select target conditions, based on their prevalence, severity, treatment options, and availability of a confirmatory test.1 Similar screening programs (based on genomics) may be emerging for the adult population, approximately 1% of whom are predisposed to a serious hereditary condition that may be preventable or ameliorated through early diagnosis.2 Large scale genomic sequencing initiatives comprising over 100,000 people have been announced3,4 and screening of the general adult population for hereditary cancer has recently been proposed.5 President Obama also announced a U.S. initiative to recruit a cohort of one million people in order to advance the cause of “Precision Medicine,” echoing the UK’s effort to sequence whole genomes of 100,000 patients focusing on cancer and rare diseases. Finally, we are witnessing the emergence of direct to consumer companies marketing the opportunity for genomic screening to healthy individuals, thus potentially initiating a vast uncontrolled experiment in such an approach.
Human genetic variation is ubiquitous, with ~3 million nucleotide variants per individual genome. The vast majority of variants have no health implications, but certain rare variants cause heritable monogenic conditions. Some variants have undisputed pathogenicity in these disorders, whereas most have limited or no evidence of pathogenicity and all individuals have novel variants that are essentially “private” to their family. Importantly, many variants previously claimed as causal for monogenic disorders have conflicting assertions regarding pathogenicity, have been disputed by subsequent evidence,10-13 or have been determined to have less penetrance than other disease-causing variants in the same gene.14 Genetic variants identified in clinical sequencing are typically classified into 5 categories with respect to their etiologic role in monogenic disorders: Pathogenic, Likely Pathogenic, Uncertain Significance, Likely Benign, and Benign.15 The Pathogenic designation implies virtually complete certainty that the variant is causal for the disorder; however, there is no universal agreement on what “likely” means. One proposal suggested that the Likely Pathogenic designation should imply 95-99% confidence in the pathogenicity of the variant,16 but quantitating confidence in variant pathogenicity is difficult and few standardized methods exist.17 For most conditions there are no “gold standard” confirmatory tests that can adjudicate the pathogenicity of genetic variants.
In screening, the test performance (sensitivity and specificity) and the prevalence of the disorder determine the predictive value of a screen positive result. If genomic screening is misapplied in the general population, false positive results could lead to overtreatment, overt harm and monetary waste. Thus, it is imperative to understand the performance of sequencing and how to optimize thresholds for returning results in the novel context of population screening, which are likely to be dramatically different than in a clinical diagnostic context.
Because of their low population prevalence, some monogenic disorders would require screening >10,000 people in order to detect a single true positive result. In such conditions, positive predictive value (PPV) is highly dependent on specificity, such that for a condition with 0.01% population frequency, reduction from 100% specificity to 99.94% specificity decreases PPV to 10%. This effect is similar but less pronounced in conditions with higher population frequencies. Although the technical sensitivity and technical specificity of sequencing (whether a genetic variant is truly present or not) can be measured relatively easily, there has so far been no effort to determine the clinical sensitivity or clinical specificity (whether the condition is truly present or not) of sequencing in the general population. Thus, it is completely unknown how genomic screening will fare in terms of predictive value.
The Center for Genomics and Society at the University of North Carolina at Chapel Hill is conducting an exploratory project, “GeneScreen,” to examine the feasibility and ethical considerations of screening 1,000 adults from the general population using massively parallel targeted sequencing of 17 genes responsible for 11 conditions that are among the most clinically actionable monogenic disorders in adults. A central obstacle to such screening is to clearly define criteria for variant selection and return, so as to avoid unacceptable numbers of false positives that would lead to unnecessary intervention and negative emotional, physical, or social consequences. Here, we evaluate different algorithms for selecting potentially pathogenic variants. The most stringent algorithm chooses variants that correspond closely to those that would be classified as “Pathogenic” in human review; the least stringent algorithm selects many additional variants that would be classified as having “Uncertain Significance.” We estimate the specificity of these algorithms in 478 exomes from a diverse cohort of well-phenotyped participants in a separate clinical sequencing exploratory research project, allowing us to simulate expected findings in the general population. This represents the first attempt to empirically measure the performance of genomic sequencing for population screening.
SUBJECTS AND METHODS
Exome sample
Exome data from 478 participants in the IRB-approved North Carolina Clinical Genomic Evaluation by Next-gen Exome Sequencing (NCGENES) project were analyzed for the 17 GeneScreen genes (APC, BRCA1, BRCA2, HFE, FBN1, KCNH2, KCNQ1, LDLR, MLH1, MSH2, MSH6, MUTYH, PMS2, RET, RYR1, SCN5A, SERPINA1). The NCGENES participants were previously sequenced in a diagnostic context for a variety of phenotypes, including cancer, aortic aneurysm, and arrhythmia (manuscript in preparation). Any participants who were sequenced for a primary indication overlapping with a condition described in this manuscript were excluded from the analysis for those conditions. Exome variants were loaded into a PostgresSQL database (v.9.0.3) for annotation and facilitation of queries.18
Variant selection and specificity calculations
Variants classified as pathogenic or likely pathogenic in the National Center for Biotechnology Information ClinVar database19 (downloaded 2/14) and variants labeled as “DM” (Disease Mutation) in the Human Gene Mutation Database20 (HGMD; 2014, v2) were collected as a source of potentially pathogenic variants. Recognizing that many variants in these databases may be erroneously classified, variants were excluded if pathogenicity assertions were discordant.
Population allele frequency estimates were determined using the Exome Aggregation Consortium (ExAC), a resource composed of 63,358 unrelated individuals sequenced through a variety of studies.21 Each gene was analyzed using one of three different minor allele frequency thresholds: 1%, 0.1%, or 0.01%. The minor allele frequency threshold chosen for each gene was based on the maximum expected population frequency of pathogenic alleles for the associated condition. In addition, variants in genes associated with conditions having dominant inheritance were eliminated if the minor allele frequency in the exome sample was >0.5%. CADD scores22 were retrieved for novel missense variants, with a Phred score of 13 as the threshold for deleteriousness. Conserved functional domains within proteins were obtained from the RefSeq database.23
Five variant selection algorithms (VSA1-5) were applied to the 17 genes, with each successive algorithm choosing more potentially pathogenic variants that could qualify as a “positive” screening result:
VSA-1 includes rare variants classified as “Pathogenic” in ClinVar. Many (but not all) of these variants would be considered “Pathogenic” in human review.
VSA-2 adds rare predicted truncating variants (nonsense, frameshift, canonical splice-site). These additional variants would largely be considered “Likely Pathogenic” in human review.
VSA-3 adds rare variants classified as “Likely Pathogenic” in ClinVar and/or as a “Disease Mutation (DM)” in HGMD. Many of these variants would be considered “Likely Pathogenic” or to have “Uncertain Significance” in human review.
VSA-4 adds rare missense variants with CADD scores >13 that are located within a conserved functional domain. Most of these variants would be considered to have “Uncertain Significance” in human review.
VSA-5 adds all rare missense variants, regardless of CADD score or location. Most of these variants would also be considered to have “Uncertain Significance”.
The algorithms take into account zygosity, such that either a homozygous variant or two potentially biallelic variants were required for a “positive” screen in MUTYH, HFE, and SERPINA1.
While each successive algorithm is expected to have increased sensitivity, the actual sensitivity cannot be directly measured in this cohort because we excluded participants with phenotypes overlapping these conditions. Given the rarity of the conditions being evaluated, we do not expect to find truly affected individuals for most of the disorders. Therefore, the number of true positives in the sample should be between 0-1 for most conditions, even including non-penetrant alleles. For estimated specificity calculations, we assumed that all “positive” screening results in this small sample were false positives, with the exception of certain variants with undisputed pathogenicity after curation of results from VSA-1 and VSA-2 (eg., HFE C282Y, Ashkenazi founder mutations in BRCA1 and BRCA2). Specificity estimates were adjusted and confidence intervals were calculated using the Wilson score interval.24
RESULTS
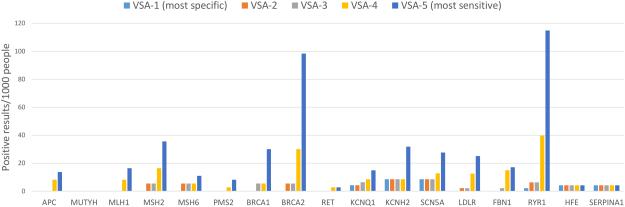
The GeneScreen project has identified 17 genes implicated in 11 monogenic disorders that are expected to be highly actionable in the adult population, in which to explore genomic screening in the general population. We simulated the performance of genomic screening of these genes in the general population by applying five algorithms for identifying potentially pathogenic variants in 478 exomes (Figure 1). VSA-1, which includes only variants classified as “pathogenic” in ClinVar, has the lowest sensitivity of the algorithms tested and returned the fewest variants overall, with specificity expected to approach 100% in ideal testing conditions if results are restricted to unquestionably pathogenic variants. Even so, some variants currently listed in ClinVar as “pathogenic” are not definitively pathogenic and thus the specificity of VSA-1 was 99.5%. The overall specificity of VSA-2 was also 99.5% and the overall specificity of VSA-3 was 99.4%. As expected, the most sensitive algorithms, which include rare missense variants, had the lowest specificity: VSA-4 had an overall specificity of 98.7%, and VSA-5 had an overall specificity of 97.1%. This demonstrates that the increased sensitivity afforded by inclusion of missense variants results in a concomitant decrease in overall specificity and hence would result in a substantial increase in false positive results.
Figure 1. Yield of potentially pathogenic variants using variant selection algorithms with varying sensitivity.
Five variant selection algorithms with different degrees of sensitivity and specificity were constructed. VSA-1 is the least sensitive algorithm and most specific, because it only considers a subset of variants previously defined as pathogenic. VSA-5 is the most sensitive but least specific, considering all rare truncating and missense variants (most of which would be considered “variants of uncertain significance”) to constitute a positive screen. These algorithms were applied to exome sequence data from 478 individuals in order to empirically evaluate the yield of possibly pathogenic variants in the 17 genes selected for population screening. The number of people who would screen positive per 1000 individuals screened is displayed on the vertical axis, with each gene along the horizontal axis, for all variant selection algorithms.
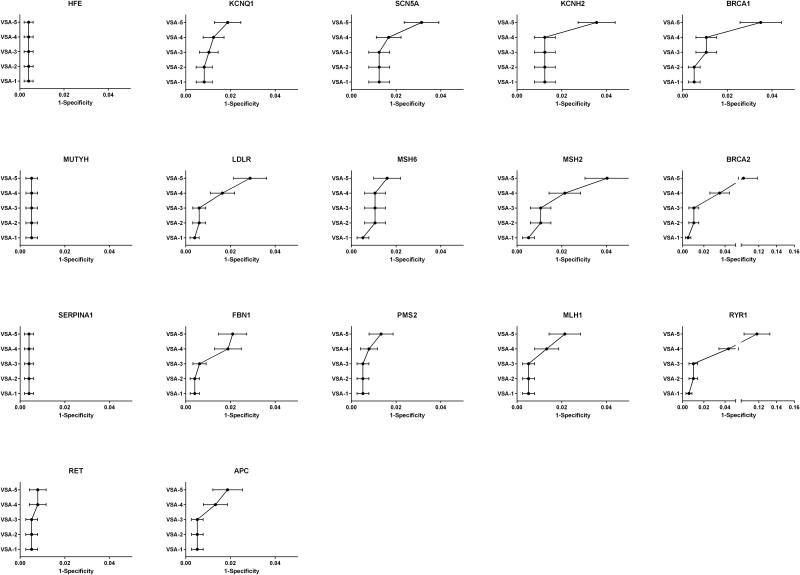
However, it is more important to consider the performance of the algorithms with respect to each gene. Two individuals were homozygous for the HFE C282Y pathogenic variant, and another two individuals were homozygous for the SERPINA1 pathogenic variant that results in the “Pi-Z” Alpha-1 Antitrypsin deficiency phenotype. These findings are interpreted to be “true positives” since their pathogenicity is undisputed. Two of the genes with very low prevalence (MUTYH and RET) had no “positive” screening results identified by any algorithm. Most of the genes had very few “positive” results identified by either VSA-1 or VSA-2, indicating that increasing the sensitivity of the algorithm by including rare predicted truncating variants does not dramatically reduce specificity. VSA-3 (which includes variants classified in ClinVar as “likely pathogenic” or HGMD as “disease mutations”) identified numerous “positive” results in several genes, including BRCA1, BRCA2, KCNQ1, LDLR, and RYR1, consistent with the well-established concern of misclassification of variants in current reference databases and highlighting the need for databases with high quality lists of pathogenic variants. Finally, a large number of “positive” results were identified in most genes by the more sensitive algorithms, VSA-4 and VSA-5, reflecting higher underlying variation in certain genes that could impose an upper limit on the possible clinical sensitivity of genomic screening in order to preserve specificity. The performance of the variant selection algorithms for each gene was plotted as 1-specificity (Figure 2).
Figure 2. False Positive Rate estimates of variant selection algorithms in 17 medically actionable genes.
Specificity estimates were calculated for five variant selection algorithms for screening of 17 genes using exome data from 478 well-phenotyped individuals. The variant selection algorithm label is displayed on the vertical axis and the associated false positive rate (1-specificity) is plotted on the horizontal axis, for each graph in the panel.
DISCUSSION
This study models genomic screening of 17 genes for 11 highly actionable monogenic disorders and demonstrates the tradeoff between sensitivity and specificity among variant selection algorithms that might be used in a large-scale screening program. Genomic screening of the general population differs significantly from the much more familiar pursuit of diagnostic genomic sequencing because a symptomatic patient has a much higher pre-test probability of having a pathogenic variant than the average individual in the general population. We have evaluated informatics algorithms that can be used to screen genomic data for potentially pathogenic variants, and for the first time attempted to measure the specificity of genomic findings in the context of population screening.
For most conditions, the true clinical sensitivity of genetic testing is not well established. In some cases, a very small number of variants account for a high proportion of cases and thus sensitivity of the most stringent variant selection algorithms will be high, whereas in other conditions a high proportion of cases are due to novel or private mutations that would not be documented in databases of known pathogenic variants, thus reducing the sensitivity of the most stringent variant selection algorithms. In most conditions it is very difficult to discern what fraction of cases are caused by recurrent mutations (which would be considered “known pathogenic”) versus novel or rare mutations that may only occur in a small fraction of cases (which might be considered “likely pathogenic”). That being said, the predictive value of a sequencing-based screening test for rare monogenic disorders hinges predominantly on specificity, not sensitivity.
The relationship between variant pathogenicity and test specificity
We suggest that the degree of confidence regarding variant pathogenicity is analogous to the specificity of that result. Tests reporting only “Pathogenic” variants approach 100% specificity, whereas tests reporting “Likely Pathogenic” variants have 90-95% specificity depending on the confidence standard applied. In the diagnostic context, reduced specificity is acceptable because of the high pre-test probability and the importance of excluding a diagnosis, when possible. However, when screening the general population for rare conditions, even a small decrease in specificity has a devastating impact on PPV. Conversely, in the case of rare conditions NPV is not significantly improved by even large changes in sensitivity, which decreases the imperative to consider novel variants that inherently reduce the specificity of the screening test. These factors are critical points to consider in the design of genomic screening algorithms for the healthy population.
For example, the prevalence of three BRCA1 and BRCA2 pathogenic founder mutations in the Ashkenazi Jewish population is 2.6%25 and the specificity for these variants is 100%, resulting in a very high PPV. However, the specificity of most other variants (including novel truncating variants) will be less than 100%. In the general population, where disease prevalence is less than 1% and a much wider range of pathogenic variants is detected,26 a decrease of specificity to even 99% would result in a PPV of 37%, which some may find unacceptable for application of such screening in the general population. This dramatic decrease in PPV, despite a very small change in specificity, is true for all rare monogenic conditions.
It should be noted that fully automated analysis of results without some level of human curation prior to confirmation and reporting is not possible at present; thus variants flagged as a possible “positive” screening result by informatics algorithms are expected to undergo scrutiny by a human analyst to further reduce reporting of clearly non-pathogenic variants. This necessity for some level of human curation will be a critical limiting factor for deployment of genomics in large-scale population screening but can be optimized by designing variant selection algorithms that maximize the sensitivity of the screen without burdening the reviewer with numerous non-pathogenic variants. In the long term, the empiric sensitivity and specificity of variant selection algorithms may be a more accurate measure of predictive value than the pathogenicity classifications used in diagnostic testing.
Balancing outcomes, interventions, and the implications of screening results
The conditions being evaluated in the GeneScreen project are heterogeneous with respect to the penetrance, expressivity of manifestations, and recommended follow-up management strategies (Table 1). Positive genomic screening results in these conditions would lead to increased clinical surveillance and other interventions. In the case of pathogenic variants in BRCA1 or BRCA2, management options include increased surveillance or prophylactic mastectomy and/or bilateral salpingoophorectomy. These recommendations are very different from management of Familial Hypercholesterolemia due to pathogenic variants in LDLR, which includes periodic monitoring of cholesterol and treatment with cholesterol-lowering medications, an efficacious intervention with dramatically less impact (physical, psychosocial and monetary). Thus, the downstream consequences of positive screening results will differ between conditions. It should be noted that in conditions with incomplete penetrance, even truly pathogenic variants may not manifest in disease and therefore would constitute overdiagnosis in some individuals who would not develop disease in their lifetime. This concern is similar in many ways to a false positive result, in that the downstream consequences would not be expected to benefit these individuals and would only expose them to harm.
Table 1.
Characteristics of screened conditions and proposed variant selection strategy
| Category | Condition (Gene) | Interventions | Suggested false positive tolerance |
Proposed Variant Selection Algorithm a |
|---|---|---|---|---|
| Cancer | Familial Adenomatous Polyposis (APC) |
Colonoscopy, endoscopy screening, thyroid ultrasound, surgery |
Low | VSA-3 |
| MUTYH-associated polyposis (MUTYH) |
Colonoscopy, endoscopy | Low | VSA-4 | |
| Lynch Syndrome (MLH1) | Colonoscopy, endoscopy, endometrial biopsy, possible surgery (prophylactic hysterectomy and salpingo- oophorectomy) |
Low | VSA-3 | |
| Lynch Syndrome (MSH2) | VSA-3 | |||
| Lynch Syndrome (MSH6) | VSA-3 | |||
| Lynch Syndrome (PMS2) | VSA-3 | |||
| Familial Breast/Ovarian cancer (BRCA1) |
Breast imaging, prophylactic mastectomy and/or salpingo- oophorectomy |
Very low | VSA-2 | |
| Familial Breast/Ovarian cancer (BRCA2) |
VSA-2 | |||
| MEN2A/2B (RET) | Prophylactic thyroidectomy, serum metanephrine blood test |
Low | VSA-3 | |
| Cardiovascular | Long QT Syndrome (KCNQ1) |
Cardiology consultation, electrocardiogram (ECG), beta- blocker medication if ECG positive; implantable cardioverter-defibrillator if symptomatic |
Low | VSA-3 |
| Long QT Syndrome (KCNH2) |
VSA-4 | |||
| Long QT Syndrome (SCN5A) | VSA-3 | |||
| Familial Hypercholesterolemia (LDLR) |
Lipid biochemical screening, pharmacotherapy if needed |
High | VSA-3 | |
| Marfan Syndrome (FBN1) | Echocardiography, ophthalmologic screening |
High | VSA-3 | |
| Other | Malignant Hyperthermia (RYR1) |
Avoidance of specific anesthetics |
Medium | VSA-3 |
| Hereditary Hemochromatosis (HFE) |
Ferritin biochemical screening, phlebotomy |
High | VSA-4 | |
| Alpha-1 Antitrypsin Deficiency (SERPINA1) |
Avoidance of smoke exposure | High | VSA-4 |
Selected based on estimated specificity and estimated proportion of pathogenic variants that are truncating vs. missense
The negative consequences of genomic screening can be minimized if any of the following conditions exists: 1) test performance maximizes PPV and thus results in few false positives, 2) low-risk confirmatory follow-up tests can reduce the number of false positives, or 3) downstream consequences of false positives are relatively insignificant. Thus, a genomic screening project could consider customizing PPV thresholds for different conditions based on the above criteria and available information about the mutational spectrum and proportion of cases attributable to known versus novel mutations (Table 1). For example, for BRCA1 or BRCA2 the false positive rate will need to be very low, because risk-reducing surgery is an important management step and no confirmatory tests exist to ensure that a genetic variant is indeed pathogenic. Thus, only carefully curated lists of pathogenic variants and, perhaps, rare truncating variants (the equivalent of VSA-2) should be reported in a screening context. In other cases, inclusion of rare, potentially damaging missense variants (VSA-4) may be acceptable, depending on the specificity of variant selection algorithms for the gene, the spectrum of pathogenic mutations observed, and the false positive tolerance based on clinical implications of a positive screening result.
The argument that population genomic screening should utilize gene-specific false positive thresholds requires precise measurement of the clinical sensitivity and clinical specificity of variant selection algorithms (as well as the prevalence of the condition) in order to optimize PPV. However, such estimates will likely require a dataset of 100,000+ individuals with extensive phenotypic data, which does not yet exist, and the specificity estimates provided here should be considered provisional. The population being sequenced also needs to be considered – existing datasets fail to comprehensively ascertain the pathogenic and benign variants that are present in diverse populations, which will impact variant selection algorithms and pathogenicity assessments. In addition, databases of clinically relevant variants are known to have entries with misattributed pathogenicity, and will need to be greatly improved before they can be relied upon for screening.
Conclusions and Future Directions
Screening the general population for deleterious variants in highly selected genes holds great promise to decrease morbidity and mortality for millions of individuals. However, we must be cognizant of our limited ability to interpret the pathogenicity of rare genomic variation, the implications this has for standards used to analyze and report genomic findings, and the downstream consequences of screening in the general population. Otherwise, great harm could result. Much additional research is required to assess the overall cost of screening efforts, including the downstream effects of false positive tests, in order to evaluate the overall economic impact of genomic screening.27 A host of factors will need to be studied, including cost-effectiveness, adverse consequences of interventions, insurance impact, education and consent materials that address the complexity of potential harms and benefits of screening, implications for reproductive choices, potential psychosocial impacts, and the benefits and harms of familial cascade testing.
In this study we have addressed a small subset of highly actionable genetic conditions, that we think are currently the most promising candidates for genomic screening in the general adult population. However, these results also have fundamental implications for any effort that considers genome-scale sequencing in healthy individuals. Although genomic sequencing is inherently appealing for personalized medical care, it should be initiated cautiously with rigorous attention to the predictive value of genetic variation, and with careful consideration of the implications of false positive results across the broad range of genetic conditions that could potentially be identified.
ACKNOWLEDGEMENTS
This study was supported by NHGRI 2P50 HG004488, NHGRI 1U01 HG006487, and the Howard Holderness Distinguished Medical Scholars Foundation.
Footnotes
The authors report no conflicts of interest. M.C.A performed data analysis. J.S.B had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
GeneScreen Investigators include:
Daniel K. Nelson, MS, CIP, National Health and Environmental Effects Research Laboratory, Environmental Protection Agency
Debra G. Skinner, PhD, Frank Porter Graham Child Development Institute, University of North Carolina at Chapel Hill
John M. Conley, J.D., Ph.D., School of Law, University of North Carolina School at Chapel Hill
Christine Rini, PhD, Department of Health Behavior, Gillings School of Global Public Health, University of North Carolina at Chapel Hill
Marcia Van Riper, RN, PhD, FANN, School of Nursing, University of North Carolina at Chapel Hill
Gabriel Lázaro-Muñoz, PhD, JD, MBE, Center for Genomics and Society, University of North Carolina at Chapel Hill
Julianne M. O’Daniel., MS, CGC, Department of Genetics, University of North Carolina at Chapel Hill
R. Jean Cadigan, PhD, Department of Social Medicine, University of North Carolina at Chapel Hill
Rebecca L. Walker, PhD, Department of Social Medicine, University of North Carolina at Chapel Hill
Ann Katherine M. Foreman, MS, CGC, Department of Genetics, University of North Carolina at Chapel Hill
Eric Juengst, PhD, Department of Social Medicine, University of North Carolina at Chapel Hill
Myra I. Roche, M.S., C.G.C., Department of Pediatrics, University of North Carolina at Chapel Hill
Kirk Wilhelmsen, MD, PhD, Department of Genetics, University of North Carolina at Chapel Hill
REFERENCES
- 1.Kemper AR, Green NS, Calonge N, et al. Decision-making process for conditions nominated to the recommended uniform screening panel: statement of the US Department of Health and Human Services Secretary's Advisory Committee on Heritable Disorders in Newborns and Children. Genet Med. 2014;16(2):183–7. doi: 10.1038/gim.2013.98. [DOI] [PubMed] [Google Scholar]
- 2.Evans JP, Berg JS, Olshan AF, Magnuson T, Rimer BK. We screen newborns, don't we?: realizing the promise of public health genomics. Genet Med. 2013;15(5):332–4. doi: 10.1038/gim.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. http://www.geisinger.org/for-researchers/research-at-geisinger/genomic-study/
- 4. http://www.genomicsengland.co.uk/
- 5.King MC, Levy-lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312(11):1091–2. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
- 6. http://www.whitehouse.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative.
- 7.Hunter DJ, Khoury MJ, Drazen JM. Letting the genome out of the bottle--will we get our wish? N Engl J Med. 2008;358(2):105–7. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- 8.Prince AE, Berg JS, Evans JP, Jonas DE, Henderson G. Genomic screening of the general adult population: key concepts for assessing net benefit with systematic evidence reviews. Genet Med. 2014 doi: 10.1038/gim.2014.129. doi: 10.1038/gim.2014.129. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med. 2012;14(4):399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue Y, Chen Y, Ayub Q, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91(6):1022–32. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133(1):1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo HI, Woo YM, Kim S, Lee ST, Ki CS, Kim JW. Challenges in assessing pathogenicity based on frequency of variants in mismatch repair genes: an extreme case of a MSH2 variant and a meta-analysis. Gene. 2014;546(2):421–4. doi: 10.1016/j.gene.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 13.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet. 2013;93(4):631–40. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132(10):1077–130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–91. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson BA, Spurdle AB, Plazzer JP. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46(2):107–15. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bizon C, Ahalt S, Fecho K, Nassar N, Schmitt C, Scott E, Wilhelmsen K. Technologies for Genomic Medicine: CANVAS and AnnoBot, Solutions for Genomic Variant Annotation. RENCI, University of North Carolina at Chapel Hill; 2014. http://dx.doi.org/10.7921/G0QN64N3. [Google Scholar]
- 19.Landrum MJ, Lee JM, Riley GR, et al. 2014;42(Database issue):D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenson PD, Ball EV, Mort M, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21(6):577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 21.Exome Aggregation Consortium (ExAC) Cambridge, MA: (URL: http://exac.broadinstitute.org) accessed 10/2014. [Google Scholar]
- 22.Kircher M, Witten DM, Jain P, O'roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt KD, Brown GR, Hiatt SM, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42(1):D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson EB. Probable inference, the law of succession, and statistical inference. J Am Stat Assoc. 1927;22:209–12. [Google Scholar]
- 25.Risch HA, McLaughlin JR, Cole DE, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98(23):1694–706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 26.Rebbeck T, Mitra N, Wan F, et al. Association of Type and Location of BRCA1 and BRCA2 Mutations with Risk of Breast and Ovarian Cancer. JAMA. 2015;313(13):1347–1361. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feero WG, Wicklund C, Veenstra DL. The economics of genomic medicine: insights from the IOM Roundtable on Translating Genomic-Based Research for Health. JAMA. 2013;309(12):1235–6. doi: 10.1001/jama.2013.113. [DOI] [PubMed] [Google Scholar]