Summary
Technology development in biological research often aims to either increase the number of cellular features that can be surveyed simultaneously or enhance the resolution at which such observations are possible. For decades flow cytometry has balanced these goals to fill a critical need by enabling the measurement of multiple features in single cells, commonly to examine complex or hierarchical cellular systems. Recently, a format for flow cytometry has been developed that leverages the precision of mass spectrometry. This fusion of the two technologies, termed mass cytometry, provides measurement of over 40 simultaneous cellular parameters at single-cell resolution, significantly augmenting the ability of cytometry to evaluate complex cellular systems and processes. In this Primer, we review the current state of mass cytometry, providing an overview of the instrumentation, its present capabilities, and methods of data analysis as well as thoughts on future developments and applications.
Introduction
Biological research across fields has shed light on the complexity of cellular systems, recognizing the unique features of individual cells within populations once assumed to be homogeneous. As a result, increasing effort has been invested to develop methods capable of quantifying cellular features at single-cell resolution. While the single-cell mindset is increasingly prevalent, it stems from a long history of investigation. Studies of single cells utilizing microscopy to discern features of cellular organization and behavior date back centuries. Similarly, investigations of inherently diverse cellular networks, such as that which exists within the immune system, have for decades relied heavily on high-throughput single-cell analysis platforms such as flow cytometry, in many respects paving the road for the current single-cell revolution in modern biology.
Simultaneously, the research community has sought to develop methods by which multiple aspects of cellular processes can be assessed or quantified simultaneously, ushering in the age of “-omics” technologies. These approaches have aimed to capture a wealth of knowledge contained at a particular level of cellular behavior—genomic, transcriptomic, proteomic, metabolomic, etc.—from any biological sample. Approaches intended to multiplex such measurements have, in turn, required development of new methods in data analysis to integrate computational and statistical tools with biological research.
While the two aforementioned goals—increased resolution and parameterization—have long inspired the development of research technologies, only recently have the tools in each arena become sufficiently mature to begin bridging the gap between them. The vision of a technology capable of multiplexing single-cell measurements on an “-omics” scale is coming to fruition in a variety of venues. Advances in single-cell genomics, transcriptomics, proteomics, functional assays and imaging each offer attractive alternatives for capturing multi-dimensional information that clarifies cellular identity and function. Here we focus on one such method, mass cytometry, which uniquely enables the quantification of over 40 parameters on single cells with the throughput required to survey millions of cells from an individual sample (Bandura et al., 2009; Bendall et al., 2011; Ornatsky et al., 2010). These characteristics enable investigating complex cellular systems as what they are—coordinated systems—by observing the diversity of cellular phenotypes and behaviors in a single sample.
Filling the Gap: Single-Cell Resolution with High Parameterization
When deciding how to address a biological question, researchers are often faced with a dilemma: should we (A) cast a broad net and capture as much information as possible at a particular level of cellular behavior or (B) take a highly-targeted approach to reveal a more limited number of cellular features with higher resolution? The tools available for either option have never been better. We are now able to sequence the entire genome or transcriptome of a given sample routinely, and advances in microfluidics have enables studies of single-cell transcriptomes in up to thousands of cells (Klein et al., 2015; Macosko et al., 2015). Alternatively, modern imaging technologies enable tracking single molecules in cells or individual cells even within a living organism.
However, a gap still remains when considering all these alternatives—one that mass cytometry is currently able to fill: resolution at the level of single cells, parameterization of over 40 simultaneous dimensions, and throughput enabling the measurement of millions of cells from an experimental sample. Throughput at this scale is essential for thorough characterization of complex cellular samples, where rare cell populations with essential biological function would otherwise be missed. The deep parameterization is sufficient to identify the major cell subsets in a sample with sufficient parameters “left over” for studies of cellular behavior. For example, quiescent hematopoietic stem cells comprise only 1 in 25,000 mononuclear cells in bone marrow of young adults according to a recent study (Pang et al., 2011), and a subset thereof may have unique biological activity. Moreover, the variance within a cell type may provide biological insights, as in the frequency of cells responding to a stimulus (Bendall et al., 2014) as measured by phosphorylation of signaling proteins. The true nature of this distribution would be obscured in the absence of sufficient sampling. Another advantage of the method compared to other modalities is that mass cytometry is not restricted to investigating one level of cellular metabolism—protein levels, posttranslational modifications, and proteolysis products can all be quantified from a single experiment (Bendall et al., 2012; Bjornson et al., 2013). Simultaneous measurement of mRNA transcripts by mass cytometry has been demonstrated (Frei et al., 2016), DNA synthesis can be monitored by incorporation of modified nucleotides (iodo-deoxy-uridine) (Behbehani et al., 2012), and activity-based probes can be utilized to quantify aspects of the cellular state such as hypoxia or enzymatic activity (Edgar et al., 2014). This platform thus opens new possibilities in biosciences, providing a tool capable of capturing diverse aspects of cellular behavior simultaneously in millions of individual cells.
In this Primer, we provide an overview of the current state of the art in mass cytometry. Below we’ll first describe the basic principles and reagents used in mass cytometry experiments, followed by an overview of the biological questions mass cytometry can answer. We then address the current limitations of the technology. With the technical foundation established, we’ll turn our attention to designing effective mass cytometry experiments, followed by an overview of data analysis and current computational tools that facilitate data interpretation. We conclude with a perspective on the future directions of the field.
Basic Principles
At its core, mass cytometry is a fusion of two experimental platforms: flow cytometry and elemental mass spectrometry (Fig. 1). The current instrumentation for mass cytometry is called Cytometry by Time-Of-Flight (CyTOF) and is described in detail elsewhere (Bandura et al., 2009; Bjornson et al., 2013), but we provide a brief overview here. The motivation behind this fusion of technologies was to increase the number of cellular parameters that could be quantified simultaneously, a goal that has similarly propelled the development of new reagents for fluorescence-based flow cytometry for decades (Baumgarth and Roederer, 2000). The advantage of utilizing mass spectrometry as a means of quantification lies in the ability to distinguish between different reporters. Conventional flow cytometry utilizes fluorophores as reporters, quantifying their light emission as a proxy for molecular expression. However, fluorophore emission spectra overlap, making them more difficult to distinguish from one another, especially as more parameters are measured in a single experiment. In contrast, elemental mass spectrometry is able to discriminate isotopes of different atomic weights with high accuracy. This fundamental difference enables significantly more cellular features to be assayed simultaneously using a mass-based platform. Thus, rather than coupling probes (often antibodies) to fluorophores, mass cytometry experiments utilize probes coupled to unique stable, heavy-metal isotopes. Thus, the quantity of reporter ions in a particular mass channel becomes a proxy for molecular expression with little signal overlap between parameters.
Figure 1. Workflow of a typical mass cytometry experiment.
Single cells are acquired, and a viability stain is applied to mark dead cells for exclusion from analyses. Fixation can optionally be applied at this point to preserve the cell state. Multiple samples can be barcoded with unique combinations of heavy metal tags, enabling them to be pooled together prior to staining to minimize technical variability at this step. After pooling samples into one tube, cells are then incubated with antibodies targeted against proteins of interest. Cell permeabilization can be performed if intracellular targets are to be measured. Cells are nebulized into droplets as they are introduced into the mass cytometer. They then travel into an inductively-coupled argon plasma (ICP), in which covalent bonds are broken and ions are liberated. The ion cloud is filtered by a quadrupole to remove common biological elements and enrich the heavy metal reporter ions to be quantified by time-of-flight mass spectrometry. Ion signals are integrated on a per-cell basis, resulting in single-cell measurements for downstream analysis. Data are compiled in an FCS file that can then be parsed and plotted in a variety of ways.
When performing a mass cytometry experiment, cells of interest are first incubated (or “stained”) with a cocktail of affinity reagents (the most common being antibodies). These affinity reagents have been previously conjugated to a polymer chain of chelating groups that bind purified, stable heavy metal isotopes (Lou et al., 2007; Ornatsky et al., 2008). These probes bind targets of interest on and/or within the cell, enabling the attached metal ions to serve as reporters for the expression level of the target. Cells are then passed in a single-cell suspension into a nebulizer, which places cells into droplets for introduction into the mass cytometer.
Upon entering the instrument, cells travel through an argon plasma, in which covalent bonds are broken to produce free atoms, which become charged in the process. The resulting ion cloud is passed through a quadrupole to discard common biological elements, enriching for heavy-metal reporter ions, which are separated by their mass-to-charge ratio in a time-of-flight mass spectrometer. The ion counts are converted to electrical signals and ultimately into a data matrix in which every column represents a distinct isotope measured and each row represents a single mass scan of the detector. Because the ion cloud from a single cell occupies 10–40 scans of the detector (a parameter termed “event length”), these signals are integrated into single-cell events for later analysis.
Mass Cytometry for High-Dimensional Imaging
Recently, antibodies labeled with mass tags have found an additional application in new high-dimensional imaging modalities. While not the focus of this Primer, we review them briefly here. One such method, termed imaging mass cytometry pairs laser ablation of tissue sections stained with metal-labeled antibodies to the CyTOF mass cytometer (Giesen et al., 2014). This method has the advantage of utilizing the same equipment as mass cytometry on suspension samples. Another approach, called multiplexed ion beam imaging (MIBI), uses an ion beam to liberate metal ion reporters, which are quantified by mass spectrometry (Angelo et al., 2014). While requiring more specialized equipment, this latter method offers increased speed, sensitivity and resolution. We anticipate that advances in methods such as these will transform our understanding of tissue architectures by enabling high-dimensional, quantitative analyses in situ.
Observing, Quantifying and Interrogating Cellular Processes
Mass cytometry uniquely enables investigation of cell identity and behavior at the level of proteins and the properties (e.g. isoforms or posttranslational modifications) of proteins, which are largely the key executors of biological processes. Determination of cell identity is often accomplished by measuring levels of transmembrane proteins expressed on the cell surface (Ornatsky et al., 2008). When used as so-called “markers,” these proteins can reveal the lineage and maturation state of individual cells, as these proteins often have restricted patterns of expression across cell types. In the biological story told by a mass cytometry experiment, these markers can be used to define the identities of our characters.
While cell-surface proteins are often used as markers, these molecules execute critical biological processes, and their quantification can provide insight into cellular behavior. These include molecules such as cell-signaling receptors, adhesion molecules, and receptor ligands. As an example, activated dendritic cells upregulate the expression of MHC molecules as well as co-stimulatory receptors such as CD80 and CD86. Therefore, identifying changes in the expression of these cell-surface proteins reveals biological insight regarding the behavior of these cells. As another example, the antigen specificity of a T cell can be revealed by using peptide:MHC tetramers that bind to a specific T cell receptor, revealing clonal expansion and cellular differentiation during an infection (Newell et al., 2013). These proteins describe how our characters are equipped to interact with their surroundings, reflecting the context and setting.
Looking inside the cell, mass cytometry is, for instance, able to enumerate the expression of transcription factors that drive gene expression programs (Spitzer et al., 2015; Zunder et al., 2015b). Moreover, the technology is capable of resolving cell signaling programs by measuring the phosphorylated (or otherwise post-translationally modified) forms of the proteins that comprise these cascades in manners that reflect biological mechanism or clinical outcomes (Bendall et al., 2011; Bodenmiller et al., 2012; Gaudilliere et al., 2014; Krishnaswamy et al., 2014; Mingueneau et al., 2013; 2014). Methods for quantifying RNA transcript levels by flow cytometry are also translatable to the mass cytometry platform, enabling elucidation of cellular behavior at another level of metabolism (Frei et al., 2016). These types of measurements reveal how the characters are each processing and reacting to information at a given point in time, allowing mass cytometry to become an objective narrator of the biological story told by a sample.
Mechanisms of intercellular communication, such as production of cytokines and growth factors, are additionally quantifiable within single cells (Newell et al., 2012). A method for robustly identifying phases of the cell cycle and monitoring cellular proliferation by incorporation of iodo-deoxyuridine (IdU, analogous to the fluorescent BrdU) has been described (Behbehani et al., 2012). By interrogating these cellular programs, researchers gain insight as to how a cell is responding to its environment, ascertaining the actions that each cell takes and developing the plot.
Thus, the majority of the tools available for interrogating cellular programs by flow cytometry have now been translated across to the mass cytometry platform. However, increased parameterization enables the assessment of many of these processes in diverse cell types simultaneously. The suite of cellular processes that can be enumerated by mass cytometry provides the potential to reveal the behavior of individual cells in a more holistic manner. This allows researchers to interrogate a cellular program of interest in great depth, surveying many molecular players that together mediate a cellular decision. By monitoring these processes simultaneously, mass cytometry can reveal co-regulation and crosstalk between cellular programs. Thus, it is now possible to observe cell signaling, proliferation and cell death programs in many cell types simultaneously.
To illustrate some of the above with an example, a recent study of B cell development in the healthy human bone marrow identified coordination points of cell signaling, proliferation and cell death in distinct stages of maturation (Bendall et al., 2014). These coordination points were only discernable because of the high-dimensional data that mass cytometry provides. For instance, the unambiguous identification of a checkpoint between the pre-B I and pre-B II cell stages required simultaneous quantification of 19 cell-surface proteins, 2 intracellular enzymes (TdT and RAG1), a proliferation marker (Ki67), a phosphorylated cell signaling protein (p-STAT5), and a modified form of a protein involved in apoptosis (cleaved PARP).
While B cell development has been studied for decades, this study was also able to reveal new cell subsets because proteins that were not previously known to discriminate early B cell progenitors (TdT vs. CD24) were measured simultaneously along with the markers required to exclude more and less mature cell types (Bendall et al., 2014). In hindsight, this observation could have been made using fewer parameters, but it was previously not seen because this particular combination of proteins required would not have been intuitive based on the canonical model of human B cell development. As with RNA-seq and its precursor technology of gene expression arrays, mass cytometry enables a hypothesis generation approach to biological inquiries of complex cell populations.
Thus, the dimensionality afforded by mass cytometry results in a more complete story of how a system is structured at the cellular level and regulates its behavior. As with a plot twist, such as the physiological perturbations caused by pathology, mass cytometry is similarly adept at identifying cellular rewiring, as showcased in a recent study of acute myeloid leukemia (Levine et al., 2015).
Current Limitations
While a profusion of cellular processes can be simultaneously investigated using mass cytometry, the technological platform does have some important limitations to consider when designing an experiment. Because cells are atomized and ionized, it remains infeasible to recover living cells after analysis. Moreover, due to the dynamics of ion flight in the mass spectrometer, the throughput of mass cytometry lags behind that of fluorescence-based instruments. Additionally, the sensitivity of mass reporters falls shy of few, more quantum-efficient fluorophores (such as phycoerythrin), making it more difficult to measure molecular features that are expressed at very low levels using mass cytometry. That said, the sensitivity range of ions across the mass range is 3–4 fold in difference, whereas fluorophores must contend with a vast range (50 fold) encumbered by serious issues in spectral output (which can be partly remedied by fluorescence compensation).
As with all new technologies, standards are being established for the comparison of data across laboratories and instruments. A recent study compared the ionization efficiency across CyTOF 2 mass cytometers, demonstrating that quantitative comparisons between instruments is significantly improved by normalization (Tricot et al., 2015). Because each instrument had a characteristic efficiency profile, comparison of data from the same instrument can be accomplished by incorporating polystyrene beads for data normalization, as described in more detail below (Finck et al., 2013). The authors of this former study concluded that another normalization reagent containing all measured reporter ions would facilitate highly quantitative inter-instrument comparisons (Tricot et al., 2015).
Perhaps most important are limitations shared by fluorescence-based flow cytometry. Some mediators of cellular behavior, such as many small molecule metabolites, are difficult or impossible to measure by CyTOF or fluorescence based cytometry because there is no easy technical approach that maintains small molecules and a binding agent associated with the cell. While certain cellular attributes can be measured (such as pH or ion concentration) with environmentally sensitive chemicals or modified fluorescence proteins, the development of mass cytometric analogues for such purposes is more difficult to contemplate. While one activity-based probe for mass cytometry has been developed (Edgar et al., 2014), further elaboration in this area could expand the realm of cellular processes available for investigation using these both cytometry platforms. Finally, the static nature of either mass cytometry or fluorescence based flow cytometry precludes serial measurements of the same cells, limiting measures of individual single cells over time.
High-quality affinity reagents with minimal cross-reactivity are required (as for all methods using antibodies or other affinity reagents), but numerous validated antibodies are commercially available, and probe design for mRNA detection is facilitated by user-friendly tools (Frei et al., 2016). Despite the use of affinity reagents, the cost of mass cytometry per cell is very favorable to other single-cell analysis modalities. The cost of reagents, disposables, and data acquisition is approximately 0.005 cents per cell when acquired by mass cytometry. In contrast, an estimated value for cells measured by single-cell RNA-sequencing using the Fluidigm C1 system and unique molecular identifiers (Islam et al., 2013) comes in at $22 per cell, (even when reagents are made in house; Heger, GenomeWeb, 2014). This amounts to a 4400-fold higher cost compared to mass cytometry. Even on a per-parameter, per-cell basis, assuming 4,000 genes are measured per cell, this amounts to a 44-fold cost differential in favor of mass cytometry.
Because mass cytometry currently focuses on the use of affinity-based reporters, the targets to be measured must be determined prior to sample acquisition. Therefore, the information collected about a sample is limited to the information content captured by the dimensions measured. Given this, careful consideration should be made as to which markers might provide the most information content to answer the biological question at hand. Even despite this fact, an advantage of high-parameter analysis is that unexpected juxtapositions of these markers often reveal novel biological or clinical findings (Bendall et al., 2011; 2014; Gaudilliere et al., 2014; Krishnaswamy et al., 2014; Spitzer et al., 2015). For example, one application of mass cytometry comes in classifying types of cells, which can also be thought of as “information content” clusters. Cell populations are essentially groups of cells defined by the co-expression of markers, which reflect the networks of genes that determine such protein expression patterns. Even given prior knowledge and careful panel design, cells expressing unique combinations of proteins have been discerned that would not have been intuitive based on the literature findings (Bendall et al., 2011; 2014; Spitzer et al., 2015).
While this limitation (and opportunity) is clear from years of work in high dimensional flow cytometry, other single cell modalities have their own inherent, and subtle, biases that can cause data to be difficult to interpret. For instance, single-cell RNA-sequencing (RNA-seq) is capable in theory of quantifying genome-wide transcript expression, but the depth of sequencing and efficiency of mRNA capture biases towards the most highly abundant transcripts (which are not always the most important biologically). For instance, traditional RNA-seq methods can require up to 50 mRNA copies per cell for reproducible detection in every cell (Hashimshony et al., 2012). Technical manipulation of cells prior to lysis and reverse transcription for RNA-seq can introduce technical artifacts that can limit clear interpretation of the results (Dvinge et al., 2014). In contrast, like flow cytometry, mass cytometry can be performed on fixed cells—essentially preserved in a manner that limits biologically related artifacts driven by technical procedures. While a method for single-cell RNA-sequencing of fixed cells has been recently reported, it currently enables detection of only the 3,000–4,000 most highly-abundant transcripts (Thomsen et al., 2015). Improvement in unbiased RNA amplification technologies for single cells, targeted RNA-seq, or merging the measurement of multiplexed RNA on a per cell basis with the throughput of mass cytometry as per Frei et al. might overcome some of these limitations.
Beyond the limitations above, the next important challenge for mass cytometry has been the interpretation of the complex datasets that result (a fortunate problem to which we turn our attention below). In order to contextualize this, we will first provide an overview of experimental design for mass cytometry.
Experimental Design and Reagent Development
Mass cytometry can be a powerful tool for interrogating complex biological systems. However, each mass cytometry experiment is a time-consuming endeavor. Maximizing the information content generated requires considerable planning. Here, we delineate the questions we usually pose to guide our own experimental and technical strategies.
The most critical aspect of a successful mass cytometry experiment is establishing clear goals at the onset: What question(s) ought the experiment answer? Is it a specific biological mechanism one is after? Are you interested in a “systems” understanding how the individual signaling components within a cell are behaving, or are you more interested in the intercellular dynamics of a complex cellular system? Are you measuring the behavior of one sample, or are you collecting a series of samples from a cohort of patients? Answering these questions up front will have great implications regarding effective experimental design. Because so many different aspects of biology can be interrogated using mass cytometry, it becomes essential to clarify the experimental intent from the onset.
Having set clear goals, we then identify the types of cell samples required to answer the desired question(s). Will we need primary patient samples, or will a cell line suffice? Are we interested in how cellular behavior changes over time? Are we attempting to quantify differences amongst or between groups of samples? Do the biological processes under investigation require activation or exogenous perturbation of the samples? The answers to these and other questions will begin to determine the scope of the experiment, which can range from a few, well-controlled samples to dozens of patient specimens. Understanding the scale of the experiment will critically inform the remainder of the planning.
We next identify the panel of cellular features that we will measure. A helpful place to start is to delineate the major landmarks, or the types of cells, to identify in the samples. Depending on the experimental goals, these may include features that determine the identity of cells as well as the behavioral state of the cells (which are not always mutually exclusive attributes). Do the experimental goals require the identification of every subpopulation of immune cell, would we be happy to focus on a particular set of cell subsets, or are we studying a cell line where cell-surface markers are less helpful? Mass cytometry experiments can range from identifying nearly every type and sub-type of cell in a given sample (Spitzer et al., 2015) all the way to dedicating just a few parameters that reveal broad classes of cells while using the remaining parameters to measure behavioral states (Bodenmiller et al., 2012).
If the experimental goal is to characterize one cellular process exhaustively, we can dedicate most parameters to enumerating cellular features that shed light on that process of interest, perhaps all the relevant members of a signaling cascade or a cell death program. If the goal is to more broadly characterize cellular behavior, however, we must decide which processes we will assess and choose cellular features to measure that best describe those processes. For example, if a study aims to provide a granular understanding of the cell cycle state, we would include several parameters that enable delineation of G1, S, G2, and M phases (Behbehani et al., 2012). If, on the other hand, cell division is only one small aspect of the study, a broad proliferation marker, like Ki-67, usually suffices. For flow cytometry experts accustomed to working with 12–15 markers, choosing a few dozen parameters for measurement may seem luxurious. However, narrowing down which parameters to measure is rarely obvious and critically defines the types of conclusions that can be drawn.
We usually start by creating a wish list of markers without consideration of practicality. We narrow the list by prioritizing the cell types in which we are most interested (this defines the surface markers). We then prioritize the intracellular attributes most relevant. Next, we determine which markers would require new reagent development and which already have high-quality reagents, and then we determine whether the staining conditions for the markers are compatible with each other. If sample supply is not limited, it is always possible to create multi-panel designs to accommodate the desired biology. For instance, if one is studying immune infiltrates of tumors, it makes sense to create a panel focused on tumor cells and, separately, an immune targeted panel. The flexible reuse of signaling systems in biology can often enable the same set of intracellular markers to be used in multiple panels, but subtle considerations of which markers ought to be in which panel have to be made.
Mass cytometry is relatively new to the scene, and as such commercial reagents for use “off the shelf” are less prevalent that those for fluorescence based approaches. While more reagents are introduced regularly, highly customized antibody panels often require in-house conjugations of purified antibodies to polymer chains binding heavy metal ions of choice (Bjornson et al., 2013; Lou et al., 2007). While the chemistry involved is a conventional method for labeling proteins (maleimide coupling through –SH groups), it remains essential that all newly-conjugated antibodies be subjected to rigorous validation and titration to confirm binding to the target of interest and ascertain the appropriate staining concentration in the system of interest. Because the instrument sensitivity varies across the mass range of isotope reporters, and each metal isotope is available at slightly different purities, choosing the best channel for each parameter requires some advance planning. For instance, while metals with several stable isotopes can be purified highly (95–99%), the small residual levels of other isotopes will result in signal bleed from one channel into another of the same element. Moreover, several reporter metal ions oxidize at a low frequency (1–2%), gaining an apparent mass of 16amu and resulting in signal bleed into that corresponding channel. However, because the interferences between isotopes pale in comparison to the spectral overlap between some fluorophores (often >50% in multicolor panels), complications with multicolor flow cytometry due to as spectral compensation artifacts are simply not an issue on a mass cytometry platform. These considerations require some navigation, but when evaluated carefully, it is a straightforward procedure to enable a 30- to 45-dimensional panel of cellular features that can be quantified on nearly any sample of interest that can be rendered into a single cell suspension.
A key consideration is controlling for experimental noise between sample runs. Minor differences in the number of cells per sample or pipetting error when adding affinity reagents can potentially introduce small amounts of technical variance, as is also true for fluorescence-based flow cytometry. In order to make the most precise comparisons between critical samples, methods for barcoding individual conditions are used that enable pooling samples for downstream staining and acquisition (Behbehani et al., 2014; Bodenmiller et al., 2012; Mei et al., 2015; Zunder et al., 2015a). These strategies use distinct combinations of a panel of metals, and each of these combinations serves as a unique barcode for an individual sample. After staining samples with these barcode metals, multiple samples can be pooled together into the same tube and then stained with cocktails of antibodies. After data collection, each respective sample is deconvoluted prior to analysis by identifying every cell with that particular barcode. Therefore, samples that require the most precise comparisons should be barcoded together to reduce technical variability. In addition to sample barcoding, a data normalization approach has been developed based on bead standards. These beads contain metal ions, and they are mixed with the cell sample and thus sampled continuously along with the cells. By monitoring the signal captured from these normalization beads over time, it becomes possible to correct for variability in the sensitivity of the instrument over the course of an experiment or between experiments (Finck et al., 2013). An open-source algorithm accompanying Finck et al. normalizes the data from each cell by computing an average of the signal intensities from beads, providing an effective and reproducible approach to data normalization that accommodates inter-sample and intra-sample variability (Finck et al., 2013).
Taking these factors into consideration, it becomes possible to design a thorough experiment to evaluate cellular attributes or behaviors in millions of individual cells from each sample of interest. However, deciding which of many analytical approaches to use in order to best derive biological meaning out of the data remains a key next step. We are often asked—which informatics technique is best? The answer depends on the biological question at hand and often is several.
An Iterative Approach to Data Analysis
As with other types of high-dimensional “-omics” data, mass cytometry provides opportunities to view the resulting data from many perspectives (Table 1, Fig. 2). On the one hand, mass cytometry data sets can be treated exactly the way flow cytometry data has been for decades. It remains possible to ask questions in the data by identifying specific cells of interest by partitioning cells in one-dimensional histograms or two-dimensional scatter plots (a process called “gating”) and quantifying a feature of interest, such as a population frequency or a measure of behavioral state. However, this would limit the value of the multiparameter approach.
Table 1.
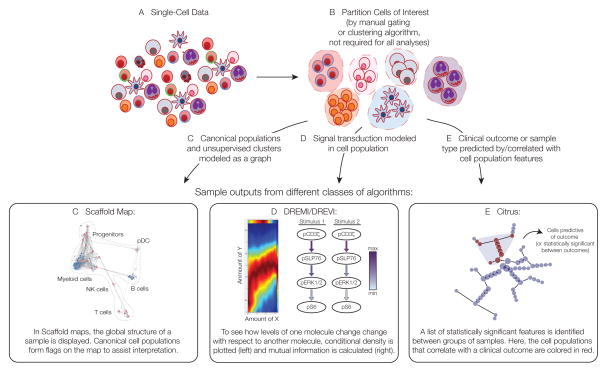
Algorithms developed or applied to the analysis of mass cytometry data. Methods are grouped by the biological questions they seek to answer, including the partitioning of cells into subsets, the visualization of the global structure of a sample, the progression of cells through a continuum (such as differentiation), the diversity of cells in a sample, the signaling networks in cell samples, and cellular features that correlate with or predict outcome. It is important to note that these goals are not always mutually exclusive—for instance, SPADE and Scaffold Maps aim to reveal the global structure of a sample but utilize clustering algorithms as a means of partitioning cells to achieve this goal.
| Type of Question | Tools Available | Unique Features | References |
|---|---|---|---|
|
| |||
| Cell Population Identification | Manual Analysis – Gating Clustering Algorithms | Easy interpretation: grounded in prior knowledge | Pervasive in the literature |
| Many available each with unique advantages | Newly developed methods for mass cytometry analysis:
|
||
|
| |||
| Global Data Structure | SPADE | Unsupervised analysis, density-dependent downsampling to preserve rare cell events | Qiu et al., Nat. Biotechnol. (2011) |
| viSNE | Single-cell resolution on resulting images, which resemble a scatter plot | Amir et al., Nat. Biotechnol. (2013) | |
| Scaffold Maps | Integrates prior knowledge for interpretability, new data can be compared to an existing reference | Spitzer et al., Science (2015) | |
|
| |||
| Cellular Progression | Wanderlust | Defines most likely linear path from a known starting cell to a known ending cell | Bendall et al., Cell (2014) |
| FLOW-MAP | Enables analysis of time course data, no assumptions about differentiation path or directionality | Zunder et al., Cell Stem Cell (2014) | |
|
| |||
| Cellular Diversity | Inverse Simpson Index | Defines the diversity of a cell population based on heterogeneity of protein expression | Strauss-Albee et al., Sci. Trans. Med. (2015) |
|
| |||
| Signaling Network Inference | DREMI/DREVI | Reveals the dependence of one molecular feature on the levels of another | Krishnaswamy et al., Science (2014) |
|
| |||
| Correlative/Predictive Features of Clinical Outcome or Sample Type | Citrus | Identifies significant features between groups of samples, build predictive model of clinical outcome | Bruggner et al., PNAS (2014) |
Figure 2. Computational methods developed for mass cytometry data analysis.
Several classes of tools have been recently developed to assist in the interpretation of mass cytometry data. Here, we focus on those methods developed specifically for this purpose. Sample results are shown from different classes of algorithms designed to assess the global structure of a sample (Scaffold maps), the relationship between two molecules in single cells (DREMI/DREVI), or the cellular/molecular features that correlate with or best predict a clinical outcome or sample type (Citrus), respectively.
Utilizing analysis methods that take advantage of the increased parameterization of mass cytometry allow researchers to test more hypotheses within a single data set. Rather than conducting independent studies to assess whether certain cell types change their behavior, mass cytometry can enable all cell subsets to be studied at once. Instead of focusing an experiment entirely on understanding whether a given subset of cells is proliferating, it is possible to monitor cell death, cytokine production or cell signaling simultaneously.
Simply thinking about cytometry data from the vantage of performing multiple experiments simultaneously would limit the opportunities provided by data structured in this manner. It is key to realize that simultaneous measurements provide a wholly different kind of insight (Chester and Maecker, 2015). Measuring two parameters in an experiment really provides information regarding parameter 1, parameter 2 and how parameters 1 and 2 change together (Fig. 3). In principle, this type of information is available from any dataset in which more than one measurement is taken across samples, including any multicolor fluorescence-based flow cytometry experiment. However, when working with data in higher dimensions, the problem of extracting and interpreting the meaningful information “tied up” in multiple dimensions becomes more pressing—and more exciting to consider!
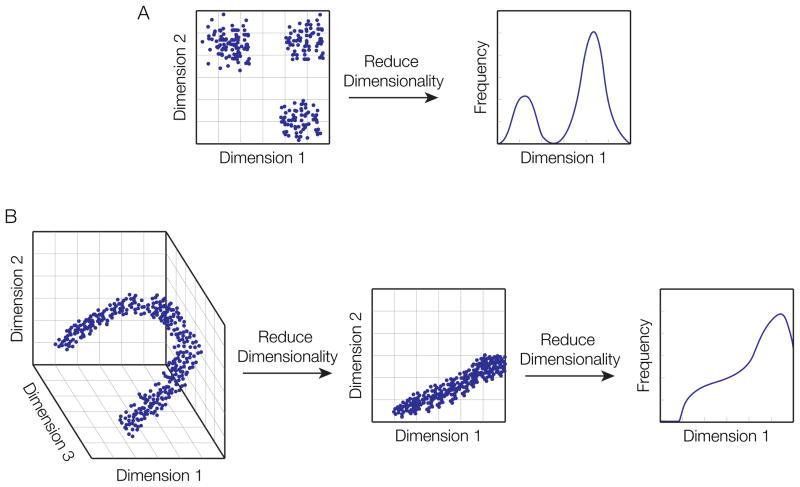
Figure 3. The challenge of visualizing high-dimensional data without information loss.
High dimensional data is challenging to visualize due to the amount of information that must be captured in an effective representation. Here are two examples of how information content is lost by compressing the dimensionality of data. A) 2-dimensional data is compressed into 1 dimension: Left) the frequency of a property called dimension 2 is plotted against the frequency of a property called dimension 1, revealing 3 distinct cell populations. Right) Plotting just the frequency of the Dimension 1 property loses this view. B) 3-dimensional data showing the range of cells that have 3 different properties (dimensions) and how they relate to each other (left) is compressed into 2 dimensions (middle) and 1 dimension (right). The trajectory of cells is entirely lost in the lower-dimensional representations.
As the number of analytical tools available for application to mass cytometry datasets continues to increase, researchers now have choices: which method(s) is most appropriate for the biological question at hand? Due to the high-dimensional nature of these datasets, new types of questions can be asked. Does the method I need even currently exist? In many cases in our lab we have found either the tools do not exist, or due to computational limitations, an algorithm that works for a 10-dimensional flow cytometry dataset may not be practical for a 40-dimensional dataset. Therefore, especially in these early days of the technological platform, an iterative approach to data analysis will likely prove necessary. Which approach is best for the question in mind? Our answer has been, “try them each and interpret for yourself.” Just as there are multiple strategies for aligning sequencing reads to a genome, there are now a number of ways to visualize and analyze mass cytometry data. The fact is that each approach makes certain assumptions about the nature of the underlying data and has the potential to showcase different facets thereof, and will inspire different insights from the same original data (Diggins et al., 2015). In the next sections we enumerate some of the techniques developed to date that we have found most useful.
Visualization Strategies to Facilitate Dynamic Analysis
One instance in which the “curse of dimensionality” becomes immediately apparent is visualizing data: How can we see all of our cells on one page to view the structure of a system? Conventional methods limit the researcher to viewing data in up to 3 dimensions, which are often insufficient for meaningful interpretation and identification of many cells of interest. To represent more information in a space interpretable by humans, the data must be compressed by reducing its dimensionality. Attempts to do so require extracting information from high-dimensional space in order to preserve the organization of the original data. Several approaches have been proposed to address this challenge, each with distinct advantages (or drawbacks). They often draw on principles from extensive prior work on dimensionality reduction in statistics/machine learning.
One older approach for dimensionality reduction that has been applied to mass cytometry data is principal component analysis (PCA), which attempts to capture the most variance within a dataset by creating linear combinations of dimensions to define new compound variables (Bendall et al., 2011; Newell et al., 2012). PCA does provide a visualization space that is somewhat comprehensible, as the axes are derived from the original parameters measured. However, PCA operates under an assumption that the underlying data are parametric, which is not always the case, and the method does not guarantee that the first 2 or 3 compound variables will enable an effective visualization in a tractable space. Moreover, the dimensions of largest variance in the data may not truly answer the question at hand, as interesting biological differences are often subtle ones. Therefore, PCA does provide a convenient method for visualizing mass cytometry data, but it does have several constraints of which one must be aware.
The first algorithm specifically developed to reduce the dimensionality of mass cytometry data into a 2-dimensional projection was termed Spanning-Tree Progression Analysis of Density-Normalized Events (SPADE) (Bendall et al., 2011; Qiu et al., 2011). The method leverages a previously developed concept in cytometry analysis, grouping common cells together by applying a clustering algorithm (Aghaeepour et al., 2013; Chan et al., 2008; Pyne et al., 2009). SPADE implements a density-dependent down-sampling to avoid obscuring rare cell populations, and then clusters similar cells. Clustering is only a first component of this algorithm, used to partition cells into groups that are more easily visualized in the later steps. This is achieved by connecting clusters together in a minimum-spanning tree, a graph in which each cluster is connected to its 2 nearest neighbors while minimizing the total edge length. The cell events are then up-sampled to re-capture the original data density. SPADE therefore enables the entirety of a high-dimensional cytometry dataset to be visualized in one planar image. Applying the algorithm to data from healthy human bone marrow exhibited its ability to showcase the structure of the hematopoietic system (Bendall et al., 2011). Importantly, having contained a given subset of cells within a cluster, the population size or relative expression of any given set of markers (or ratios thereof) can be represented by the size of the node in the graph or its color.
Public implementations of the SPADE algorithm provide researchers a means to interact with the data in user interfaces such as that incorporated into the analysis suite Cytobank (www.cytobank.org) (Chen and Kotecha, 2014), visualizing cellular features of interest across all the cells in a dataset. However, a limitation of the method is the rigid connectivity of the structure, which can prevent similar cell clusters from positioning themselves correctly in a structure that truly represents the underlying biology. The stochastic nature of the density-dependent down-sampling also prevents the graph from being deterministic, meaning that its results can vary between iterations. Moreover, the coordinates of a cluster in the visualization have no meaning, so the user can manipulate the organization. More recently developed algorithms have sought to address these limitations.
A newer strategy for dimensionality reduction of mass cytometry data, Visualization of t-Distributed Stochastic Neighbor Embedding (viSNE), illustrates the structure of high-dimensional data without clustering cells into mutually exclusive groups (Amir et al., 2013). Instead, this approach grants each individual cell a unique location in a 2-dimensional projection that maximizes the similarity—and distinctiveness—of the cell states in the system. The resulting output is a visualization that resembles a dot plot. However, this data representation comes with a tradeoff. First, applying the approach to very large numbers of cells is currently computationally unfeasible. In addition, subtle differences in cell population densities can be obscured. A method has also been developed to identify local maxima from viSNE plots to find putative cell populations for later characterization (Shekhar et al., 2014). While viSNE provides single-cell resolution, and can robustly group similar cells in a 2-dimensional plane, it sacrifices certain features that may be required to address biological questions pertaining to global system structure or cell lineage relationships.
Most recently, we have implemented a new strategy for visualizing global structure that incorporates prior knowledge as a guide for the researcher (Spitzer et al., 2015). This algorithm, termed Single-cell analysis by fixed force- and landmark-directed (Scaffold) maps, is based on yet another strategy, force-directed graphs (Eades, 1984; Fruchterman and Reingold, 1991). Cells are first clustered to enable visualizing the entirety of the data set. Clusters are then spatialized in a 2-dimensional plane by two forces until the energy of the system is minimized. The first is a repulsive force: all nodes are repelled by one another as if they were equivalent poles of magnets. The second is an attractive force: if two clusters are sufficiently similar to one another, they are connected by a spring (i.e., an edge) that pulls them together in the final layout. To incorporate prior knowledge into these maps, manually-gated landmark populations can be included alongside unsupervised cell clusters in the final layout. This enables the landmark nodes to be fixed in place before overlaying data from distinct samples onto the reference structure. This approach uniquely allows an uncharacterized sample to be viewed with respect to a characterized reference for rapid comparison of their global structure. Because each sample can be clustered individually, new data can be integrated and compared to an existing reference framework, which will enable the collation of datasets over time into a repository moving forward. Alternatively, samples can also be clustered together to enable extremely precise comparisons. The approach is capable of discerning immune organization across distinct runs of instruments, between fluorescence based cytometry and mass cytometry datasets, and even across species (Spitzer et al., 2015).
Force-directed layouts can similarly be implemented in the absence of manually-identified landmarks to visualize the landscape of a cellular compartment (Spitzer et al., 2015; Zunder et al., 2015b). These graphs enable visualizing clusters without the constraint of a minimum-spanning tree, allowing clusters to form as many edges as the data should dictate. The result is a more robust final structure in which all sufficiently similar clusters form connections, and the length of these edges reflects their similarity. This flexible framework enables the incorporation of time course data (FLOW-MAP) (Zunder et al., 2015b), and an approach to extract groups of similar cells from a graph using community detection (PhenoGraph) (Levine et al., 2015) is also available. These graphs can similarly be colored by individual parameters for interactive data analysis.
Although the arena of mass cytometry data visualization has seen an emergence of several tools with which to interrogate structure in datasets, researchers ought to consider which method seems best suited for their particular questions. Notwithstanding, these tools provide great opportunities for revealing the organization in high-dimensional data. We speculate that improvements made to these algorithms, as well as the development of novel strategies, will continue to sharpen the visualization toolkit of the cytometry community, enabling the discernment of biological structure from complex high-dimensional data.
Analytical Methods for Characterizing Biological Processes and Cellular Diversity
In addition to the suite of visualization tools described above, a number of strategies for discerning, organizing and characterizing biological processes from mass cytometry data have additionally been developed. These methods uniformly take advantage of high-dimensional data content to model dynamic events such as cell-signaling cascades and cell death programs from cytometric data. One such strategy utilizes Bayesian network analysis to reconstruct cell signaling pathways from phospho-specific flow cytometry data (Sachs et al., 2005). While this method has not yet been applied to mass cytometry data, its increased parameterization and reduced crosstalk between reporters should provide a distinct advantage over fluorescence-based methods for this type of analysis.
More recently, a method has been developed to ascertain the effect of one signaling protein’s activity on that of another (Krishnaswamy et al., 2014; Mingueneau et al., 2014). This approach conditions across the activity range of one signaling molecule in the dataset and asks how a second signaling molecule behaves across this range at the single-cell level. The strategy is capable of highlighting key aspects of signal transduction behavior and cross talk between co-regulated pathways. While these strategies have heretofore been applied to cell signaling, additional cellular behaviors, such as mediators of proliferation, cell death or intercellular communication (i.e. cytokines, growth factors, etc.), could be similarly assessed.
Another type of question that a mass cytometry experiment can address is to define a cellular progression, such as a differentiation trajectory from a primitive to a mature cell type. A recent example examined the path of B cell lymphopoiesis in human bone marrow (Bendall et al., 2014). This study provided a generalizable algorithm for discerning the most likely path from a defined starting point (such as a stem cell) to an ending point (such as a mature cell) in single cell data called Wanderlust. The method, which defines the cellular progression as an average of k-nearest neighbor graphs through the data, enables identifying cellular events that take place over the course of this cellular trajectory. For example, cell-signaling states, proliferative bursts and coordinated points of cell death can be identified along the cellular differentiation pathway. One current limitation is that the algorithm makes the assumption of a linear differentiation scheme (i.e., no branch points are permitted), providing opportunities for future developments.
Rather than modeling a differentiation scheme, another question to which mass cytometry data can provide an answer is that of cellular diversity. Several studies have investigated variability within a particular cell population in order to understand the suite of possible fates a cell can adopt. Adopting tools from ecology, the diversity of human natural killer cells has been quantified using the Inverse Simpson Index (Horowitz et al., 2013). Intriguingly, this score of NK cell diversity serves as a prognostic metric of HIV acquisition (Strauss-Albee et al., 2015), demonstrating the utility of high-dimensional single-cell observations. Moreover, the application of visualization tools described above can also provide insight regarding the phenotypic variability of cell populations (Becher et al., 2014; Spitzer et al., 2015; Wong et al., 2015).
Transforming Data into Statistical Inference
While these methods aim to reveal biological mechanism from mass cytometry data, yet another approach identifies cellular features that correlate with a desired outcome, such as patient survival. The algorithm is termed Cluster Identification, Characterization & Regression (Citrus) (Bruggner et al., 2014) and combines hierarchical clustering of cell events with machine learning approaches to identify statistically significant features between groups of samples or to build a predictive model for a particular sample type (Bair and Tibshirani, 2004). The result is a robust method for identifying groups of cells with behavioral characteristics that correlate with an annotated feature of the samples of interest, such as time to recovery or overall survival. Recently applied to immune behavior during hip replacement surgery, the method identified cell signaling features predictive of patient recovery from trauma (Gaudilliere et al., 2014). Other approaches for building predictive models from fluorescence-based flow cytometry data (Aghaeepour et al., 2013) will likely find applications in the analysis of mass cytometry data as well. A final approach is the use of correlation clusters to map “modules” of interacting features across a complex immune landscape (Hotson et al., 2016). This approach can be used to discover novel interacting cellular components in a complex cellular system.
From Present to Future
While many methods for utilizing mass cytometry to investigate biological systems have been developed, the applications thereof are just at their beginning. Many important and unanswered questions in immunology could find their answers through the implementation of these approaches. One analytical strategy that we believe is particularly amenable to future development is the immune system reference framework that utilizes the Scaffold maps algorithm (Spitzer et al., 2015). These concepts are specifically designed to provide a foundation on which future studies can be layered for detailed comparison. By creating an analytical platform that is extensible, the reference framework will enable meta-analyses across multiple cytometry experiments, and ultimately single-cell data from other platforms, analogous to what has been achieved with transcriptomics data. In this way, the effects on the immune system of any perturbation or change in context can be discerned and compared to results from other studies for additional insight. Additionally, the concept should allow a data-driven encyclopedia of immunology to be collated in a format that facilitates analysis moving forward. While currently built on data from the mouse, the addition of human data as well as studies in other organisms will expand its utility. Studies to assess optimal strategies for comparing datasets are currently underway.
In the last few years, mass cytometry has transitioned from a promising emerging technology to a developed and accepted platform for high-dimensional single-cell analysis. It is now possible to discern cellular populations of interest while simultaneously interrogating many facets of cellular behavior from a single experiment. This potential expands the utility of cytometry for revealing mechanism at the level of the fundamental biological unit—the cell—with the throughput required to capture the emergent properties of integrated cellular systems. The development of new technologies has historically allowed researchers to ask questions from perspectives previously unattainable. Mass cytometry will likely do the same.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Matthew H. Spitzer, Email: matthew.spitzer@ucsf.edu.
Garry P. Nolan, Email: gnolan@stanford.edu.
References
- Aghaeepour N, Finak G, Hoos H, Mosmann TR, Brinkman R, Gottardo R, Scheuermann RH FlowCAP Consortium, DREAM Consortium. Critical assessment of automated flow cytometry data analysis techniques. Nature Methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir E-AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnology. 2013 doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:e108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J Immunol Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KWW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nature Immunology. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbehani GK, Thom C, Zunder ER, Finck R, Gaudilliere B, Fragiadakis GK, Fantl WJ, Nolan GP. Transient partial permeabilization with saponin enables cellular barcoding prior to surface marker staining. Cytometry A. 2014;85:1011–1019. doi: 10.1002/cyto.a.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Davis KL, Amir EAD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir EAD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:677–678. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Current Opinion in Immunology. 2013;25:484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature Biotechnology. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci USa. 2014;111:E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry A. 2008;73:701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TJ, Kotecha N. Cytobank: Providing an Analytics Platform for Community Cytometry Data Analysis and Collaboration. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 127–157. [DOI] [PubMed] [Google Scholar]
- Chester C, Maecker HT. Algorithmic Tools for Mining High-Dimensional Cytometry Data. J Immunol. 2015;195:773–779. doi: 10.4049/jimmunol.1500633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins KE, Ferrell PB, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Ries RE, Ilagan JO, Stirewalt DL, Meshinchi S, Bradley RK. Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci USa. 2014;111:16802–16807. doi: 10.1073/pnas.1413374111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eades PA. A heuristic for graph drawing. 1984:149–160. [Google Scholar]
- Edgar LJ, Vellanki RN, Halupa A, Hedley D, Wouters BG, Nitz M. Identification of Hypoxic Cells Using an Organotellurium Tag Compatible with Mass Cytometry. Angew Chem Int Ed. 2014 doi: 10.1002/anie.201405233. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei AP, Bava F-A, Zunder ER, Hsieh EWY, Chen S-Y, Nolan GP, Gherardini PF. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nature Methods. 2016 doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchterman TM, Reingold EM. Graph drawing by force-directed placement. Software: Practice and Experience. 1991;21:1129–1164. [Google Scholar]
- Gaudilliere B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, Silva J, Ganio EA, Yeh CG, Maloney WJ, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Science Translational Medicine. 2014;6:255ra131. doi: 10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. CellReports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Science Translational Medicine. 2013;5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotson AN, Gopinath S, Nicolau M, Khasanova A, Finck R, Monack D, Nolan GP. Coordinate actions of innate immune responses oppose those of the adaptive immune system during Salmonella infection of mice. Sci Signal. 2016;9:ra4–ra4. doi: 10.1126/scisignal.aaa9303. [DOI] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nature Methods. 2013;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, Pe’er D, Nolan GP. Conditional density-based analysis of T cell signaling in single-cell data. Science. 2014;346:1250689–1250689. doi: 10.1126/science.1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015:1–15. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei HE, Leipold MD, Maecker HT. Platinum-conjugated antibodies for application in mass cytometry. Cytometry. 2015 doi: 10.1002/cyto.a.22778. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Cruse R, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, Boehmer von H, Mathis D, et al. The transcriptional landscape of αβ T cell differentiation. Nature Immunology. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M, Krishnaswamy S, Spitzer MH, Bendall SC, Stone EL, Hedrick SM, Pe’er D, Mathis D, Nolan GP, Benoist C. Single-cell mass cytometry of TCR signaling: Amplification of small initial differences results in low ERK activation in NOD mice. Proceedings of the National Academy of Sciences. 2014;111:16466–16471. doi: 10.1073/pnas.1419337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nature Biotechnology. 2013 doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky OI, Lou X, Nitz M, Sheldrick WS, Baranov VI, Bandura DR, Tanner SD. Study of Cell Antigens and Intracellular DNA by Identification of Element-Containing Labels and Metallointercalators Using Inductively Coupled Plasma Mass Spectrometry. Anal Chem. 2008;80:2539–2547. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USa. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne S, Hu X, Wang K, Rossin E, Lin T-I, Maier LM, Baecher-Allan C, McLachlan GJ, Tamayo P, Hafler DA, et al. Automated high-dimensional flow cytometric data analysis. Proc Natl Acad Sci USa. 2009;106:8519–8524. doi: 10.1073/pnas.0903028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nature Biotechnology. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc Natl Acad Sci USa. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, Finck R, Carmi Y, Zunder ER, Fantl WJ, et al. An interactive reference framework for modeling a dynamic immune system. Science. 2015;349:1259425–1259425. doi: 10.1126/science.1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Science Translational Medicine. 2015;7:297ra115–297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, et al. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nature Methods. 2015:1–10. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricot S, Meyrand M, Sammicheli C, Elhmouzi-Younes J, Corneau A, Bertholet S, Malissen M, Le Grand R, Nuti S, Luche H, et al. Evaluating the efficiency of isotope transmission for improved panel design and a comparison of the detection sensitivities of mass cytometer instruments. Cytometry. 2015;87:357–368. doi: 10.1002/cyto.a.22648. [DOI] [PubMed] [Google Scholar]
- Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HTK, De Lafaille MAC, Poidinger M, Newell EW. Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. CellReports. 2015:1–28. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir EAD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015a;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A Continuous Molecular Roadmap to iPSC Reprogramming through Progression Analysis of Single-Cell Mass Cytometry. Cell Stem Cell. 2015b;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]