Summary
The twisting of DNA due to the movement of RNA polymerases is the basis of numerous classic experiments in molecular biology. Recent mouse genetic models indicate that chromosomal breakage is common at sites of transcriptional turbulence. Two key studies on this point mapped breakpoints to sites of either convergent or divergent transcription, but arrived at different conclusions as to which is more detrimental and why. The issue turns on whether DNA strand separation is the basis for the chromosomal instability or collision of RNA polymerases?
Introduction
The association between transcription, DNA point mutations, and DNA recombination has been known for some time (Jinks-Robertson and Bhagwat, 2014; Thomas and Rothstein, 1989; White et al., 1992; Wierdl et al., 1996). Also, it was apparent early-on that there exists a clear link between transcription and topological tension (Sternglanz et al., 1981). In fact, certain regions of the eukaryotic genome require active topoisomerases for transcription as well as replication (Brill et al., 1987; Christman et al., 1988). Currently, RNA-seq technology has allowed for large-scale examination of transcription, with results forming the theory that a large portion of the genome is undergoing extensive transcription, the majority of it non-coding (Berretta and Morillon, 2009; Consortium, 2012). As such, the cause and consequence of transcription-induced genome rearrangements and the role of topological tension is worth revisiting.
Two recent papers have shown that closely-spaced transcriptional promoters can result in DNA double-strand breaks (DBSs) that lead to translocations or other large scale structural changes in mammalian B cells (Meng et al., 2014; Pefanis et al., 2014). The close proximity of the promoters described in these papers may be increasing the torsional stress in a localized area of the genome, which could be a causal factor in generating DNA DSBs. This observation can be understood in terms of the twin domain model of superhelicity (Liu and Wang, 1987) where two nearby RNA polymerase II (RNAP2) complexes moving in opposite directions near each other can create either positive or negative supercoiling depending on if they are moving toward (convergent) or away (divergent) from each other, respectively.
Here, we review recent work concerning how promoter orientation can result in genome rearrangements and consider the results in the broader historical context of the relationship between transcription and torsional stress. We also consider specific cases of how this is relevant to lymphoid and other malignancies and anticipate the next experimental steps necessary to gain further insights into this phenomenon.
Divergent Transcription as a Source of DNA Damage
A surprising observation upon examination of annotated maps of the human genome was the number of closely-spaced genes that had promoters arranged in a divergent orientation, meaning the genes are arranged so that transcription complexes move in opposite directions upon expression. For example, out of 120 DNA repair genes examined, 50 (42%) were expressed divergently with transcription start sites (TSS) less than 1 kb apart (Adachi and Lieber, 2002). Interestingly, among those 50, 40 had TSS that were less than 300 bp apart, illustrating that, while the human genome is not as compact as lower eukaryotes such as S. cerevisiae, some genes are located quite near each other, and much closer than previously was anticipated.
A full examination of all the genes on gene-rich chromosome 22 and gene-sparse chromosome 21 was conducted to determine if divergent promoters were a common feature of human genes and not necessarily limited to genes within specific functional groups. In fact, 18% (56/319) of the genes on chromosome 22 and 22% (31/144) of the genes on chromosome 21 are arranged divergently with less than 1 kb between them. A smaller fraction of gene pairs within a zone on chromosome 22 are arranged either convergently or in tandem, suggesting a bias for divergent genes to be found close together. These results were confirmed by work from another group that examined the number of divergent promoters within 1 kb of each other in the entire genome and found the number to be ~11% with a bimodal distribution (Trinklein et al., 2004), suggesting that divergent promoters make up a class of genes.
One possible explanation of the higher-than-expected percentage of divergent promoters is that genes arranged divergently are co-regulated. Indeed, CpG islands are a common feature between two divergent promoters (Adachi and Lieber, 2002; Trinklein et al., 2004). Trinklein et al. found that the vast majority of divergent human promoters tested for function by a luciferase assay fired concurrently, with some variations among different cell lines.
Since a substantial percentage of human genes appear to be expressed divergently, we were curious if this arrangement posed a risk. Converging or diverging RNAP2 complexes can each distort the structure of DNA (Figure 1), and this distortion may result in a DSB either due to physical strain or due to nucleases that cleave at specific structures. This would be difficult to directly test in human cells as it would require both an outcome following a break between two promoters that is selectable in a genetic assay and a way to control the firing of the two promoters. Testing our hypothesis in the simple haploid model organism, S. cerevisiae, was a first logical step as both of the aforementioned parameters are easily met.
Figure 1. Divergent and convergent transcription increase supercoiling of opposing signs.
(A) Two promoters are arranged divergently (purple arrows). An RNAP2 complex (light and dark green objects) is recruited to each promoter and begins transcribing. The movement of the two complexes away from each other causes negative supercoiling (underwinding) between the two polymerases, represented by the blue-colored helix with “−” signs. This underwinding generates some degree of single-strandedness in the DNA between the two promoters. (B) Two promoters are arranged convergently. Upon transcription, the movement of two RNAP2 complexes towards each other causes positive supercoiling (overwinding) between the two polymerases, represented by the red-colored helix with “+” signs.
The degree to which convergent or divergent promoters increased instability was measured by a gross chromosomal rearrangement (GCR) assay. GCRs are typically the result of a DSB causing the loss of two genetic markers from the non-essential, telomere proximal arm of a chromosome (Chen and Kolodner, 1999), thus the outcome does not rely on formation of a specific repair product. We found that two, strong, closely-spaced promoters arranged divergently did, in fact, result in an increase in the GCR rate (Pannunzio and Lieber, 2016). Interestingly, we found that the collision of sense and antisense RNAP2 complexes emanating from convergent promoters did not increase GCRs as measured in the assay. While single subunit bacteriophage RNA polymerases are small enough to bypass each other when transcribing convergently (Ma and McAllister, 2009), both in vitro and in vivo assays suggest that when two eukaryotic RNAP2 complexes collide, each pauses until one or both are removed (Garcia-Rubio and Aguilera, 2012; Hobson et al., 2012). Therefore, physical strain or stalling of two converging complexes does not seem to present a danger for forming a GCR in wild-type cells; in contrast, the DNA structure left in the wake of two RNAP2 complexes traveling on the DNA in opposite directions does (Figure 1A), suggesting that the underwound DNA topology may be a key potentiator of DNA DSBs.
DNA Topology Discoveries in Bacteria
Much of the work in the eukaryotic studies can by understood in the light of classic work performed in E. coli where links between transcription and topological tension were first described (Sternglanz et al., 1981). In 1987, Liu and Wang postulated their twin domain model of supercoiling to interpret several pieces of data that where being presented at the time concerning the topological state of the pBR322 plasmid (Liu and Wang, 1987). In one study, inhibition of E. coli DNA gyrase, which relaxes positive supercoiling by introducing negative supercoils, resulted in a positively supercoiled plasmid (Lockshon and Morris, 1983). In other work, by Pruss and Drlica, mutation of the E. coli topoisomerase I gene, topA, which removes negative, but not positive supercoils, yields a plasmid with a higher density of negative supercoils (Pruss and Drlica, 1986). Since the activities of topA and DNA gyrase act to counterbalance each other, these results would seem to make sense; but it was the degree of supercoiling measured in each case that was puzzling, as it was higher than expected from loss of each topoisomerase alone (Liu and Wang, 1987).
Pruss and Drlica also made several variants of pBR322 that removed portions of the tet gene, conferring tetracycline resistance. Interestingly, in several of these variants the high level of negative supercoiling was reduced. Therefore, it was not just the absence of topoisomerase I or DNA gyrase activity that was causing increased supercoiling, but cis features on the plasmid were actively producing both positive and negative supercoils simultaneously. When the cell is deficient in one of the balancing topoisomerase activities, either positive or negative supercoils accumulate. Importantly, the deleted regions of the tet gene that led to a decrease in supercoiling were the promoter and upstream portions of the gene, but not downstream segments, meaning that by preventing transcription of the tet gene, only normal levels of supercoiling were measured, implicating transcription as a major source of both positive and negative supercoiling. But how was transcription exerting this effect?
In 1987, Liu and Wang put forth the twin domain model of supercoiling to clarify these results. The premise was that, as RNA polymerase moves along the DNA during transcription, the ability of the large complex to rotate around the helix would be limited, therefore, the DNA must turn around its helical axis to accommodate the movement. If the DNA were anchored in place at the ends, there would be no way to relieve this turning of the DNA. Therefore, without a topoisomerase to change the linking number, positive supercoiling occurs in front of the traveling polymerase and negative supercoiling occurs behind, accounting for the results observed upon loss of either topoisomerase I or DNA gyrase.
On a plasmid though, supercoiling from a single transcription unit should be dissipated across the plasmid as the polymerase moves, so why do the supercoils accumulate? The twin domain model also describes the effect of having two RNA polymerases moving away from each other from divergent promoters. In this case, the DNA could not turn to accommodate the movement, and negative supercoiling would accrue behind the two complexes and positive supercoiling ahead of each, creating two zones on the plasmid with opposing superhelical tension. Indeed, in pBR322 the tet gene is one of a pair of genes, the other being amp, that is transcribed divergently.
In further confirmation of the twin domain model, it was later shown that transcription-induced supercoiling occurs even in Top+ E. coli cells. The formation of structure-specific cruciform structures can be used as in indicator of negative supercoiling as they do not form in linear or relaxed DNA (Sinden, 1994). The results of Krasilnikov, et al (Krasilnikov et al., 1999) show that negative supercoiling can occur nearly a kilobase away from a TSS. Moreover, the intensity of cruciform formation is elevated when the structure forming sequence is positioned between two divergent promoters. Therefore, negative supercoiling between two divergent promoters occurs even with functional topoisomerase activity.
Support for the Twin Domain Model in Eukaryotes: Yeast and Mammals
A link between transcription and supercoiling was also explored and confirmed in eukaryotes. An important note, though, is that the complement of topoisomerases involved in transcription in eukaryotes, both yeast and mammals, differ from that in bacteria. Instead of having one topoisomerase to deal with positive supercoiling and another for negative supercoiling, the two major topoisomerases in eukaryotes involved in transcription, topoisomerase I, encoded by the TOP1 gene and topoisomerase II, encoded by the TOP2 gene, can relieve both positive and negative supercoiling (Table 1). The topA homolog in eukaryotes, TOP3, appears to be more involved in DNA recombination. Details for each of the different topoisomerases is reviewed elsewhere (Nitiss, 1998; Wang, 2002).
Table 1.
Prokaryotic and Eukaryotic Topoisomerases
| Protein | Gene | Type | Method of Action | |
|---|---|---|---|---|
| Prokaryote | TopI | topA | IA | single-strand break, removes negative supercoils |
| Gyrase | gyrA, gyrB | II | double-strand break, removes positive supercoils | |
| Eukaryote | Top1 | TOP1 | IB | single-strand break, removes positive and negative supercoils |
| Top2 | TOP2 | II | double-strand break, removes positive and negative supercoils | |
| Top3 | TOP3 | IA | single-strand break, removes single-stranded negative supercoils |
Similar to the work in bacteria, plasmids from yeast were shown to have increased supercoiling when isolated from top1 single and top1 top2 double mutants (Brill and Sternglanz, 1988). Further evidence of the generation of both positive and negative supercoiling was reported by expressing the bacterial topA gene in a yeast top1 top2 double mutant (Giaever and Wang, 1988). The plasmid or 2μm DNA extracted from these cells was positively supercoiled. As the gene product of topA only introduces positive supercoils in regions of negative supercoiling, the DNA in a top1 top2 double mutant must be excessively negatively supercoiled. Incidentally, this also means that the topA homolog in yeast, Top3, which, at the time, had not been discovered, must not be very active on negatively supercoiled DNA. In both works, the increase is positive and negative supercoiling is dependent on transcription. Therefore, in eukaryotes, as in bacteria, transcription is the key driver of excessive supercoiling. Interestingly, not only are cells with a complete deletion of the TOP1 gene viable, but also there is no measurable effect on mRNA synthesis, indicating that the RNAP2 complexes can efficiently progress though most topological barriers (Thrash et al., 1985; Wang et al., 1998), though perhaps not completely free of consequence.
Is this increased supercoiling measurable on chromosomal DNA, though? Experiments to determine the extent of supercoiling in vivo on eukaryotic chromosomes were initiated as far back as 1980 when DNA was probed for negative supercoils with trimethylpsoralen (Sinden et al., 1980). In bacteria, introduction of nicks produced a measurable relaxation of the normally supercoiled state of the DNA. This was not the case in eukaryotic cells, however, as the DNA remained in its supercoiled state after excessive nicking, the hypothesis being that in eukaryotes, with the DNA wrapped around nucleosomes, the nucleosomes may be able to maintain the in vivo superhelical state.
It is possible, however, that localized changes in superhelical tension occur, especially near active transcriptional promoters, and these transient changes could be detected in a more sensitive assay. More recent experiments suggest this is the case as increased negative supercoiling between two divergent promoters was measured on a fragment of DNA captured from a chromatinized episome in human cells with functional topoisomerases (Kouzine et al., 2008). Thus, the twin domain model appears to be equally applicable in lower and higher eukaryotes.
Genetic Instability Through the Lens of the Twin Domain Model
The decades of work on the subject make the link between transcription and torsional stress clear. A key point is that the region between two divergent promoters is subjected to increased negative superhelical tension. That underwound DNA was detected in human cells with functional topoisomerases would explain the increase in GCRs with divergent promoters in wild-type cells in our assay. Negative supercoiling would occur between the promoters and possibly form a cleavable non-B-form DNA structure even with active Top1 and Top2 enzymes present. Convergent promoters have no effect in the assay and is likely explained by evidence that two RNAP2 complexes that collide head-on will stall until they are removed (Hobson et al., 2012). While this likely affects transcription of the genes in the vicinity of the stall site (Garcia-Rubio and Aguilera, 2012), recombination is not detected in wild-type cells to a significant degree.
More evidence implicating DNA topology came from performing the GCR assay in cells defective for either Top1, or Top2 (Pannunzio and Lieber, 2016). As stated previously, both topoisomerases relax both positive and negative supercoiling, but by different mechanisms. Top1 creates a transient nick on one strand of the DNA double helix. Top2 relieves torsional stress by creating a DSB in an ATP-dependent process, allowing an intact DNA double helix to pass through the gap created (Table 1). In S. cerevisiae, the TOP1 gene can be completely deleted, however TOP2 is essential, therefore temperature-sensitive mutant alleles of TOP2 must be utilized (Nitiss et al., 1993).
In our survey of the necessity of these two proteins in preventing damage, we found that loss of Top1 had no significant effect on the breakage rate at either convergent or divergent promoters. This was surprising given the known role of Top1 in transcription (Choder, 1991; Di Mauro et al., 1993; Fleischmann et al., 1984; Kroeger and Rowe, 1992; Nitiss, 1998; Yadav et al., 2014) and suggests that in the context of our particular genetic assay, perhaps Top2 is able to compensate for loss of Top1. The largest increase in breakage rate was measured in cells bearing the top2 mutant; but, in contrast to wild-type cells, it was the convergent promoters that produced the largest effect, not the divergent promoters.
These results provide two important clues. First, the topological tension conferred by convergent transcription can greatly increase DNA instability and requires Top2 for resolution. Second, while Top2 may be able to offset the loss of Top1 in terms of preventing rearrangements, the reverse does not seem to be true, considering the enormous increase in GCRs in a top2 mutant. A partial explanation for this is provided by Kouzine et al. who show that both Top1 and Top2 are deployed at active promoters, but that Top2 appears to be more critical at promoters undergoing high levels of expression (Kouzine et al., 2013). This may explain the results we obtain using the GAL1 and GAL2 promoters, which are known to be robust (Johnston and Davis, 1984; Kuras et al., 2003; Pannunzio and Lieber, 2016).
Importantly, since convergent transcription only increased the GCR rate in a top2 mutant, the cause of the DSB is likely not the physical collision of two converging RNAP2 complexes -- as this would also occur in wild-type cells -- but rather due to changes in DNA structure that increase the likelihood of a break. In the case of a defective Top2 enzyme, we speculate that as positive supercoiling builds up to overwind the DNA, a nuclease likely recognizes the abnormal structure and cleaves, generating a DSB. This removes the topological tension, but without the bridging function of Top2 to hold the broken ends in place, the non-essential chromosome arm is lost. In further support of this is that the top2 mutation causes reduced viability even at the semi-permissible temperature, likely due to breaks elsewhere in the genome that result in the loss of essential genes.
Non-B-DNA Structures as a Consequence of Topology and Transcription
Up until this point, we have only discussed changes in DNA topology in terms of the activity of two promoters that transcribe in either a convergent or divergent manner without regard to the actual sequences that RNAP2 is transcribing. Most likely, the sequence composition between relatively closely-spaced divergent or convergent promoters could exacerbate the effects of negative or positive supercoiling, respectively, to a substantial degree. While much more work testing such sequences between convergent or divergent promoters is required, the effects observed with transcription from a single promoter through these sequences are worth briefly noting.
One of the best examples of transcription-induced rearrangements are the physiologic breaks that occur during class switch recombination (CSR) at the mammalian immunoglobulin heavy chain (IgH) locus of B cells (Shinkura et al., 2003; Yu et al., 2003). Here, DSBs generated within switch regions change the class of antibody produced by the cell from IgM to IgG, IgA, or IgE. Switch regions are comprised of 25–80 bp degenerate repeats that span several kilobases and are enriched in guanine bases on the nontemplate strand (Dunnick et al., 1993; Kinoshita and Honjo, 2000; Stavnezer, 1996). DSBs are initiated within switch regions when transcription from a sterile promotor leads to the formation of kilobase-long R-loop structures where the G-rich RNA strand exiting the RNAP2 complex reanneals to the template DNA strand, leaving the nontemplate strand single-stranded (Roy and Lieber, 2009; Roy et al., 2010; Yu et al., 2003). Following this, several DNA repair factors act to break and religate the DNA to allow for expression of different constant regions to change the antibody class (Lieber, 2010). The presence of R-loops at switch regions has been rigorously confirmed and it is unlikely that they are of such length and abundance elsewhere in the genome due to the sequence requirements for R-loop formation (Zhang et al., 2015a; Zhang et al., 2014a; Zhang et al., 2015b; Zhang et al., 2014b).
As few as four repeat units of a switch region can be ectopically transcribed and form R-loops, indicating it is the sequence itself that is integral in forming and stabilizing the structure (Roy et al., 2008; Yu et al., 2003). Also, R-loops readily form in cloned switch sequences on a supercoiled plasmid due to the negative supercoiling favoring strand separation (Yu et al., 2003; Yu et al., 2005), reiterating the close relationship between transcription and DNA topology. In the experiments of Yadav et al (Yadav et al., 2014), a GCR assay was employed in yeast to examine the effects of transcription through a 760 bp fragment of the switch mu region (Sμ) from the mouse IgH locus containing approximately 17 repeat units. The region was inserted downstream of an inducible promoter. Similar to other studies (Ruiz et al., 2011), Sμ alone had no effect in wild-type cells, perhaps as yeast lack specific CSR factors. Interestingly, loss of TOP1 increased the GCR rate in an orientation-specific manner (Yadav et al., 2014). A separate study showed that a high rate of transcription also increased the rate of gene conversions initiated by Sμ sequences in a top1Δ background (Kim and Jinks-Robertson, 2011). This indicates that a defect in the ability to relieve supercoiling that accumulates as a consequence of transcription through unique DNA regions can result in rearrangements. A TOP2 defect was not tested, but it is possible that it would also show a stimulation in recombination. While it has been suggested that the negative supercoiling can result in the displaced G-rich DNA strand forming G-quadraplex (G4) structures, it is important to point out that G4 structures are not required for physiological R-loop formation as stable R-loops form in the presence of Cs+ and Li+ cations that destabilize G4 structures (Roy et al., 2008). Still, the experiments with Sμ provide an excellent example of transcribed sequences that are sensitive to topological tension.
In addition to R-loops, other sequence-specific distortions of the dsDNA helix from the typical B-form confirmation can also occur (Calladine et al., 2004; Mirkin, 2006; Sinden, 1994) and these sequences may be sensitive to transcription and changes in topological tension. For example, when dsDNA sequences that contain a short direct repeat become separated due to transcription, they can adopt a slipped mispaired structure as they reanneal that can lead to contractions or expansions of DNA regions (Sinden and Wells, 1992). Negative supercoiling may promote breathing of the dsDNA within these regions, increasing the probability of the slipped mispaired structure. Furthermore, inverted and mirror repeat sequences can form cruciform and triplex DNA, respectively, upon strand separation and both require the energy of supercoiling to stably form (Sinden, 1994). Sequences that generate these non-B-form structures could be scattered throughout the genome and, now that we are beginning to map regions of non-coding RNA (ncRNA) expression, could be relevant to genome instability considering how entwined transcription is to DNA topology.
Relevance of Convergent and Divergent Transcription in the Genetic Instability of Cancer
Several recent studies have demonstrated that instances of divergent transcription are not limited to the expression of two genes in close proximity, as promoters that generate ncRNA can occur outside of the TSS for genes and even within the gene body (Mayer et al., 2015; Meng et al., 2014; Pefanis et al., 2014). The ncRNAs that are expressed appear to differ between different cell-types and even during different stages of development of the same cell type. For example, a pre-B cell may not have the same ncRNA profile as a mature B cell. Therefore, divergent and even closely-overlapping convergent promoters can arise in a tissue and stage-specific manner due to the transcription of ncRNA, (Lu et al., 2005; Schotte et al., 2011). If the expression of ncRNA increases torsional stress to the point where DSBs are generated, this may have intriguing implications for why specific translocations are associated with some human cancers but not others.
Two recent studies in mouse B cells have implicated transcriptional dynamics in human lymphoid malignancies (Meng et al., 2014; Pefanis et al., 2014). Each group proposed a different hypothetical mechanism for the generation of DNA DSBs and disagreed on which is more critical, divergent or convergent transcription. Meng et al demonstrate that clusters of enhancers, termed super-enhancers, appear to be responsible for allowing rouge transcription to occur (Meng et al., 2014). When super-enhancers overlap with the body of a gene, antisense transcription with respect to the TSS of the gene, can generate zones of convergent transcription that the authors suggest will lead to stalling of RNAP2 and that stalled transcription can eventually recruit DNA modifying enzymes that result in DSBs. In Pefanis et al. divergent, and some convergent, transcription is revealed following disruption of the RNA exosome (Pefanis et al., 2014). The RNA exosome appears to regulate some ncRNA expression and degrades antisense transcripts associated with TSSs that can generate divergent transcription. They suggest that ssDNA is generated by negative supercoiling between divergently travelling promoters and this ssDNA could be used as substrate for enzymes that mutate or cut DNA (Bransteitter et al., 2003).
While our yeast data supports divergent transcription as a cause of DSBs, this must be reconciled with the fact that the role of convergent transcription seems important as well. Our lab has noted the specific example of the human BCL6 gene where the majority of breakpoints are clustered in a 2 kb zone within the 11 kb intron 1 (Lu et al., 2015). Analysis of this region revealed that a long non-coding RNA (lncRNA) expressed within intron 1 converges on the BCL6 promoter. Remarkably, >87% of the breakpoints occurred within this region of convergent transcription. While a super-enhancer is also reported to be in this region (Meng et al., 2014), the breakpoint cluster maps more precisely with the lncRNA than with the super-enhancer that is spread out over a threefold larger area (Lu et al., 2015).
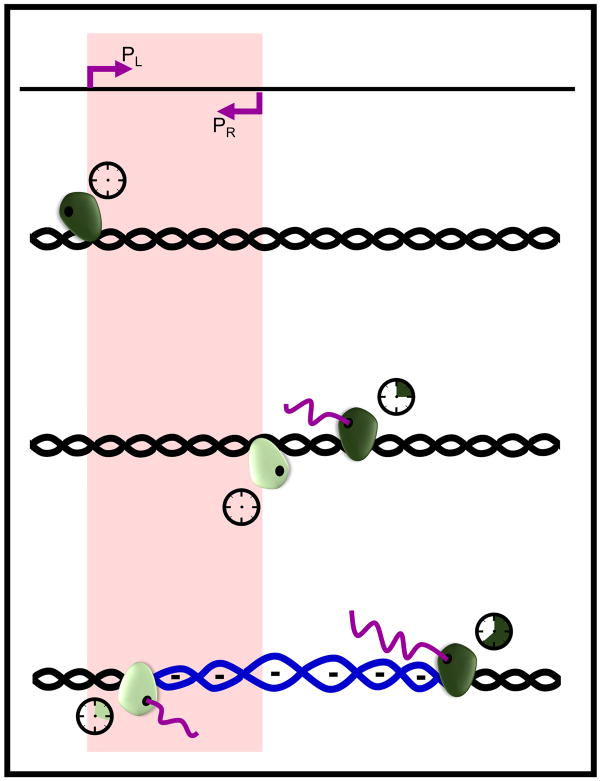
Divergent transcription still provides the most logical explanation for the formation of ssDNA that can act as a substrate for the DNA modifying enzymes since, as discussed above, the negative supercoiling that allows for its formation is known to occur in wild-type cells. Also, considering convergent transcription, it is unclear how close two promoters can be and still allow for simultaneous expression that would lead to RNAP2 collision. In vitro reconstitution assays have shown that two RNAP2 complexes can load onto a 150 bp substrate and transcribe towards each other until they collide (Hobson et al., 2012), but the size of the RNAP2 complex (Kornberg, 2007) would likely prevent simultaneous initiation of convergent transcription at shorter distances as two DNA strands cannot be transcribed at the same time. Consider, however, that even though two promoters can be arranged convergently, it is the actual movement of the RNAP2 that is important. Provided there is sufficient distance between the promoters, simultaneous firing of promoters may lead to a collision, (Figure 1b), but asynchronous firing would not. In the latter case, an RNAP2 complex would transcribe past the TSS of an opposing convergent promoter prior to the firing of that promoter. Once the second promoter fires and a separate RNAP2 begins transcribing, the end result is two RNAP2 complexes moving divergently even though each initiated from promoters arranged convergently (Figure 2). Therefore, it may be asynchronous firing of promoters that leads to translocations in cases such as BCL6 following increased negative supercoiling from two RNAP2 complexes moving apart.
Figure 2. Asynchronous firing of two promoters arranged convergently can result in divergent transcription.
Two promoters (PL and PR) are arranged convergently within a zone indicated by the red shading. An RNP2 complex (dark green object) is recruited to PL. In the middle panel, the dark green RNAP2 has had time to transcribe beyond PR (indicated by the clock traveling with the RNAP2 and the production of a purple RNA strand) when a second RNAP2 complex (light green) is recruited PR. In the bottom panel, the movement of the two RNAP2 complexes that began transcribing at different times has resulted in negative supercoiling (underwinding).
Concluding Remarks
The results of several recent studies linking transcription with DNA DSBs (Meng et al., 2014; Pannunzio and Lieber, 2016; Pefanis et al., 2014) fit with the twin domain model of supercoiling, first proposed in 1987. Divergent promoters on linear eukaryotic promoters are prone to more frequent breakage. The effect of topoisomerase mutants further confirms the link between transcription and topological tension. With the yeast system, there is potential to explore the combined effects of distance between promoters and promoter strength. Furthermore, there are a number of sequences that are known to break or that can adopt a unique non-B-form structures when over- or underwound and would be of interest to test (Sinden, 1994; Tsai et al., 2009; Tsai and Lieber, 2010).
These concepts also touch on a recent debate about the directionality of promoter sequences, namely if transcription moves unidirectionally or bidirectionally (Andersson et al., 2015; Duttke et al., 2015). It is important to note that the divergent transcription we describe in the yeast assay refers to the promoter regions of two genes, whereas the term is also used to describe transcription moving in both directions from a single promoter region. In the latter case, the RNAP2 complexes would still be moving divergently with the potential to underwind the DNA between. There is evidence that this occurs in a large number of human genes (Andersson et al., 2015). The magnitude of negative supercoiling would depend on the frequency of bidirectional promoter firing and the processivity of RNAP2, since the antisense transcript appears to be degraded in normal cells (Pefanis et al., 2014). Determining whether these events impact the generation of DSBs, would also be worth testing.
Finally, there is strong evidence that transcription may play a role in the formation of clinically relevant B cell translocations. Considering the relatively rare frequency of these harmful translocations and the defined stage in which they occur (Tsai et al., 2008), it would seem that a number of events would need to occur within a precise window of time in order to generate a break. New technology allowing us to better track ongoing transcription within different cell types and advances in whole-genome sequencing to detect translocations gives us greater insight into when and where these breaks may form (Meng et al., 2014; Pefanis et al., 2014). In turn, we can simulate various conditions in functional assays, like our yeast genetic system, to quantify the risk of DNA breakage due to the twisting of the DNA helix undergoing torsional stress to better understand how breaks are forming.
Acknowledgments
We thank Dr. Ray Mosteller for comments on the manuscript. Work was supported by NIH grants to M.R.L. Additional support for N.R.P. was provided by a generous memorial endowment established by Barbara Knight through the ARCS Foundation, Inc., Los Angeles founder chapter, John H. Richardson Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Andersson R, Chen Y, Core L, Lis JT, Sandelin A, Jensen TH. Human Gene Promoters Are Intrinsically Bidirectional. Mol Cell. 2015;60:346–347. doi: 10.1016/j.molcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Brill SJ, Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Calladine CR, Drew HR, Luisi BF, Travers AA. Understanding DNA: The Molecule and How it Works. 3. San Diego: Elsevier Academic Press; 2004. [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mauro E, Camilloni G, Verdone L, Caserta M. DNA topoisomerase I controls the kinetics of promoter activation and DNA topology in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:6702–6710. doi: 10.1128/mcb.13.11.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick WA, Hertz GZ, Scappino L, Gritzmacher C. DNA sequence at immunoglobulin switch region recombination sites. Nucl Acid Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Human promoters are intrinsically directional. Mol Cell. 2015;57:674–684. doi: 10.1016/j.molcel.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann G, Pflugfelder G, Steiner EK, Javaherian K, Howard GC, Wang JC, Elgin SC. Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci U S A. 1984;81:6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio ML, Aguilera A. Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes. Nucleic Acids Res. 2012;40:1050–1064. doi: 10.1093/nar/gkr840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, Bhagwat AS. Transcription-associated mutagenesis. Annu Rev Genet. 2014;48:341–359. doi: 10.1146/annurev-genet-120213-092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst) 2011;10:953–960. doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Honjo T. Unique and unprecedented recombination mechanisms in class switching. Curr Opin Immunol. 2000;12:195–198. doi: 10.1016/s0952-7915(99)00072-2. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- Kroeger PE, Rowe TC. Analysis of topoisomerase I and II cleavage sites on the Drosophila actin and Hsp70 heat shock genes. Biochemistry. 1992;31:2492–2501. doi: 10.1021/bi00124a008. [DOI] [PubMed] [Google Scholar]
- Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D, Morris DR. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983;11:2999–3017. doi: 10.1093/nar/11.10.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu Z, Pannunzio NR, Greisman HA, Casero D, Parekh C, Lieber MR. Convergent BCL6 and lncRNA promoters demarcate the major breakpoint region for BCL6 translocations. Blood. 2015 doi: 10.1182/blood-2015-07-657999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, McAllister WT. In a head-on collision, two RNA polymerases approaching one another on the same DNA may pass by one another. J Mol Biol. 2009;391:808–812. doi: 10.1016/j.jmb.2009.06.060. [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, et al. Convergent Transcription at Intragenic Super-Enhancers Targets AID-Initiated Genomic Instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim Biophys Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- Nitiss JL, Liu YX, Hsiung Y. A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: a genetic system for determining the targets of topoisomerase II inhibitors. Cancer Res. 1993;53:89–93. [PubMed] [Google Scholar]
- Pannunzio NR, Lieber MR. Dissecting the Roles of Divergent and Convergent Transcription in Chromosome Instability. Cell Rep. 2016 doi: 10.1016/j.celrep.2015.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss GJ, Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986;83:8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition Between the RNA Transcript and the Nontemplate DNA Strand During R-Loop Formation In Vitro: A Nick Can Serve as a Strong R-loop Initiation Site. Mol Cell Biol. 2010;30:146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A. AID induces double-strand breaks at immunoglobulin switch regions and c-MYC causing chromosomal translocations in yeast THO mutants. PLoS Genet. 2011;7:e1002009. doi: 10.1371/journal.pgen.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389–1399. doi: 10.1038/leu.2011.105. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. The influence of transcriptional orientation on endogenous switch region function. Nature Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- Sinden RR. DNA Structure and Function. San Diego: Academic Press; 1994. [Google Scholar]
- Sinden RR, Carlson JO, Pettijohn DE. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Sinden RR, Wells RD. DNA structure, mutations and human genetic diseases. Curr Opin Biotech. 1992;3:612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- Stavnezer J. Antibody Class Switching. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- Sternglanz R, DiNardo S, Voelkel KA, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang JC. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thrash C, Bankier AT, Barrell BG, Sternglanz R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc Natl Acad Sci U S A. 1985;82:4374–4378. doi: 10.1073/pnas.82.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N, Aldred SF, Hartman S, Schroeder D, Otillar R, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Engelhart AE, Hatmal MM, Houston SI, Hud NV, Haworth IS, Lieber MR. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem. 2009;284:7157–7164. doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Lieber MR. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics. 2010;11(Suppl 1):S1. doi: 10.1186/1471-2164-11-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- White MA, Detloff P, Strand M, Petes TD. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P, Harcy V, Argueso JL, Dominska M, Jinks-Robertson S, Kim N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 2014;10:e1004839. doi: 10.1371/journal.pgen.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Fine-structure analysis of activation-induced deaminase accessbility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Hsieh CL, Okitsu CY, Han L, Yu K, Lieber MR. Effect of CpG dinucleotides within IgH switch region repeats on immunoglobulin class switch recombination. Mol Immunol. 2015a;66:284–289. doi: 10.1016/j.molimm.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Han L, Hsieh CL, Yu K, Lieber MR. The Strength of an Ig Switch Region Is Determined by Its Ability to Drive R Loop Formation and Its Number of WGCW Sites. Cell Rep. 2014a;8:557–569. doi: 10.1016/j.celrep.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh C-L, Yu K, Lieber MR. Complexities Due to Single-Stranded RNA During Antibody Detection of Genomic RNA:DNA Hybrids. BMC research notes. 2015b doi: 10.1186/s13104-015-1092-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K, Lieber MR. The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res. 2014b;42:13186–13193. doi: 10.1093/nar/gku1100. [DOI] [PMC free article] [PubMed] [Google Scholar]