SUMMARY
Inappropriate activation of the receptor tyrosine kinase EGFR contributes to a variety of human malignancies. Here we show a mechanism to induce vulnerability to an existing first line treatment for EGFR driven cancers. We find that inhibiting the palmitoyltransferase DHHC20 creates a dependence on EGFR signaling for cancer cell survival. The loss of palmitoylation increases sustained EGFR signal activation and sensitizes cells to EGFR tyrosine kinase inhibition. Our work shows that the reversible modification of EGFR with palmitate “pins” the unstructured C-terminal tail to the plasma membrane; impeding EGFR activation. We identify by mass spectrometry palmitoylated cysteine residues within the C-terminal tail where mutation of the cysteine residues to alanine is sufficient to activate EGFR signaling promoting cell migration and transformation. Our results reveal that the targeting of a peripheral modulator of EGFR signaling, DHHC20, causes a loss of signal regulation and susceptibility to EGFR inhibitor induced-cell death.
INTRODUCTION
Receptor tyrosine kinases (RTK) are widely deregulated in cancer and increased RTK signaling contributes to a variety of human malignancies. Members of the ErbB family play critical roles in responding to extracellular cues and initiating downstream signaling cascades through effector pathways (Lemmon et al., 2014; Roskoski, 2014; Walton et al., 1990). The epidermal growth factor receptor, EGFR, is one of four members of the ErbB family and is known to facilitate tumorigenesis and cancer progression. EGFR is structurally comprised of an extracellular ligand binding domain, a transmembrane region, a tyrosine kinase domain, and an unstructured C-terminal tail that harbors receptor auto-phosphorylation sites (Seshacharyulu et al., 2012). Ligands including the epidermal growth factor, EGF, bind to the extracellular domain of EGFR causing a conformational change that facilitates homo- and heterodimerization with members of the ErbB family (Seshacharyulu et al., 2012). Dimerization induces activation of the tyrosine kinase activity of the receptor leading to auto-phosphorylation of C-terminal tyrosines. The phosphorylated tyrosine residues serve as docking sites for adaptor proteins that link the receptor to downstream signaling pathways including Ras-Raf-MEK-ERK, PI3K-AKT, Src and JAK-STAT culminating in the regulation of cell migration, proliferation and survival (Seshacharyulu et al., 2012). Spatial and temporal control of EGFR signaling is mediated by receptor endocytosis. In response to EGF, EGFR is trafficked through early and late endosomes in route to lysosomes for signal termination and receptor degradation.
Dysregulation and inappropriate activation of EGFR is a common event in cancer and increased expression and mutations in EGFR that enhance signaling and resistance to therapy have been identified in breast and lung cancer (Roskoski, 2014). While EGFR mutations commonly reside within the extracellular (EGFR-vIII) and kinase domains (L858R, T790M), recent studies identified EGFR mutations within the C-terminal tail (Pines et al., 2010). Deletion of EGFR exons 25-27 was found in lung cancer and glioblastoma multiforme (Cho et al., 2011; Ekstrand et al., 1992; Imielinski et al., 2012). Ectopic expression of EGFR lacking exons 25 -26 promotes cell transformation and increases EGFR and AKT activation (Imielinski et al., 2012); however the mechanisms involved remain unknown.
Palmitoylation is the reversible modification of cysteine residues with a 16-carbon fatty acid which regulates protein localization, trafficking, stability and protein-protein interactions (Aicart-Ramos et al., 2011). Palmitoylation is regulated by two classes of enzymes, the DHHC domain containing protein acyl-transferases (PAT) which mediate the addition of palmitate to target substrates, and the acyl-protein thioesterases (APT) which remove palmitate (Conibear and Davis, 2010). Twenty three PATs have been identified in mammals and alterations in the expression and function of several PATs have been observed in cancer (Conibear and Davis, 2010; Greaves and Chamberlain, 2011; McCormick et al., 2008).
The role of palmitoylation in cancer has mostly focused on the palmitoylation of H-Ras and N-Ras which facilitates Ras localization to the plasma membrane and is required for activity (Swarthout et al., 2005). Analysis of The Cancer Genome Atlas (TCGA) reveals that alterations in DHHC20 expression including deletions, amplifications and mutations occur in cancers of the breast, lung and prostate. We found that the C-terminal tail of EGFR is palmitoylated by DHHC20. Inhibiting DHHC20 increases EGFR activation and increases the dependency on EGFR signaling for cell survival. We identify cysteine residues 1025, 1034 and 1122 as palmitoylation sites within the C-terminal tail. Mutation of 1025 or 1122 to alanine attenuates EGFR palmitoylation, activates EGFR signaling and increases cell migration and anchorage independent growth. Finally, our results reveal a mechanism for EGFR activation caused by mutations in the C-terminal tail of EGFR previously identified in lung cancer.
RESULTS
Silencing DHHC20 increases EGFR mediated cell responses
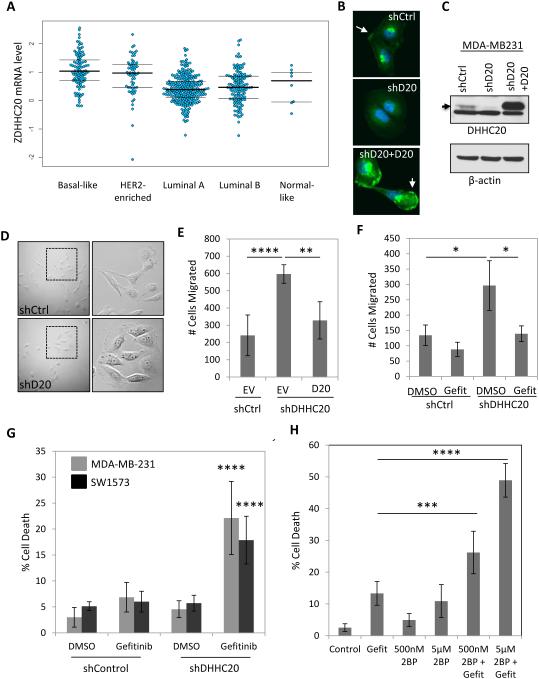
The palmitoyltransferase DHHC20 is expressed in multiple human breast and lung cancer cell lines and was found to suppress metastatic behavior in melanoma cells (Figure S1) (Wang et al., 2015). Analysis of the TCGA database revealed DHHC20 mRNA is elevated in basal and HER2-enriched breast carcinoma compared to luminal A and B tumors (Figure 1A). In the triple negative breast adenocarcinoma cell line MDA-MB-231 DHHC20 is localized to the plasma membrane and punctate structures adjacent to the nucleus when observed by immunofluorescence (IF) microscopy (Figure 1B). The levels of DHHC20 staining and the abundance of a specific 32kDa band on an SDS PAGE gel were decreased by DHHC20 shRNA (Figure 1 B-C). Expression of a shRNA resistant isoform of DHHC20 showed a consistent staining pattern by IF and the same molecular weight band that was reduced by shRNA, confirming this is the correct size and localization of DHHC20 protein (Figure 1 B-C). Inhibition of DHHC20 induced a change in cell morphology from an elongated spindle shape to a more spread morphology with extensive membrane ruffling in MDA-MB-231 cells (Figure 1D). The change in morphology correlated with a 3-fold increase in chemotaxis towards media containing 10% FBS that was suppressed by stable expression of shRNA resistant DHHC20 (Figure 1E). MDA-MB-231 cells have been shown to express high levels of EGFR which signals to downstream pathways important in cell migration (Price et al., 1999). We therefore asked if EGFR inhibition was sufficient to block the increase in chemotaxis. Treatment with the EGFR inhibitor Gefitinib significantly reduced chemotaxis in cells expressing DHHC20 shRNA (shDHHC20) by approximately 50%, but had a minimal effect on the chemotaxis of control shRNA cells (Figure 1F).
Figure 1. Silencing DHHC20 increases EGFR-dependent cell migration and enhances Gefitinib-induced cytotoxicity.
(A) DHHC20 mRNA expression is altered in human breast cancer subtypes. Global patterns of ZDHHC20 expression were identified in human breast invasive carcinoma in the TCGA database; Basal-like (n=98), HER2-enriched (n=58), Luminal A (n=230), Luminal B (n=125) and Normal-like (n=8). (B-C) DHHC20 expression measured by immunoblotting and immunofluorescence microscopy is inhibited by shRNA in MDA-MB-231 cells. Expression of shRNA resistant DHHC20 produces a product with the same molecular weight and cellular localization as the endogenous protein. (B) Immunofluorescence staining of DHHC20 (green) and DAPI (blue) show expression of DHHC20 at the plasma membrane (arrow) and perinuclear region. (C) Immunoblotting with a DHHC20 specific antibody shows inhibition of the DHHC20 band (arrow). (D) Silencing DHHC20 in MDA-MB-231 cells induces cell spreading with increased membrane ruffling. (E) Knockdown of DHHC20 increases chemotaxis towards FBS and this is rescued by expression of shRNA resistant DHHC20 (mean +/−StDev). (F) Increased chemotaxis of shDHHC20 MDA-MB-231 cells is inhibited by treatment with Gefitinib (10μM) (mean +/−StDev). (G) Gefitinib treatment (10 μM) increases cytotoxicity in DHHC20 silenced MDA-MB-231 (gray bars) and SW1573 (black bars) cells (mean +/−StDev). (H) Inhibiting palmitoylation with 2BP increases Gefitinib induced cytotoxicity. MDA-MB-231 cells were treated with 10μM Gefitinib alone or in combination with 500nM or 5μM 2BP (mean +/−StDev).
The relatively short exposure to Gefitininb that inhibited cell migration did not affect cell viability in the MDA-MB-231 cells that are normally insensitive to Gefitinib (Figure S1B). We next asked if silencing DHHC20 increased the cytotoxic effects of Gefitinib at longer time points. Treating MDA-MB-231 cells with Gefitinib for 72 hours increased cell death in DHHC20 shRNA cells (22.1%) compared to control shRNA cells (6.9%) (Figure 1G). Similar results were observed using SW1573 lung adenocarcinoma cells (Figure 1G, Figure S1C). The small molecule 2-bromo-palmitate (2BP) inhibits palmitoyltransferases (Figure S2) (Jennings et al., 2009). We therefore asked if 2BP produced an effect on Gefitinib sensitivity similar to silencing DHHC20. Treatment of MDA-MB-231 cells with 500nM 2BP resulted in 4.9% cell death after 72 hours. When treated in combination with 10μM Gefitinib the percentage of cell death increased (26.2%) compared to Gefitinib alone (13.3%) (Figure 1H). Increasing the concentration of 2BP to 5μM in combination with Gefitinib elevated the percentage of dead cells (48.9%) compared to 5μM 2BP alone (10.9%) (Figure 1H).
Inhibition of DHHC20-mediated palmitoylation increases EGF-induced EGFR activation
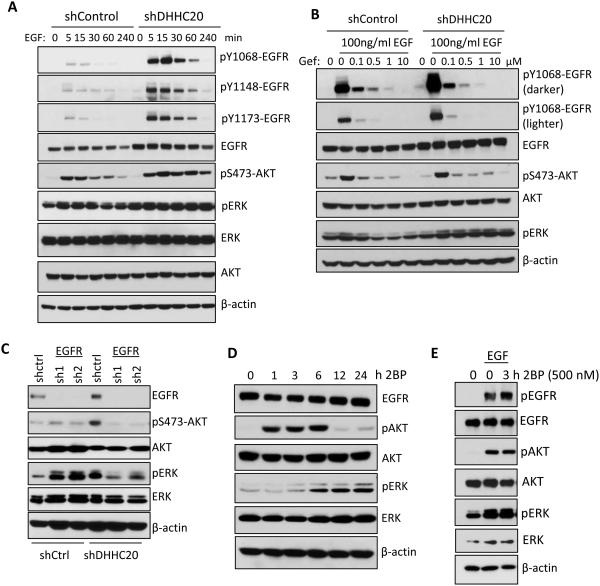
The increased sensitivity to Gefitinib shown by silencing DHHC20 suggests that decreased DHHC20 expression causes a dependence on EGFR signaling. Under resting conditions the activation state of EGFR in shDHHC20 cells measured by phosphorylation of EGFR (tyrosines1068, 1148 and 1173) and the downstream signaling component AKT (serine 473) was similar to control cells (Figure 2A). However, upon EGF stimulation silencing DHHC20 dramatically increased the amplitude and duration of EGFR activation (Figure 2A). Furthermore, the level of phosphorylated AKT in shDHHC20 cells was elevated and sustained through the four hour time course of EGF treatment compared to shControl cells (Figure 2A). In contrast to AKT phosphorylation the levels of phosphorylated ERK were higher in resting DHHC20 shRNA expressing cells relative to the shControl cells and did not increase further with EGF stimulation (Figure 2A). These results are rescued by expression of shRNA resistant DHHC20 (Figure S3A). Similar results were observed in SW1573 lung adenocarcinoma cells (Figure S3B).
Figure 2. DHHC20 knockdown increases EGFR activation and signaling.
(A) Silencing DHHC20 in MDA-MB-231 cells increases EGFR expression and EGF (100ng/ml) induced phosphoryation of EGFR and AKT. (B) Gefitinib inhibits activation of EGFR and AKT. MDA-MB-231 cells were serum starved in the presence of 10μM Gefitinib, treated with EGF (100ng/ml) for 15 minutes and activation of EGFR, AKT, and ERK was determined by SDS-PAGE. (C) The increased phosphorylation of ERK in DHHC20 silenced MDA-MB-231 cells is inhibited by EGFR shRNA. (D) MDA-MB-231 cells were treated with 500nM 2BP and the activation of AKT and ERK was analyzed by SDS-PAGE. (E) 2BP increases EGF-induced EGFR activation in MDA-MB-231 cells treated with 500nM 2BP for 3 hours and then stimulated with EGF for15 minutes.
We next asked if inhibition of DHHC20 increased the sensitivity to EGF stimulation compared to control cells. Cells were treated with varying concentrations of EGF and phosphorylation of EGFR and AKT was measured. While phosphorylated EGFR was detected in shControl cells at EGF concentrations as low as 10ng/ml, EGFR was activated in shDHHC20 cells at 5ng/ml of EGF (Figure S3C). Although the level of EGFR was elevated by 2-fold in the DHHC20 shRNA cells the ratio of phosphorylated EGFR/total EGFR was increased by nearly 6-fold indicating a higher level of receptor activation (Figure S3C). The elevated level of EGFR activation in shDHHC20 cells suggested higher doses of Gefitinib might be required to inhibit EGFR signaling. Although the shDHHC20 cells had a much higher amount of activated EGFR, treatment with 0.1μM Gefitinib reduced EGFR and AKT phosphorylation to the same level in both the control and the DHHC20 shRNA expressing cells (Figure 2B). The increase in cell death with Gefitinib treatment in shDHHC20 cells may be the result of the greater overall decrease in EGFR signaling. Unexpectedly, ERK phosphorylation was not inhibited by Gefitinib suggesting ERK is activated through an alternative signaling pathway. However, inhibition of EGFR expression by shRNA reduced ERK activation in DHHC20 silenced cells demonstrating it is dependent on EGFR (Figure 2C).
We next asked if acute inhibition of palmitoylation with 2BP increases EGFR activation and downstream signaling. After 1 hour AKT activation rapidly increased in cells treated with 500nM 2BP which returned to basal levels by 12 hours (Figure 2D). In contrast, ERK activation in response to 2BP was induced later, at 6 hours, and was maintained throughout the 24 hour time course (Figure 2D). In the presence of EGF, treatment with 2BP increased the phosphorylation of EGFR compared to vehicle control (Figure 2E).
Inhibition of DHHC20 expression disrupts EGFR endocytic trafficking
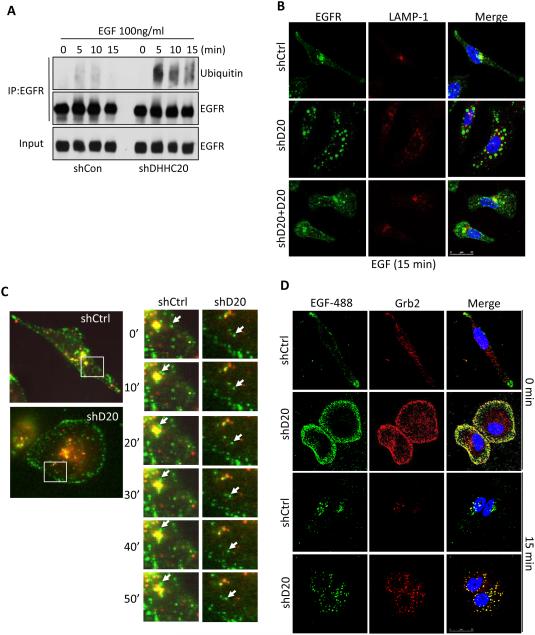
Ubiquitylation of EGFR serves as an endosomal trafficking signal to facilitate trafficking to the lysosome for degradation (Katzmann et al., 2002). Cells expressing shDHHC20 had higher levels of ubiquitylated EGFR compared to shControl cells (Figure 3A). Immunofluorescence staining was used to examine the effect of inhibiting DHHC20 expression on endocytic trafficking of EGFR. In resting cells EGFR is predominantly localized at the plasma membrane in both shControl and shDHHC20 cells (data not shown). Upon EGF stimulation of shControl cells EGFR was internalized and clustered around the perinuclear region of the cell containing the lysosomal membrane protein LAMP-1 (Figure 3B). In contrast, in shDHHC20 cells the internalized EGFR positive vesicles maintained a peripheral distribution and failed to localize with LAMP-1 positive vesicles after 15 minutes of EGF stimulation (Figure 3B). Expression of shRNA resistant DHHC20 partially restored the wild type perinuclear localization and lysosomal targeting of EGFR in response to EGF (Figure 3B). In shDHHC20 cells, EGFR containing vesicles localized to the cell periphery and lacked markers of early (EEA1, Rab5) or late (Rab7) endosomes within 15 minutes of EGF stimulation (Figure S4A-F). Additionally, EGFR localization did not overlap with the recycling endosome marker Rab11 indicating that the aberrant EGFR trafficking was not the result of increased receptor recycling (Figure S4G). Live imaging of fluorescently tagged EGF indicates EGF is internalized in DHHC20 cells similar to control cells. After internalization, the localization of the EGF containing endosomes in the DHHC20 shRNA expressing cells is static at the periphery of the cell (Figure 3C). This is in contrast to control cells where EGF is rapidly trafficked to lysosomes (Figure 3C). To determine if the altered trafficking is specific to EGFR, cells were labelled with fluorescently tagged transferrin. We found that unlike EGF the trafficking of the endocytosed transferrin is similar between DHHC20 silenced cells and control cells and the accumulation of enlarged endosomes that formed with EGF did not form with transferrin (Figure S5).
Figure 3. Silencing DHHC20 expression disrupts EGFR endocytic trafficking.
(A) Ubiquitination of EGFR is increased in shDHHC20 MDA-MB-231 cells treated with EGF (100ng/ml) and EGFR was immunoprecipitated using anti-EGFR (sc-120). (B) Silencing DHHC20 decreases EGF-induced EGFR (green) trafficking to LAMP-1 (red) positive lysosomes, which are rescued by exogenous expression of DHHC20. DAPI (blue). Colocalization of EGFR and LAMP-1 was determined by Mander’s Overlap Coefficient; shControl (0.403), shDHHC20 (0.242), shDHHC20+DHHC20 (0.395). (C) Live cell imaging shows that trafficking of Alexa-fluor488 labeled EGF (green) to lysosomes (red) is disrupted in shDHHC20 MDA-MB-231 cells. (D) Silencing DHHC20 increases Grb2 (red) localization to Alexa-fluor488 labeled EGF (green) labelled endosomes in MDA-MB-231 DAPI (blue). Colocalization between EGFR and LAMP-1 was determined by Mander’s Overlap Coefficient; shControl (0.627), shDHHC20 (0.723).
After internalization EGFR is thought to continue to signal until the receptor is sequestered into multivesicular bodies (MVB). Endosome specific signaling to ERK and AKT has been demonstrated for EGFR following the endosomal recruitment of scaffold proteins such as Grb2 (Murphy et al., 2009). We asked if the increased amount of EGFR in the endosomal pool actively signals in shDHHC20 cells by examining endogenous Grb2 localization by IF. After 15 minutes of EGF treatment the EGF containing endosomes in the DHHC20 silenced cells localized with very high levels of Grb2 compared to control cells that contained low levels of endosomally localized Grb2 (Figure 3D). These findings suggest that the accumulation of endosomal EGFR likely contributes to the sustained signaling responses observed in shDHHC20 expressing cells.
DHHC20 palmitoylates EGFR within the C-terminal tail
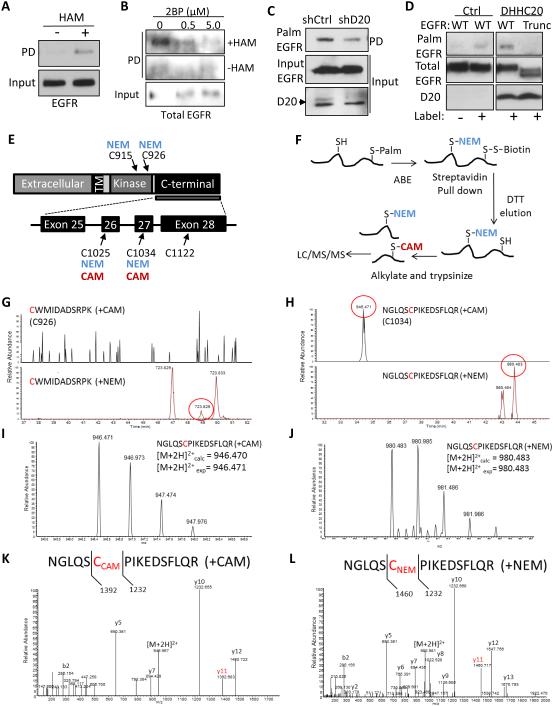
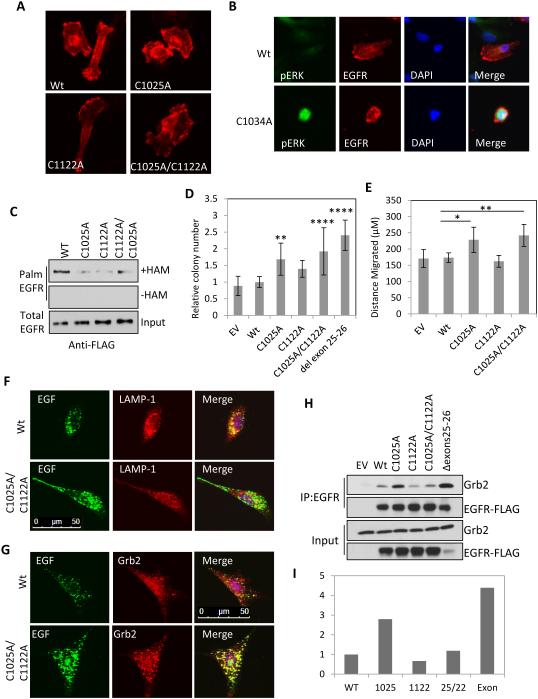
Since EGFR was recently reported to be palmitoylated, we asked if EGFR is the target of DHHC20 (Bollu et al., 2015). To detect palmitoylated EGFR we performed an in vitro acyl-biotinyl exchange (ABE) assay on MDA-MB-231 cells. The ABE assay removes palmitate from cysteine residues with hydroxylamine followed by substitution with biotin. In the presence of hydroxylamine, EGFR was detected in the streptavidin pull-down fraction compared to the negative control without hydroxylamine indicating the presence of palmitoylated EGFR (Figure 4A). Treatment with 2BP effectively inhibited EGFR palmitoylation (Figure 4B). Palmitoylation of EGFR in MDA-MB-231 cells was also observed by metabolically labelling cells with palmitic acid azide as a second approach to confirm EGFR palmitoylation (Figure 4C). Silencing DHHC20 reduced EGFR palmitoylation indicating that DHHC20 is required for wild type levels of EGFR palmitoylation (Figure 4C). Overexpression of DHHC20 was sufficient to increase EGFR palmitoylation in HEK293T cells expressing wild type (WT) EGFR compared to vector control cells (Figure 4D). Consistent with palmitoylation inhibiting EGFR signaling, when protein depalmitoylation is blocked with the small molecule Palmostatin B EGFR palmitoylation is increased and EGFR and AKT phosphorylation is inhibited (Figure S6A, B).
Figure 4. DHHC20 palmitoylates EGFR within the C-terminal tail.
(A) EGFR palmitoylation in MDA-MB-231 cells determined using an acyl-biotinyl exchange (ABE) assay. (B) The palmitoyltransferase inhibitor 2BP reduces EGFR palmitoylation. MDA-MB-231 cells were treated with 2BP for 24 hours and EGFR palmitoylation was determined by ABE. (C) Silencing DHHC20 decreases EGFR palmitoylation in MDA-MB-231 cells determined by metabolic labeling. (D) DHHC20 palmitoylates the C-terminal tail of EGFR. HEK293T cells transiently expressing full length EGFR (WT) or a C-terminal tail truncation mutant (Trunc) and either empty vector control (EV) or DHHC20. (E) Schematic of EGFR including the exons and cysteine residues located within the C-terminal tail and kinase domain. Numbering corresponds to human EGFR excluding the signal sequence. (F) Experimental strategy for detecting EGFR palmitoylation by mass spectrometry. (G) Selected ion chromatograms of the [M+2H]2+ peptide (CWMIDADSRPK) with both potential carbamidomethylation (CAM, +57 Da) and N-Ethylmaleimide (NEM, +125 Da) modifications of cysteine 926. As can be seen in the chromatograms, only a peak for the CWMIDADSRPK (+NEM) peptide was found, and no peaks for CWMIDADSRPK (+CAM) were observed. (H) Selected ion chromatograms of the [M+2H]2+ peptide (NGLQSCPIKEDSFLQR) with both potential carbamidomethylation (CAM, +57 Da) and N-Ethylmaleimide (NEM, +125 Da) modifications of cysteine 1034. As can be seen in the chromatograms, both peaks for NGLQSCPIKEDSFLQR (+NEM) and NGLQSCPIKEDSFLQR (+CAM) were observed (~ 10 min retention time shift between the species). (I) Full mass spectrum of the parent ion corresponding to the NGLQSCPIKEDSFLQR (+CAM), and accurate mass confirms the correct assignment. (J) Full mass spectrum of the parent ion corresponding to the NGLQSCPIKEDSFLQR (+NEM), and accurate mass confirms the correct assignment. (K) MS/MS spectrum of the [M+2H]2+ peptide NGLQSCPIKEDSFLQR (+CAM), with highlighted fragment ion indicating the CAM modification on the Cys residue 1034. (L) MS/MS spectrum of the [M+2H]2+ peptide NGLQSCPIKEDSFLQR (+NEM), with highlighted fragment ion indicating the NEM modification on the cysteine residue 1034.
EGFR contains nine cysteine residues within the intracellular domain, six of which are located within the tyrosine kinase domain and three within the C-terminal tail (Figure 4E). An EGFR truncation mutation which deletes amino acids 1024-1186 including the three cysteine residues within the C-terminal tail completely abolished EGFR palmitoylation in HEK293T cells ectopically expressing DHHC20, indicating that the palmitoylated cysteine residues are located within the C-terminal tail (Figure 4D).
To identify the specific palmitoylated residues by mass spectrometry (MS) the palmitoylated peptides were purified by ABE followed by alkylation with iodoacetamide (Figure 4F). When analyzed by MS the intracellular palmitoylated cysteine residues were detected as carbamidomethyl (CAM) modified and the unmodified cysteine residues in the cytosolic domains that were blocked with NEM were detected as NEM modified based on the mass difference between NEM and CAM (Figure 4E, F). Cysteine residues C915, C926 in the kinase domain were detected only as NEM modified indicating the sites are not palmitoylated (Figure 4E, G). However, peptide ions were identified with both NEM and CAM modifications for both C1025 and C1034 in the C-terminal tail (Figure 4H-L, data not shown). This indicates both C1025 and C1034 sites are both palmitoylated and unpalmitoylated. The large size of the tryptic peptide containing cysteine residue C1122 prevented the identification of the peptide by MS.
Mutation of palmitoylated cysteine residues increases EGFR interaction with Grb2
To determine the molecular mechanism by which EGFR palmitoylation regulates its signaling activity cysteine residues 1025, 1034 and 1122 were mutated to alanine either alone or in combination. When expressed in MDA-MB-231 cells the C1025A, C1122A and double cysteine mutants correctly localized to the cell membrane and were indistinguishable from WT EGFR (Figure 5A). In contrast to the other mutants the C1034A mutation could be detected by indirect IF microscopy in only a small number of rounded cells that contained very high levels of phosphorylated ERK compared to cells expressing wild type EGFR (Figure 5B). The morphology and high levels of phosphorylated ERK in the C1034A expressing cells could indicate a block in mitosis. As a result the C1034A mutant could not be detected by immunoblotting and we were therefore unable to examine this mutation in palmitoylation and signaling assays. In all the subsequent experiments we discuss the C1025 and C1122 sites with the knowledge that C1034 is possibly if not likely palmitoylated. Mutation of either C1025 or C1122 markedly reduced EGFR palmitoylation compared to the wild type receptor in HEK293T cells, but mutation of both cysteine residues was not sufficient to completely eliminate palmitoylation indicating these residues are at minimum required for wild type EGFR palmitoylation levels (Figure 5C). This raised the question of whether there are mutations in cancer that delete any of the inhibitory cysteine residues that would indicate a biological function for EGFR palmitoylation.
Figure 5. Mutation in C1025 and C1122 activates EGFR, promotes cellular transformation and increases cell migration.
(A) Mutation of cysteine residues 1025 and 1122 of EGFR-FLAG (red) does not alter receptor localization in MDA-MB-231 cells. (B) Expression of C1034A mutant EGFR-FLAG (red) in MDA-MB-231 cells increases ERK phosphorylation (green), DAPI (blue). (C) Mutation of EGFR cysteine residues 1025, 1122 and 1025/1122 decreases EGFR palmitoylation in HEK293T cells. (D) Mutation of EGFR C1025, C1025/C1122 or deletion of exons 25-26 deletion increases colony formation of NIH 3T3 cells grown in soft agar. Expression levels of each mutant are shown in Figure S7A (mean +/− StDev). (E) Mutation of EGFR C1025 and C1025/C1122 increases migration of NIH 3T3 cells (mean +/− SEM). (F) Mutation of EGFR C1025/C1122 delays EGFR endocytosis and trafficking of EGF-alexafluor488 (green) labelled endosomes to lysosomes stained for LAMP-1 (red). Cells expressing the EGFR C1025/C1122 mutant have increased localization of Grb2 (red) to EGF-alexafluor488 (green) labelled endosomes. (H) The C1025A mutation or the exon 25-26 deletion increases the interaction of EGFR-FLAG with endogenous Grb2 in HEK293T cells. EGFR was immunoprecipitated with anti-EGFR (sc-120 AF488). (I) Quantification of co-immunopreciptated Grb2 by densitometric analysis of immunoblots in (H). Y-axis indicates arbitrary units normalized to WT EGFR.
A mutation identified in lung cancer that deletes exons 25-26 increases EGFR phosphorylation, downstream signaling to AKT, and anchorage independent growth (Figure 5D) (Imielinski et al., 2012). Since this mutation removes cysteine1025, we asked if mutating this residue is sufficient to increase anchorage independent growth. When NIH 3T3 cells expressing WT or the EGFR cysteine mutants were grown in soft agar the exon 25-26 deletion increased the number of colonies by 2.4 fold compared to cells expressing WT EGFR (Figure 5D). The C1025 and C1025/C1122 mutants significantly increased colony formation compared to WT EGFR by 1.7 and 1.9 fold respectively even though the C1025/C1122 mutant is expressed at a much lower level than WT (Figure 5D, Figure S7A). The C1122 single mutant did not have a significant effect on colony formation. Furthermore cells expressing EGFR C1025A or C1025A/C1122A migrate significantly faster than cells expressing WT EGFR (Figure 5E). We conclude that the loss of palmitoylation at these cysteine residues is important for the increased migration observed with DHHC20 shRNA and the cell transformation previously described for the exon 25-26 deletion seen in lung cancer patients. However, the sites are not equivalent in promoting EGFR mediated cell behavior since C1025A has a stronger effect on colony formation and migration than C1122A.
To determine if the accumulation of EGFR and the high endosomal localization of Grb2 observed in DHHC20 silenced cells was caused specifically by decreased EGFR palmitoylation the EGFR C1025/1122A mutant was examined in NIH 3T3 cells. Similar to what was observed in shDHHC20 cells the C1025/1122A mutant receptor accumulated in peripherally localized endosomes that did not colocalize with LAMP-1 (Figure 5F). Furthermore, there was an increase in Grb2 staining in cells expressing C1025/1122A compared to cells expressing the WT receptor (Figure 5G).
To study the mechanism of receptor activation in greater detail we examined activation mediated Grb2 binding to the EGFR mutants. Immunoprecipitation of WT and mutant EGFR revealed an increase in the interaction of the C1025A mutant with Grb2 compared to WT EGFR (Figure 5H-I). However, EGFR C1122A and C1025A/C1122A did not increase the interaction with Grb2 indicating that palmitoylation of C1025 is unique in its ability to attenuate Grb2 binding. Grb2 binding with the exon 25-26 deletion mutant was higher compared to WT EGFR and is strikingly similar to what was observed with the C1025A mutant, providing further evidence that the pathway activation caused by the exon 25-26 deletion is through decreased palmitoylation at C1025 (Figure 5H-I).
EGFR palmitoylation promotes the turnover of activated EGFR and association of the C-terminal tail with the plasma membrane
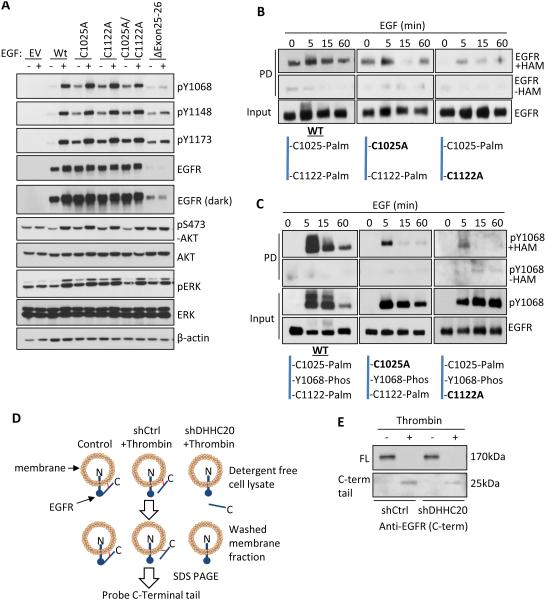
We examined the activation of the palmitoylation defective EGFR mutants to further understand the mechanism by which palmitoylation suppresses EGFR activation. We first asked if the cysteine point mutations phenocopy the increase in EGFR activation observed with DHHC20 shRNA. When transiently expressed in NIH 3T3 cells EGFR mutants C1025A, C1122A and C1025A/C1122A increased the basal phosphorylation of EGFR at Tyr1068, Tyr1148, and Tyr1173 and phosphorylation of AKT compared to the WT receptor under serum starved conditions (Figure 6A). When cells expressing mutant EGFR were treated with 10μM Gefitinib the activation of AKT was inhibited, but ERK activation was unaffected (Figure S7B). Taken together, the high basal EGFR tyrosine phosphorylation observed in the EGFR mutants in conjunction with Grb2 binding being specific to the C1025 mutant indicates that tyrosine phosphorylation is not sufficient to promote Grb2 binding.
Figure 6. Palmitoylation regulates the activity of EGFR in response to EGF.
(A) Mutation of EGFR palmitoylated cysteine residues increases EGFR activation independent of EGF in NIH 3T3 cells compared to WT EGFR. (B) EGFR palmitoylation increases upon EGF (100ng/ml) stimulation in MDA-MB-231 cells. (C) Dually phosphorylated and palmitoylated EGFR and EGFR mutants C1025A and C1122A turns over more rapidly than total EGFR in response to EGF (100ng/ml). (D) Experimental strategy of the thrombin cleavage assay. (E) Palmitoylation pins the C-terminal tail of EGFR to the cell membrane. HEK293T cells transiently expressing EGFR-FLAG containing a thrombin cleavage site (LVPRGS) at Gly959 were lysed in the presence or absence of thrombin protease. The membrane fraction was isolated by centrifugation and EGFR was immunoprecipitated from the membrane fraction. The presence of membrane bound full length (170kDa) EGFR and the cleaved C-terminal tail (25kDa) was determined by SDS-PAGE.
To determine how palmitoylation of EGFR is regulated we examined the levels of palmitoylated and dually palmitoylated/phosphorylated EGFR in response to EGF stimulation. Palmitoylated WT EGFR is detected in resting cells and increases modestly after 5 minutes of EGF stimulation, returning to basal levels by 60 minutes (Figure 6B). To determine the contribution of the individual palmitoylation sites the level of palmitoylated EGFR during EGF stimulation was examined in the EGFR cysteine point mutants. In the C1025A mutant palmitoylated C1122 peaked at 5 minutes post-EGF treatment and subsequently decreased below basal levels at 15 minutes and returned to basal levels at 60 minutes (Figure 6B). Unlike C1122, palmitoylation of C1025 in the C1122A mutant is below detection in resting cells. After 5 minutes of EGF stimulation C1025 palmitoylation increased and then steadily decreased by 60 minutes (Figure 6B).
We next examined the phosphorylation kinetics of palmitoylated EGFR in response to EGF and found that palmitoylated EGFR is also phosphorylated at tyrosine 1068. However, in contrast to the total level of phosphorylated WT EGFR (input) which is does not decrease until 60 minutes, phosphorylation of palmitoylated WT EGFR rapidly decreased between 5 and 15 minutes (Figure 6C). This suggests the dually phosphorylated and palmitoylated receptor fraction is more rapidly turned over than the phosphorylated unpalmitoylated receptor fraction. In the C1025A mutant palmitoylation is restricted to C1122 and phosphorylation of the palmitoylated receptor peaked at 5 minutes of EGF stimulation, but then decreased down to basal levels by 15 minutes (Figure 6C). In the C1122A mutant the receptor is palmitoylated at C1025 and is only weakly phosphorylated consistent with the palmitoylation of C1025 suppressing EGFR activation. Furthermore, in the C1122A mutant the total receptor phosphorylation (input) is sustained and does not decrease by 60 minutes (Figure 6C). This indicates C1122 palmitoylation promotes receptor turnover. While we were unable to address the function of cysteine 1034 these results indicate that C1025 and C1122 are palmitoylated in response to EGF with each site having a unique effect on receptor function.
The C-terminal tail is not included in current crystal structures of EGFR and therefore little is known about how it is positioned relative to the kinase domain. Although EGFR is membrane associated, the C-terminal tail is unstructured and thought to extend away from the plasma membrane and into the cytosol. We reasoned that palmitoylation could promote peripheral association of the C-terminal tail with the plasma membrane by burying palmitate in the lipid bilayer. To test this possibility a recognition sequence for the protease thrombin was inserted in frame between the kinase domain and the C-terminal domain to allow the C-terminal tail to be cleaved from the plasma membrane. Cell lysates were treated either with or without thrombin and the membrane fraction was isolated by centrifugation and analyzed by SDS PAGE. If the C-terminal tail associates with the plasma membrane after proteolytic cleavage then it will be detected by immunoblotting in the membrane fraction with an antibody specific to the C-terminus of EGFR (Figure 6D). Only after thrombin treatment is the 25kDa C-terminal tail detected in the membrane fraction (Figure 6E). The membrane association of the cleaved C-terminal tail is dependent on palmitoylation as inhibition of DHHC20 by shRNA markedly reduces the 25kDa fragment in the membrane fraction (Figure 6E).
DISCUSSION
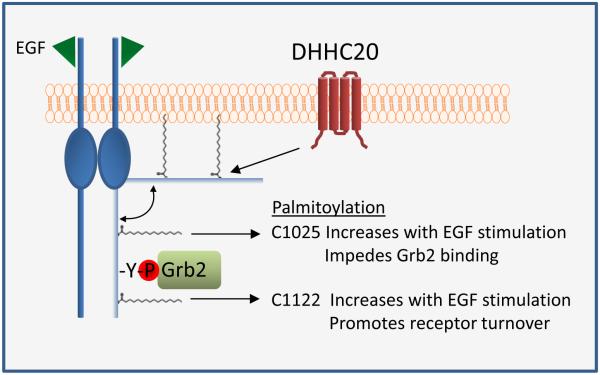
We have identified a mechanism to regulate EGFR activation through palmitoylation of the C-terminal tail of EGFR. We show that EGF induces palmitoylation of EGFR and that palmitoylation facilitates receptor inactivation through two distinct mechanisms. Mutation of cysteines 1025 and 1122 alone and in combination increase the basal phosphorylation of EGFR, ERK, and AKT. However, mutation of C1025 has the strongest increas in the binding of EGFR with the adapter protein Grb2. Furthermore, EGFR that is palmitoylated on C1025 has very low levels of EGF induced phosphorylation indicating that palmitoylation at C1025 inhibits receptor signaling. Mutation of C1122 sustains EGFR phosphorylation in response to EGF and both total and phosphorylated EGFR palmitoylated on C1122 decreases precipitously after 15 minutes of EGF stimulation consistent with a role for palmitoylation at C1122 in promoting receptor turnover. This is consistent with the effects of silencing DHHC20 on EGFR function. When DHHC20 is silenced by shRNA EGFR signaling is increased and sustained, there is increased localization of Grb2 to EGFR positive endosomes, and there are increased levels of total EGFR (Figure 7).
Figure 7. Mechanistic summary of the effects of palmitoylation on EGFR activation and signaling.
Palmitoylation of the C-terminus of EGFR promotes membrane association. Palmitoylation of EGFR at C1025 impedes the binding of Grb2 to EGFR whereas palmitoylation at C1122 increases with EGF stimulation and promotes receptor turnover.
Our findings demonstrate that palmitoylation of EGFR C-terminal cysteines reversibly “pins” the C-terminal tail to the plasma membrane to promote receptor inactivation. Disrupting the membrane association of the C-terminal tail by inhibiting palmitoylation may reduce steric hindrance or provide greater accessibility for adaptor protein binding (Figure 7). Additionally, the interaction between EGFR and endosomal ESCRT complex may require palmitoylation of the EGFR C-terminal tail to facilitate endocytic proper trafficking.
Palmitoylation is required for the endocytic trafficking and downregulation of EGFR. The delayed endocytic trafficking of EGFR observed in DHHC20 silenced cells may account for the elevated level of EGFR expression in these cells. The delay in EGFR endocytosis was recapitulated in the C1025/C1122 mutant supporting that palmitoylation of the receptor is responsible for the trafficking defect. Our findings suggest that delayed endocytic trafficking provides a platform for EGFR to signal within endosomal compartments. In support of this, other studies have shown that activated receptors accumulate in endosomes and can transmit signals that are distinct from those at the plasma membrane due to the endosomal localization of certain essential signaling components (Murphy, PNAS, 2009). Our findings reveal that the endosomal localization of Grb2 is increased when DHHC20 is silenced and when cysteines 1025 and 1122 are mutated. Based on our data we argue that the slower rate of endocytic trafficking to the lysosome in shDHHC20 cells causing increased receptor signaling on endosomal compartments.
Blocking EGFR palmitoylation genetically by silencing DHHC20 or pharmacologically by treatment with 2BP sensitizes cells to EGFR TK inhibition. While silencing DHHC20 or mutating EGFR C-terminal palmitoylated cysteines increases EGFR activation, Gefitinib effectively reduces receptor phosphorylation to the same level as control cells. This marked decrease in EGFR signaling may be the mechanism leading to the increased toxicity of Gefitinib in DHHC20 silenced cells. The clinical implications of these findings are three-fold. First, the increase in dependency of EGFR signaling when DHHC20 is inhibited by shRNA raises the possibility that patients with inactivation of DHHC20 could show increased responsiveness to EGFR inhibitor therapy. Second, inhibition of DHHC20 with a small molecule could function therapeutically in combination with EGFR inhibitors. Third, the EGFR exon 25-26 deletion mutation identified in lung cancer increased colony formation in soft agar and the interaction between EGFR and Grb2 similar to what was observed with the C1025A mutant. Therefore, loss of palmitoylation at C1025 may serve as a mechanism for the increased EGFR activation and transforming properties of the deletion mutant. Taken together, these data suggest that targeting of DHHC20 in combination with EGFR TK inhibitors may serve as an effective clinical approach to treat cancers such as TNBC that are inherently resistant to EGFR targeted therapy.
MATERIALS AND METHODS
Cell culture and transfection
MDA-MB-231, HEK293T, and NIH 3T3 cells were cultured in DMEM containing 10% FBS. SW1573 cells were maintained in RPMI containing 10% FBS. All transfections were carried out using TransIT-LT1 (Mirus) according to the manufacturer’s instructions.
Antibodies
Anti-DHHC20 and anti-flag M2 antibodies were purchased from Sigma-Aldrich. Anti-pY1068-EGFR, pY1148-EGFR, pY1173-EGFR, EGFR, pERK, ERK, pS473-AKT, AKT, β-actin, EEA1, Rab5, Rab7 and Rab11 were obtained from Cell Signaling Technologies. Anti-LAMP-1 (CD107a) was purchased from BD Pharmigen. EGFR (528) Alexa Fluor 488 and EGFR (sc-120) were purchased from Santa Cruz Biotechnology. Transferrin from Human Serum, Alexa Fluor® 488 Conjugate was purchased from ThermoFisher. Phalloidin-594 and EGFR (528) Alexa Fluor 488 were purchased from Life Technologies.
Analysis of TCGA data
The level of ZDHHC20 mRNA was plotted against the PAM50.SUBTYPE using the beeswarm package in the software program “R”. Box plots were created to visualize the median values and quartiles of each subtype. Statistics were calculated using the function aov in “R”; (p < 2e-16).
Silencing of human DHHC20 and EGFR
The oligonucleotides for shControl, shDHHC20, and shEGFR constructs were synthesized (Integrated DNA Technologies) and inserted into the pLKO.1 vector. shControl encodes the non-targeting sequence of SHC002 (Sigma), the shRNA target sequences of human DHHC20 is 5’-GAGCTCTGCGTGTTTACTATT-3’, and the shRNA sequences of EGFR are 5’ CACAGTGGAGCGAATTCCTTT 3’ (shEGFR 2.4) and 5’ GCTGGATGATAGACGCAGATA 3’ (shEGFR 3.2). MDA-MB-231 cells were transduced with lentivirus encoding shControl or shDHHC20 and selected by puromycin treatment (1 μg/ml). A shRNA resistant DHHC20 construct was used to rescue expression in shDHHC20 cells. MDA-MB-231 cells transduced with shEGFR lentivirus were harvested 72 hours post infection.
Soft Agar Colony Formation
5×103 NIH 3T3 cells were suspended in 0.4% agarose and plated onto a 0.8% solidified agarose layer. Colonies were manually counted from triplicate wells at 8 weeks.
Cell Viability
Cells were treated with Gefitinib (10μM) and/or 2BP (500nM and 5μM) for 72hrs and viability was measured by Trypan Blue staining. Quantification was done using a 1-way ANOVA with Tukey’s multiple comparison post-test.
EGFR signaling
MDA-MB-231 and SW1573 cells were serum starved for 17 hours in DMEM+0.2% BSA. Cells were stimulated with 100ng/ml of EGF and harvested in RIPA buffer containing 150mM NaF, 2mM Na3V04, 1mM PPi, 2μg/ml pepstatin A, 1μg/ml aprotinin and 1μg/ml leupeptin. Activation of EGFR, ERK and AKT was measured by SDS-PAGE.
Immunofluorescence microscopy
MDA-MB231 shControl and shDHHC20 cells were serum starved for 17 hours in DMEM+0.2% BSA, treated with 100ng/ml EGF, and fixed in formalin. Cells were permeabilized in 0.1% triton-X-100, blocked in 5% BSA and primary antibodies were added overnight. Where indicated, cells were incubated with 1μg/ml Alexa-fluor488 labeled EGF for 1 hour on ice and transferred to 37°C for 15 minutes before fixing and adding primary antibodies. Secondary Alexa-Fluor antibodies were used prior to mounting the coverslips with DAPI-containing mounting media. Cells were visualized on a Leica SP8 confocal microscope or a Leica AF6000 microscope. Images were obtained using the Hamamatsu ORCA R2 digital CCD camera at 40x magnification. Quantification was determined using Perkin Elmer’s Volocity software.
Live cell microscopy
MDA-MB-231 shControl and shDHHC20 cells were serum starved in DMEM + 0.2% BSA for 17 hours and incubated with 50nM lysotracker (red) for 30 minutes. The lysotracker was removed and cells were incubated on ice for 30 minutes with 25μg/ml Alexa-fluor488 labeled EGF (green) or 2.5ug/mL Alexa-fluor488 labeled transferrin and 15ng/mL unlabeled EGF. Cells were washed, incubated in HBSS media (HBSS + 1mg/mL glutamine + 1% FBS + 20mM HEPES, pH 7.4), and images were obtained in a 37°C humidified chamber every 10 seconds for 20 minutes using the Leica AF6000 microscope and the Hamamatsu ORCA R2 digital CCD camera at 40x magnification.
Transwell Migration Assay
MDA-MB231 cells were plated onto the apical chamber of transwell dishes and allowed to migrate for 17 hours towards DMEM containing 10% FBS. Where indicated, cells were pretreated with DMSO or 10μM Gefitinib for 30 minutes before adding the chemoattractant. Cells were washed in PBS, fixed in methanol and stained with 0.05% Crystal Violet for 40 minutes. Cells on the apical side of the membrane were removed with a Q-tip and cells that had migrated to the basolateral chamber were imaged using the Leica AF6000 microscope. Images were obtained using the Hamamatsu ORCA R2 digital CCD camera at 10x magnification and counted. Quantification was done using a 1-way ANOVA with a Tukey’s multiple comparison post-test.
Scratch Assay
NIH 3T3 cells were plated onto 6-well dishes and scratched at confluence. Images were taken at 0hrs and 8hrs using the Hamamatsu ORCA R2 digital CCD camera at 4x magnification. Images of four representative areas of each scratch were captured and 4 measurements were taken per image for a total of 12 measurements per scratch. The distance migrated was calculated using Leica Microsystems LAS AF software.
Palmitoylation assay with metabolic labelling
MDA-MB-231 shControl and shDHHC20 cells were treated with 100μM palmitic acid azide in serum free DMEM for 4 hours at 37ºC. Cells were washed in PBS and lysed in 200μl buffer (50mM Tris pH 7.5, 1% SDS, and 1μg/ml leupeptin, 1μg/ml aprotinin, 2μg/ml pepstatin A). Lysates were sonicated and centrifuged at 15,000 RPM for 10 minutes. 50μl of lysate was reacted with biotin alkyne using the Click-IT assay in a 200 μl final reaction volume. Biotinylated proteins were isolated using streptavidin agarose, washed 5 times in wash buffer (50mM Tris pH 7.5, 0.1%SDS) and palmitoylated EGFR was analyzed by SDS PAGE.
Acyl-biotinyl exchange (ABE) assay
The protocol is adapted from Wan et al., 2007. EGFR was immunoprecipitated with anti-EGFR (sc-120) and bound protein was eluted from the beads in 4%SDS buffer+50mM NEM. Protein was precipitated by methanol/chloroform (m/c) precipitation and the pellet was resuspended in 4%SDS buffer+50mM NEM. Samples were m/c precipitated, resuspended in 4%SDS buffer and incubated in either hydroxylamine buffer or control buffer. Samples were m/c precipitated and the pellets were resuspended in 4%SDS buffer containing 10μM Biotin-HPDP. 160μl of 0.2% Triton X-100 buffer +10μM Biotin-HPDP was added for 1hour and the samples were m/c precipitated and resuspended in 4%SDS buffer and 1% Triton X-100. Streptavidin agarose beads were added overnight and the samples were analyzed by SDS-PAGE.
EGFR purification for Mass spectrometry
HEK293T cells were transfected with EGFR and DHHC20 and EGFR was immunoprecipitated by standard methods using anti-EGFR sc-120. The acyl biotin exchange assay was used to label palmitoylated cysteine residues with 10μM Biotin-HPDP. The biotinylated proteins were isolated with streptavidin beads and eluted with 10μM dithiothriotol to break the Biotin-HPDP disulfide linker. The reduced cysteine residues were blocked with 4mM iodoacetamide and the proteins were digested with trypsin.
Nano-LC-MS/MS analysis and data analysis
Digestion solution was acidified by 5% formic acid, and peptides were desalted prior to LC-MS/MS analysis using in-house C18 STAGE tips as previously described (Rappsilber et al, 2003) . Peptide samples were loaded onto a 75 μm I.D. × 20 cm fused silica capillary column packed with Reprosil-Pur C18-AQ resin (3 μm; Dr. Maisch GmbH, Germany) and resolved by an EASY-nLC 1000 HPLC system (Thermo Scientific) coupled in-line with a Q-Exactive (Thermo Scientific). The HPLC gradient was 2-30% solvent B (A = 0.1% formic acid in water; B = 0.1% formic acid in acetonitrile) for 70 min, followed by 30% to 95% solvent B for 10 min, and then held at 95% solvent B for 10 min, with a constant flow-rate of 300 nL/min. Full MS spectrum scans (m/z 350-1600) were performed at a resolution of 70,000 (at 200 m/z), and the 3 most intense ions were selected for MS/MS performed with high-energy collision dissociation (HCD) with normalized collision energy of 25 at a resolution of 17,500 (at 200 m/z). Five target MS/MS (508.7633, 736.3706, 1103.0043, 1104.0523 and 1470.3366) were set in case they were missed in data-dependent acquisition mode. AGC targets of full MS and MS/MS scans were 1×106 and 5×104, respectively. Unassigned charge states and singly charged species were rejected, dynamic exclusion was set to 30 seconds, and lock mass calibration was implemented using polysiloxane ions 371.10123 and 445.12000. Mascot was used for database searching. Two trypsin miss-cleavage sites were allowed, and precursor ion and fragment ion tolerances were set to 10 ppm and 0.02 Da, respectively. Oxidation (+15.9949) on methionine, carbamidomethylation (+57.0215) and N-ethylmaleimide (125.0477) on cysteine were set as dynamic modifications. A peptide score of 20 was chosen to filter the peptide identification matches. Peptide quantification was performed on the extracted ion chromatograms (XICs) of peptides with all charge states.
Thrombin cleavage assay
HEK293T cells expressing either shControl or shDHHC20 vectors were co-transfected with DHHC20 and either EGFR Wt or EGFR containing a thrombin cleavage sequence (LVPRGS) inserted at Gly959. Cells were harvested 48 hours post-transfection in 250μl of either buffer A (10mM Tris pH 8, 1.5mM MgCl2, 10mM KCl, 100μM Palmostatin B, 150mM NaF, 2mM Na3V04, 1mM PPi, 2μg/ml pepstatin A, 1μg/ml aprotinin and 1μg/ml leupeptin) or buffer B (10mM Tris pH 8, 1.5mM MgCl2, 10mM KCl, 100μM Palmostatin B and 50 units of thrombin). Lysates were disrupted 7 times by passing through a 22 gauge needle and centrifuged at 800xg for 10 minutes. The thrombin containing samples were incubated at room temperature for 1 hour and centrifuged at 45,000 rpm for 30 minutes to isolate the membrane fraction which was then resuspended in RIPA buffer and incubated with anti-flag beads for 1 hour. The beads were washed with lysis buffer and the bound protein was eluted by boiling in SDS-loading buffer. Samples were loaded on an SDS-PAGE gel and membrane bound EGFR was detected using an EGFR antibody.
Supplementary Material
Acknowledgements
We would like to thank the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania for their assistance with measuring IF colocalization. We would also like to thank Mark Lemmon for his helpful discussion and advice. This work was supported by NIH Grants R01CA181633 (to E.S.W), T32-CA-557726-07 (to K.B.R) and by ACS Grants RSG-15-027-01 (to E.S.W), IRG –78-002-34 (to E.S.W) and DOD grant BC123187P1 (to B.A.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
E.S.W, K.B.R., A.K, and B.A.G conceived the study, designed experiments, and analyzed results. K.B.R and A.K carried out the majority of experiments; E.S, confocal microscopy and live cell imaging; W.W DNA cloning; X.C, LC-MS/MS; K.B.R and E.S.W wrote the manuscript with input from all co-authors.
References
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochimica et biophysica acta. 2011;1808:2981–2994. doi: 10.1016/j.bbamem.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Bollu LR, Katreddy RR, Blessing AM, Pham N, Zheng B, Wu X, Weihua Z. Intracellular activation of EGFR by fatty acid synthase dependent palmitoylation. Oncotarget. 2015;6:34992–35003. doi: 10.18632/oncotarget.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Pastorino S, Zeng Q, Xu X, Johnson W, Vandenberg S, Verhaak R, Cherniack AD, Watanabe H, Dutt A, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer research. 2011;71:7587–7596. doi: 10.1158/0008-5472.CAN-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Davis NG. Palmitoylation and depalmitoylation dynamics at a glance. Journal of cell science. 2010;123:4007–4010. doi: 10.1242/jcs.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends in biochemical sciences. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. Journal of lipid research. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature reviews. Molecular cell biology. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harbor perspectives in biology. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick PJ, Dumaresq-Doiron K, Pluviose AS, Pichette V, Tosato G, Lefrancois S. Palmitoylation controls recycling in lysosomal sorting and trafficking. Traffic. 2008;9:1984–1997. doi: 10.1111/j.1600-0854.2008.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Padilla BE, Hasdemir B, Cottrell GS, Bunnett NW. Endosomes: a legitimate platform for the signaling train. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines G, Kostler WJ, Yarden Y. Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS letters. 2010;584:2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JT, Tiganis T, Agarwal A, Djakiew D, Thompson EW. Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3'-kinase and phospholipase C-dependent mechanism. Cancer research. 1999;59:5475–5478. [PubMed] [Google Scholar]
- Roskoski R., Jr. ErbB/HER protein-tyrosine kinases: Structures and small molecule inhibitors. Pharmacological research : the official journal of the Italian Pharmacological Society. 2014;87:42–59. doi: 10.1016/j.phrs.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Seshacharyulu P, Ponnusamy MP, Haridas D, Jain M, Ganti AK, Batra SK. Targeting the EGFR signaling pathway in cancer therapy. Expert opinion on therapeutic targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Walton GM, Chen WS, Rosenfeld MG, Gill GN. Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. The Journal of biological chemistry. 1990;265:1750–1754. [PubMed] [Google Scholar]
- Wang W, Runkle KB, Terkowski SM, Ekaireb RI, Witze ES. Protein Depalmitoylation Is Induced by Wnt5a and Promotes Polarized Cell Behavior. The Journal of biological chemistry. 2015;290:15707–15716. doi: 10.1074/jbc.M115.639609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.