Summary
Biogeography and individuality shape the structural and functional composition of the human skin microbiome. To explore these factors’ contribution to skin microbial community stability, we generated metagenomic sequence data from longitudinal samples collected over months and years. Analyzing these samples using a multi-kingdom, reference-based approach, we found that despite the skin’s exposure to the external environment, its bacterial, fungal, and viral communities were largely stable over time. Site, individuality, and phylogeny were all determinants of this stability; foot sites exhibited the most variability, individuals differed in stability, and transience was a particular characteristic of eukaryotic viruses, which showed little site-specificity in colonization. Strain and single nucleotide variant level analysis showed that individuals maintain, rather than reacquire prevalent microbes from the environment. Longitudinal stability of skin microbial communities generates hypotheses about colonization resistance and empowers clinical studies exploring alterations observed in disease states.
Introduction
Human skin is the first line of defense against pathogens while simultaneously harboring a diverse milieu of commensals including bacteria, fungi, and viruses. These symbiotic organisms play essential roles in lipid metabolism, colonization resistance to transient organisms, and education of the immune system (Belkaid and Segre, 2014; Grice, 2015; Scharschmidt and Fischbach, 2013). Previous studies have shown a strong site-specificity to microbial community composition and function: the physiologic characteristics of a skin site, including pH, temperature, moisture, sebum content, and topography shape the local microbial community (Costello et al., 2009; Findley et al., 2013; Grice et al., 2009; Grice and Segre, 2011; Oh et al., 2014). Understanding community variability across skin sites has provided the foundation to study the corresponding site-specificity to disease predilection, for example atopic dermatitis (eczema) in the bends of the arms and legs (Kong et al., 2012) and psoriasis on the elbows and knees (Alekseyenko et al., 2013). However, it is poorly understood why disease predilection changes over human lifespans and whether fluctuations in host-intrinsic factors, such as immunity or hygiene, influence microbial community composition and function. Understanding stability determinants is critical to studies investigating if homeostatic forces contribute to a healthy skin microbial community and if alterations influence host health.
In addition to skin’s biogeography, as defined by physiologic factors such as sebaceous, moist, or dry, individual discriminatory attributes also likely contribute to skin microbial community dynamics over time. Using the high resolution and multi-kingdom analyses afforded by metagenomic shotgun sequencing, we have shown that low abundance microbial species including bacteria, fungi, and viruses can differentiate between individuals. Observing inter-kingdom dynamics is meaningful since these interactions may exacerbate disease severity (Peleg et al., 2010) or facilitate transitions from opportunistic to pathogenic. Moreover, a subspecies-level analysis of dominant skin species showed that strains can be unique to an individual, while the population heterogeneity of other species can be more specific to skin physiology (Oh et al., 2014). Distinguishing between strains of the same species is necessary because some strains are beneficial while others may be pathogenic to the host.
Shotgun sequencing data provides resolution difficult to achieve with phylogenetic marker gene analyses. Metagenomic surveys of the gut have shown that single nucleotide variants (SNVs) (Schloissnig et al., 2013) and gene copy number variations (Greenblum et al., 2015; Zhu et al., 2015) can identify individual-specific strains, which illustrate functional differences that cannot be explained by species composition alone. Longitudinal studies in the gut have found individual-specific strains persist for a year (Schloissnig et al., 2013) or more (Faith et al., 2013). These studies have leveraged a combination of compositional and functional attributes to identify a broad trend of long-term retention of one’s individual strains over time, even at the SNV level.
To understand community dynamics of healthy human skin, we investigated the temporal stability and diversity of skin microbial communities, expanding our previous metagenomic study to re-sample individuals at successive timepoints. Here, we show that community stability persists regardless of sampling time interval and despite constant exposure of skin communities to extrinsic factors. Interestingly, the nature and degree of this stability is highly individual-specific; a trend previously observed with phylogenetic marker gene sequencing across body sites (Flores et al, 2014; Gajer et al., 2012). Strain-level analysis reveals that this stability is driven primarily by the maintenance of individual strains over time, rather than through the acquisition of prevalent microbes from the environment and other individuals (Lax et al., 2014). We present new insights into how biogeography and individuality regulate stability and transience of the skin microbiome community.
Results
Skin microbes are largely stable at a community level
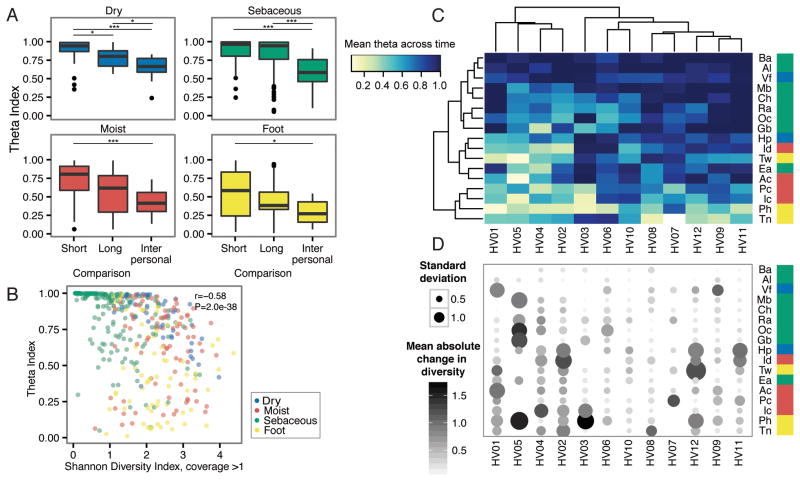
Our previous studies defined that the diversity and composition of skin microbial communities possess both site- and individual-specific qualities (Oh et al., 2014). To assess the effect of time on these characteristics, we collected samples over long (1–2 years) and short (1–2 months) time intervals. 12 healthy individuals were sampled across 17 skin sites at 3 timepoints for a total of 594 samples and 720 Gbp of shotgun microbial sequence data (Figure S1A–B). For taxonomic reconstructions, we mapped microbial reads to a multi-kingdom reference database. To assess the stability of skin microbial communities, we compared community membership and structure over short and long time intervals using the Yue-Clayton theta index, which calculates the distance between communities based on relative proportions of shared and non-shared species in each population (Yue and Clayton 2005). θ = 0 indicates dissimilar and θ = 1 identical communities. We observed that an individual’s short- and long-term community similarity significantly exceeded similarity between individuals (Figure 1A, Figure S1A), similar to observations in gut and other communities (Caporaso et al., 2011; Costello et al., 2009; Faith et al., 2013; Flores et al., 2014; Human Microbiome Project, 2012). At all sites, long-term was lower than short-term similarity at the species level; a trend also observed when comparing timescales of 1 day versus 3 months (Costello et al., 2009). Bacterial and fungal communities of sebaceous sites were the most stable regardless of time intervals. Surprisingly, dry sites including high-exposure, high-perturbation sites like the palm, were also stable regardless of time. Foot sites were the least stable, with significant differences over both short- and long-term. This may be due to a combination of behavioral and physiologic factors, including shoe-wearing habits, personal hygiene, or features such as the thickness of plantar stratum corneum (the upper layer of skin). We also examined stability at higher taxonomic levels to ask what level of taxonomic resolution contributes most to stability. We observed similar trends of long- and short-term stability to the phylum level, and differences in short and long-term stability disappeared at the kingdom level (data not shown).
Figure 1. Community Stability is Dependent on Both Site and Individuality.
Sites are color coded by characteristic and defined in Figure S1. (A) Boxplots of Yue–Clayton theta indices calculate similarity between sites aggregrated by characteristic. ‘Long’ duration indicates ~1–2 years between samplings; ‘Short’ duration averages a month (T2 to T3). For comparison, ‘Interpersonal’ values shows the average between individuals. Bonferroni-adjusted *P <0 .05 value, **P <0 .01 value, ***P < .001 value. (B) Quantification of community stability relative to community diversity by partial Spearman correlation. Diversity is measured by the Shannon Diversity Index, calculated for genomes with coverage greater than 1. (C) Hierarchical clustering (complete linkage) of mean theta indices over average of ‘long’ and ‘short’ values for each individual at each site sampled. (D) Mean change in community diversity over time, ordered as in (C) to show that community stability tracks with stability in diversity. Circle size shows the standard deviation of diversity, and color indicates magnitude of changes in diversity.
See also Figure S1.
We investigated next whether stability over time is linked to different community traits, such as diversity. Stability was modestly anti-correlated with community diversity (Figure 1B) as measured by the Shannon Diversity Index, which measures both species richness and evenness. This suggested that high-diversity sites (foot, moist) were likely to be less stable than low-diversity (primarily sebaceous) sites, consistent with previous reports (Grice et al., 2009). We then asked if skin communities typically reach and maintain species saturation over time, which could reflect a constancy in community diversity over time. Here, we examined trends of longitudinal diversity on a per-site, per-individual basis. No site exhibited statistically significant trends of diversity changes over time (P > 0.05, all sites). This suggested that community diversity is generally homeostatic in these healthy individuals, and if changes in the community occur over time, species may be replaced by similar types and numbers, preserving overall niche characteristics.
In certain individuals, we observed large shifts in diversity at multiple sites. This suggested that while skin microbial communities overall exhibit site-specific trends in stability, stability itself may be largely an individual trait, which has been observed previously in skin, gut, and vaginal communities (Flores et al., 2014; Gajer et al., 2012). To assess this, we clustered communities to identify potential individual-specific patterns in stability. Two-way hierarchical clustering of mean theta indices over time showed that sebaceous sites were stable irrespective of the individual, while foot and moist sites tended to be more variable, as previously observed (Figure 1C). However, a cluster comprised of 4/12 individuals (HV01, HV02, HV04, HV05) we defined as ‘high-variability’, because even at stereotypically stable sites (e.g., sebaceous), their communities tended to fluctuate more. This variability was even more pronounced at moist and dry sites. We then examined whether length of time between sampling was associated with this increased variability, as longer time intervals might increase stochastic drift and perturbation. Surprisingly, we observed no significant change in community composition nor magnitude of diversity as a function of days between ‘long’ sampling time intervals (Figure S1C–D). While certain sites (e.g., moist), trended towards increased variability as a function of time, we believe that individual factors may play a role in defining stability. We observed a similar trend when incorporating changes in community diversity over time (Figure 1D). ‘High-variability’ individuals also tended to have greater shifts in community diversity over time. Such shifts could reflect greater responsiveness to intrinsic or extrinsic perturbation events, for example, conditions that favor expansion of a limited set of species, bottleneck events, or large-scale selective sweeps at certain sites in these individuals.
Diverse signatures of skin microbe stability at the species level
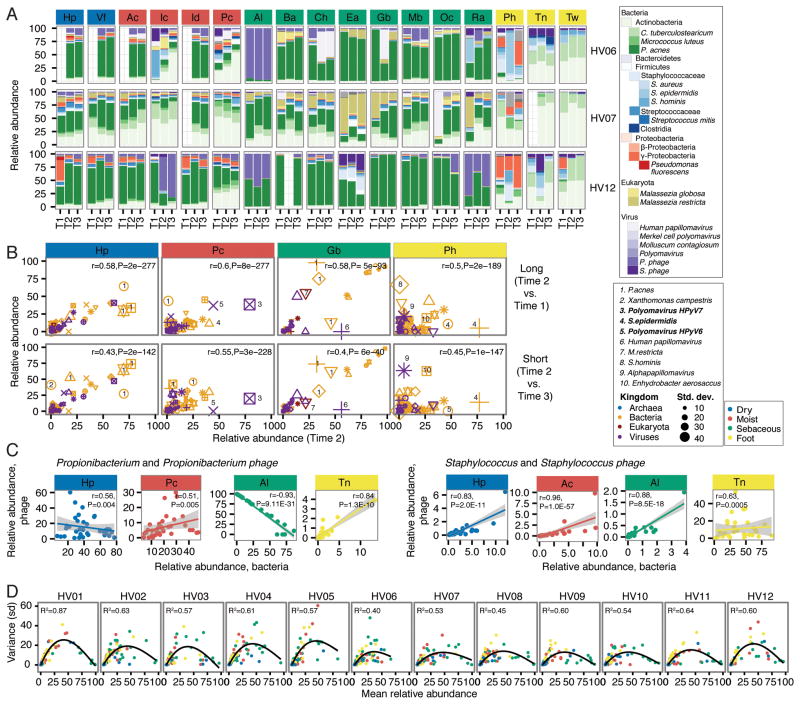
We then sought to identify if specific species or community types are intrinsically unstable in the skin niche, or if increased variability is a phenomenon where many species flux simultaneously in a community. For example, if a species is poorly adapted to a skin niche, relative abundances could vary significantly over time with growth competition. Or, in antagonistic interspecies relationships, cyclic changes in relative abundance could also occur. Community composition, describing the relative abundances of bacteria, fungi, and DNA viruses across samples confirmed that skin communities are largely stable over time (Figure 2A, Figure S2A). Of communities that tended to be less stable, we asked whether certain species were more likely to contribute to instability. Correlation analysis of relative abundances of skin species between timepoints in these sites showed that ‘high flux’ species, those that fluctuated more over time, differed by site and individual (Figure 2B). For example, in the dry hypothenar palm (Hp), we observed that Propionibacterium acnes was a high-flux organism for multiple individuals. In the moist popliteal crease, Polyomavirus HPyV7 was a high flux species for one individual, Polyomavirus HPyV6 for another, and Staphylococcus epidermidis for yet another. In the high-instability plantar heel, abundances of S. hominis, S. epidermidis, and Alphapapillomavirus changed significantly over time. While these species may flux in certain individuals and sites, most skin microbes appeared intrinsically stable. Thus, a complex interaction of host status and environmental perturbations likely influences these local changes in skin communities.
Figure 2. Community Stability is Defined by Different Species Characteristics, Depending on Site and Individual.
For all panels, sites are color coded by site characteristic. (A) Relative abundances of the most common skin bacteria, fungi, and viruses shown for 3 representative individuals. Full set of taxonomic classifications is shown in Figure S3. T1, T2, and T3 indicate order in time series. (B) Correlating taxon abundances for ‘Long’ vs ‘Short’ time series for select sites from (A) that showed higher variability. Different shapes represent different individuals; different colors represent kingdom of species, and size of point indicates species standard deviation between samples. Select abundant taxa (five most highly variable) are numbered as indicated. Partial Spearman correlations adjusting for repeated measures with associated P-values are in the upper right. (C) Phage and host dynamics at select sites. For Propionibacterium acnes and Staphylococcus, relative abundances of phage versus bacteria are plotted for each sample. A linear model was fitted to the data with shading showing the 95% confidence interval for the goodness of fit. To quantify correlation or anticorrelation, partial Spearman correlation coefficients and associated adjusted P-values were calculated. (D) The relationship between taxon abundance and variance, measured as standard deviation, is shown for taxa > 0.25% relative abundance in the population. A generalized additive model, showing a second order relationship, was fitted to the data to generate correlation coefficients (upper left). All P-values ≪0.05.
See also Figure S2.
We also examined phage-host relationships as a potential interspecies source of variability in microbial communities. While mixed phage-bacterial communities were typically stable (Figure 2A, Figure S2A), we observed instances of large-scale shifts in phage-bacteria ratios that could reflect a perturbation event, e.g., HV12. We investigated the relative abundances of the most common phages in skin (Figure 2C, Figure S2B) to search for relationships that might provide clues about bacteria-phage interactions. Both P. acnes and Propionibacterium phage are abundant in sebaceous sites, and we observed a strong anti-correlation in sebaceous communities that contain both P. acnes and its phage (Figure 2C). While the compositional (versus absolute) nature of the data means that taxon abundances are not perfectly independent, this anti-correlation together with the observed phage-host dynamics over time suggests antagonism.
We also observed multiple populations of P. acnes that had a neutral or positive correlation with Propioinibacterium phage abundance (Figure S2B, points along x-axis), potentially reflecting variability of P. acnes strains in skin communities in which some strains may be interacting with phages and others may not (Liu et al., 2015). Similar findings were also evident in dry and moist sites, most notably in the feet in which a positive correlation was observed. On the feet, Propionibacterium phages may interact with different Propionibacterium species (e.g., humerusii), or they may exist in a lysogenic versus a primarily lytic state in sebaceous sites. Relative abundances of Staphylococcus and Streptococcus phages were largely positively correlated with Staphylococcus and Streptococcus, suggesting that these may be prophages.
Finally, we examined species abundance as a predictor of stability over time (Figure 2D). We observed a second order, power-law relationship between taxon mean relative abundance across time and variance, excluding transient species (taxa that disappear from one timepoint to the next)(Ma 2015). Both low and high abundance species had low variance, and the largest variance was observed in intermediate abundance species. This second order relationship was present whether site or individual was used to track species, supporting that this intrinsic property of abundance is not determined solely by species identity. This suggests a broader ecological context in which high abundance species can be considered ‘fixed’ in the community, and a limited assortment of rare (but not transient) species can also stably persist. Overall, dominant community characteristics such as diversity and composition are largely stable over time, where primary determinants of stability are site and individual-specificity, with different species contributing to variation in the community over time.
Transience and eukaryotic dsDNA viral stability
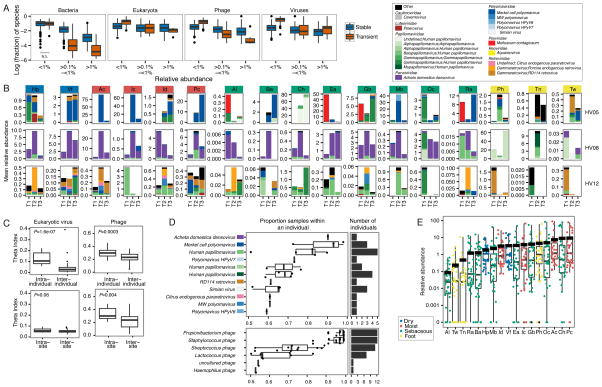
Because transient species also likely contribute to community variations over time, we next examined the fraction of the skin community that was transient, i.e. species that were below the detectable range between timepoints. Because technical limitations, including sampling depth, sequencing depth, and amplification bias, can influence detection particularly at low relative abundances, we measured transience at three abundance ranges (<0.1%, ≥0.1% to ≤ 1%, >1%) and required a species to occur in that range in at least one timepoint (Figure 3A). Of very low abundance organisms (<0.1% relative abundance), a significantly higher fraction consisted of transient organisms from all kingdom classes except bacteria, yet this could be partially attributed to reduced robustness in species classification of fungi and viruses at this abundance threshold. For higher abundance ranges, the proportion of stable species significantly exceeded the number of transient organisms for bacteria, fungi, and phages, suggesting that at higher abundances, species from these kingdoms were more likely to be stable residents.
Figure 3. Assessment of Transient vs. Non-transient Species Across Time by Kingdom, and Eukaryotic Viral Stability in the Skin.
For panels in (A and B), sites are color coded by site characteristic. (A) Fraction of species that are transient or not transient for each complete longitudinal set of sites per individual. Transience, defined as present in only one of three time points, is measured at three ranges of abundances. All comparisons were adjusted P-value < 0.05 between transient/non-transient except for Bacteria, <0.1%, which is marked not significant (N.S.). (B) Relative abundances of the most common predicted eukaryotic viruses are shown for 3 representative individuals. Full set of classifications is shown in Figure S3. T1, T2, T3 indicate order in time series. (C) Predicted eukaryotic viruses tend to be individual-specific, with significantly more sharedness, calculated with the Yue-Clayton theta coefficient, within an individual than between individuals and with little site-specificity. P-value, Wilcoxon rank-sum test shown in left corner. (D) Coreness of predicted eukaryotic viruses. Box plots and overlaid points show the proportion of samples within an individual in which a virus is observed, with a cutoff of >50% occurrence. For lineage, color code is based on panel (B) legend. Right panel shows number of individuals in which a eukaryotic virus is observed. (E) Relative abundance of eukaryotic viruses arranged by site with each individual’s value overlaid and colored by site characteristic. Thick black line indicates mean relative abundance of total (eukayotic and bacterial) DNA viruses at that site.
See also Figure S3.
Noteworthy, we observed that double stranded eukaryotic DNA viruses had a significantly greater fraction of species that were transient at the higher abundance classes (0.1–1% and >1%), in striking contrast to the stable residences of bacteria, fungi, and phages (Figure 3A). This is consistent with previous studies where viral communities were more unstable than the whole metagenome (Hannigan et al., 2015). Types of eukaryotic viruses identified in skin were relatively few (<two dozen, Figure S3A), with large fluctuations in number of species observed in some individuals. Taxonomic characterization of these viruses showed three major classes of communities (Figure 3B, Figure S3B). We observed a ‘uniform’ phenotype, in which the viral communities at all sites were predominantly a single viral type (Figure 3B, e.g., HV08). In an ‘intermediate’ phenotype, we observed a dominant species in a large number of sites but high diversity at other sites (Figure 3B, e.g., HV05). In a ‘variable’ phenotype, viral communities tended to differ markedly from site to site and over time (Figure 3B, e.g., HV12) at low abundances. Viruses’ lack of site specificity was reflected in comparisons using the theta index (Figure 3C, e.g., HV12), where eukaryotic viral communities were highly dissimilar between sites. Moreover, individuals tended to harbor a unique virome (Figure 3C), with intrapersonal similarity significantly exceeding interpersonal similarity.
Unlike bacterial and fungal species like P. acnes and Malassezia globosa, which are nearly universal both between and within individuals, we observed little evidence of a core virome in our cohort, although human polyomaviruses and papillomaviruses were identified in ~half of our subjects (Figure 3D, top). Generally, the most abundant viruses in our cohort were specific to single individuals, including retroviruses, which may be stably incorporated into the host human genome. By contrast, we observed the presence of a strong core phageome (Figure 3D, bottom), in which Propionibacterium, Staphylococcus, and Streptococcus phages were nearly universal between and within individuals. This underscores differences in host specificity in which phages are likely obligate partners with a community’s abundant bacterial species and thus have site specificity while eukaryotic viruses do not (Figure 3C).. As viral databases improve and expand, we anticipate that strain resolution of phages and eukaryotic viruses will further refine these interkingdom interactions.
Because eukaryotic viruses inhabiting the human body presumably infect human cells, we inquired if different skin sites had different abundances of eukaryotic viruses. For example, the thick skin of the foot might harbor fewer detectable viruses than a moist site, whose thinner layers may be more susceptible to skin barrier breaks. While shotgun sequencing methodologies prevent estimation of true abundances, we can gain some insight from the proportion of the community comprised of viral sequences. A significant increase in detectable viral DNA could signify a viral bloom relative to the bacterial and fungal community. Note that we observed no consistent trend with amount of human DNA recovered (data not shown), which could have skewed viruses recovered with higher or lower numbers of active human cells obtained in collection.
We created a ranked distribution of eukaryotic viral relative abundances stratified by skin site (Figure 3E). Certain sites, including alar crease, toeweb, toenail, retroauricular crease, and back, had a large fraction of samples with no viral representation and lower relative abundance than other sites. Sites with modestly higher viral relative abundances were moist skin creases like the popliteal and antecubital fossae (as well as adjacent sites on the forearm and palm), cheek, and occiput. But we observed little uniformity in what viruses tended to be higher in relative abundance at these sites; in most individuals, higher viral relative abundances could be attributed to a diversity of polyomaviruses or papillomaviruses. Interestingly, the composition and presence of eukaryotic viruses in skin communities appeared less dependent on physiologic characteristics that strongly shape bacterial and fungal communities; these dynamics may depend more on host status and topography than nutrient availability.
Individual-specific signatures are shared across time and space
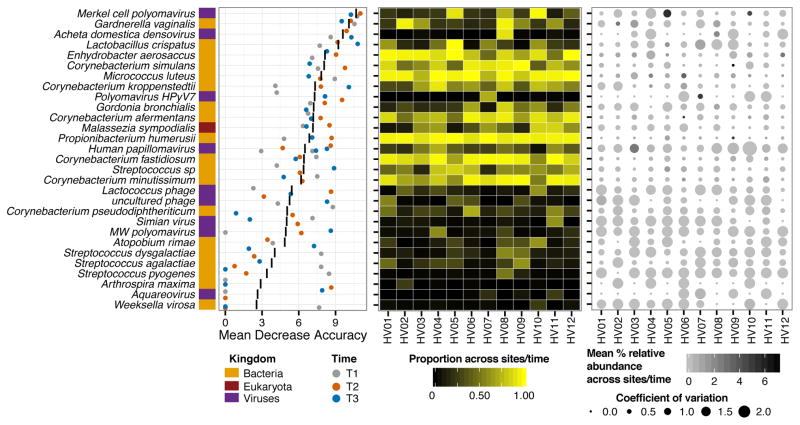
As overall stability is likely an individual trait, with the degree of stability and involved species varying between individuals, we next investigated whether individuals retain unique taxonomic signatures that would allow them to be distinguished from other individuals at different points in time. Recent research has suggested that individuals might possess distinguishing microbial fingerprints that are stable over time (Franzosa et al., 2015). Here, we used a supervised random forests algorithm (Liaw and Wiener, 2002) for each timepoint to identify key taxa that could differentiate individuals, and to identify if these taxa remained the same at each timepoint. Comparing classifier strength (mean decrease in accuracy) over the different timepoints, we observed that, surprisingly, a relatively small number of species retained their discriminatory power over time (Figure 4). Low abundance (<5% relative abundance) organisms were typically those with the highest resolving power between individuals, for example Merkel cell polyomavirus, Gardnerella vaginalis, and Acheta domestica densovirus (top half of Figure 4, right panel). These organisms also tended to be stable in relative abundance over time with low coefficients of variation (Figure 4, right), and were present at most sites within an individual across time (Figure 4, center). Species with minimal discriminatory power between individuals (bottom half of Figure 4) were characterized as lacking ability to discriminate individuals at one or more timepoints, having low prevalence across body sites, and having high variance in their relative abundances over time. The limited ability of species to discriminate individuals over time has been previously described by (Franzosa et al., 2015) who found strain-level codes were more robust to temporal variability.
Figure 4. Individual-specific Signature Taxa are Shared across Time, Sites, and are Typically Low Abundance and Highly Stable.
Left, variable importance plot of supervised random forest algorithm output. Top 30 taxa contributing to model are shown on the y-axis ranked by importance, quantified by the mean decrease in accuracy; i.e., degree to which inclusion of this predictor in model reduces classification error. Black line indicates the mean decrease in accuracy across the three timepoints; colored points indicate the score for each timepoint. Center, for each individual, proportion of body sites for which each taxa is present, averaged across timepoints. Right, variation in mean relative abundance across sites and timepoints (color) with coefficient of variation.
Individuals have distinct microbial SNV signatures that are stable over time
Shotgun metagenomic data empowers unprecedented resolution of microbial communities at the strain and SNV level. To explore stability at this high resolution, we focused on the prevalent skin bacterial commensals Propionibacterium acnes and Staphylococcus epidermidis, which have dozens of available genome sequences (Conlan et al., 2012; Tomida et al., 2013). Previously, by mapping shotgun metagenomic reads to this reference set of phylogenetically diverse genomes, we identified that skin sites harbor complex subspecies variation for both P. acnes and S. epidermidis. Strain heterogeneity could be unique to an individual (P. acnes) or specific to skin site (S. epidermidis) (Oh et al., 2014).
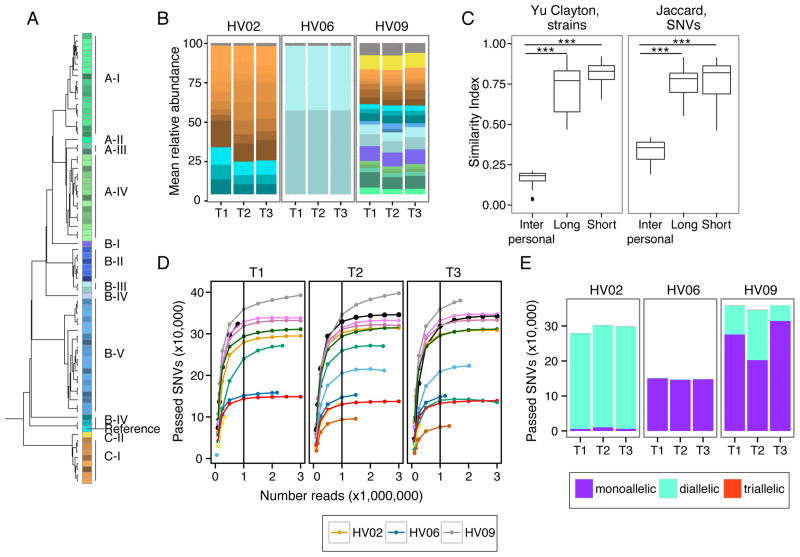
Here, we observed that P. acnes strains are remarkably stable over time across body sites (Figures 5A and S4A). We quantified this stability with the Yue-Clayton theta similarity index (Figure 5B), taking into account both strain presence/absence and relative abundance. Temporal stability, short- or long-term, surpassed the similarity between individuals, indicating that P. acnes stability likely derives from the maintenance of an individual’s strains over time and less from the acquisition of new strains from the environment or other individuals.
Figure 5. Individual Specific Strain and SNV Signatures are Stable Over Time.
(A) Dendrogram of P. acnes strain genome similarity based on core SNVs. (B) P. acnes strain relative abundance plots of 3 representative individuals’ manubrium, colors as in (A). Full set of strain classifications is shown in Figure S6B. (C) Boxplots of Yue-Clayton (left) and Jaccard (right) theta indices indicate similarity between strains (all body sites) and SNVs (manubrium and back) of P. acnes in a time series (θ = 1 is identical). ***P < .001 value, Wilcoxon rank-sum test. (D) Rarefaction curves demonstrate core SNV accumulation with read subsampling for manubrium sites. For remaining panels, SNVs are reported for samples subsampled to 1 million reads. Colors correspond to individuals as shown in Figure S1B. (D) Number of SNVs that are mono, di, and triallelic. T1, T2, and T3 indicate order in time series. HV is healthy volunteer. See also Figure S4.
To validate this hypothesis and to assess within-subject retention of strains, we also examined shared SNVs in the P. acnes genomes across the longitudinal samples. To power this analysis, we focused on two sebaceous sites, the manubrium and back, which have high sequencing depth and P. acnes abundance (Figure S4C, S4D). For these sites, we used SNVs specific to the P. acnes core genome (2,248,676 bps region of the genome shared between all 78 sequenced strains) to ensure even representation of SNVs in every metagenomic sample. We identified 83,081 variant positions in the P. acnes core or ~24,000 SNVs per sample after filtering variants for an allele frequency > 1% and > 4 read depth. To test if P. acnes sequencing depth was sufficient to identify the majority of variants, we generated rarefaction curves of SNVs discovered over increasing read depths (Figure 5C). We found that 1 million reads, 40X coverage of the P. acnes core, was sufficient for variant discovery.
The major advantage of an SNV-based approach is that given sufficient sequencing depth, temporal stability and genetic diversity can be estimated in the absence of a large number of sequenced reference strains. By comparing the sharedness of SNVs between timepoints or individuals using the Jaccard index, which measures similarity based on presence/absence of features, we found the stability of P. acnes SNVs mirrors that of P. acnes strains over time (Figure 5B). We found that regardless of duration, an individual shares significantly more SNVs with themself over time than with other individuals (P-value<0.001).
Finally, polyallelism can reflect genetic heterogeneity, i.e., the presence of multiple strains, in a community. While the number of diallelic sites does not scale linearly with the number of strains in a sample, low levels are indicative of a mono-colonized population. We focused on diallelic states, as triallelism was extremely rare (<1.0% in the population). We calculated the cumulative distribution of alternate alleles derived from the shotgun metagenomic reads as a function of distance from a defined reference genome (Figure 5D, Figure S4E). Through these analyses, we identified a putatively monocolonized individual with strikingly low diallelism (1.0% of sites), in contrast to other individuals, e.g., HV02 and HV09, in which 97.4% and 25.6% of alternate sites have reads reliably mapping to both reference and an alternate allele indicating the presence of multiple strains. For future disease studies, fluctuations in pollyallelism, for example, a dramatic decrease in the number of diallelic positions could indicate emergence of a dominant pathogenic strain.
P. acnes pangenome maximized across a multi-phyletic community
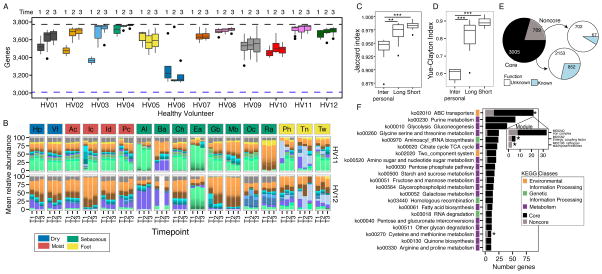
Since skin sites stably maintain the same P. acnes strains over time, we wanted to explore the community’s full gene content to generate hypotheses about evolutionary forces shaping community drift, resilience, and stability on a functional level. A species’ total functional repertoire is the ‘pangenome’, composed of core (conserved) and non-core (absent in at least one strain) genes. The P. acnes pangenome is composed of 3,774 non-redundant gene clusters, of which 1,685 were core (Figure S5A–B). Each additional genome adds 3 novel genes to the total (Figure S5C) (Tomida et al., 2013), implying that the majority of P. acnes functional capacity is captured within these 78 reference genomes. We mapped our reads to this P. acnes pangenome database, requiring 1 million reads for adequate sequencing depth (Figure S5D). Between 82.6 (3,117) and 99.9% (3,771) of the known P. acnes pangenome (Figure 6A) was represented in healthy individuals. 70% of samples had > 95% (3,585) of the pangenome.
Figure 6. P. acnes Pan-genome Reaches Functional Saturation with Distinct Strain Combinations.
(A) Boxplots show number of genes present at > 40% coverage across sites of individuals at 3 time points. Black dashed line indicates the 3,774 genes in P. acnes pangenome. Blue dashed line indicates the 3,005 genes present in every individual at every time point in at least 50% of body sites. Sites with < 1 million P. acnes reads were excluded. (B) Relative abundance plots of P. acnes strains across all body sites, color-coded by site characteristic. Strain colors defined in Figure 5A. T1, T2, and T3 indicate order in time series. (C) Boxplots of Jaccard theta indices indicate stability of gene presence over time (θ = 1 is identical). **P <0 .01, ***P < 0.001, Wilcoxon rank-sum test. (D) Boxplots of Yue-Clayton theta indices indicate the stability of gene copy numbers over time (θ = 1 means identical). (E) Pie chart indicates distribution of P. acnes genes between those present in all individuals “core” and those absent from some individuals “noncore”. Majority of core and noncore genes are pathway “unknown” when compared to a KEGG database. (F) Distribution of core and noncore genes for prevalent KEGG pathways. Colors indicate broader KEGG class of each pathway. Pathways with * are functionally enriched in noncore based on Fisher exact test with FDR <0.05. See also Figure S5.
Interestingly, combining functional capacity with strain signatures revealed that similar pangenomic capacity can be achieved with distinct strain combinations (Figure 6A, 6B). This suggests that functional niche saturation can occur through multiple combinations of a limited number of strains, rather than requiring a full phylogenetic complement. Thus, while individuals have distinct P. acnes strain signatures, their functional capacities are remarkably similar, only differing 5% between individuals, and an individual is more likely to retain those unique genes over time (Figure 6C). This percent difference between individuals increases to ~40 when relative copy numbers of genes are compared (Figure 6D), further illustrating that while individuals’ microbial communities have the same set of genes their relative abundances vary.
In addition to gene retention, we also observed an increase in pangenome size in three of the individuals (HV01, HV02, and HV03) over time (Figure 6A). Thus, although strain stability is more typical, individuals can acquire new gene content over time. Because of convergent functionality between individuals, we redefined the core genome to represent a ‘functional’ core that is characteristic of healthy communities, towards which a probiotic approach might strive. We defined that 2,982 genes were present in at least 50% of sites in all individuals at all times, exceeding the 1,860 genes that are derived using genomes alone. This functional core genome increases to 3,186 genes when we exclude HV06 whose strain community is predominated by a single P. acnes clade (Figure S4A).
To evaluate functional enrichment within the 769 genes not identified as core, we assigned KEGG pathway annotations to clusters with BLAST. Unsurprisingly, these noncore genes were statistically enriched for pathway “None” (702 of 769 genes, Figure 6E), underscoring that more extensive gene annotations are needed to better understand functional variation. After removing unannotated clusters, we found accessory genes to be enriched in functions associated with ABC transporters and cysteine and methionine metabolism (Figure 6F). Ubiquitous across bacteria, ABC transporters facilitate communication between bacteria and the environment through the active transport of substances such as ions, sugars, lipids, proteins, and drugs across membranes, contributing to nutrient sensing and other processes.
Multi-phyletic S. epidermidis communities have more variable gene content
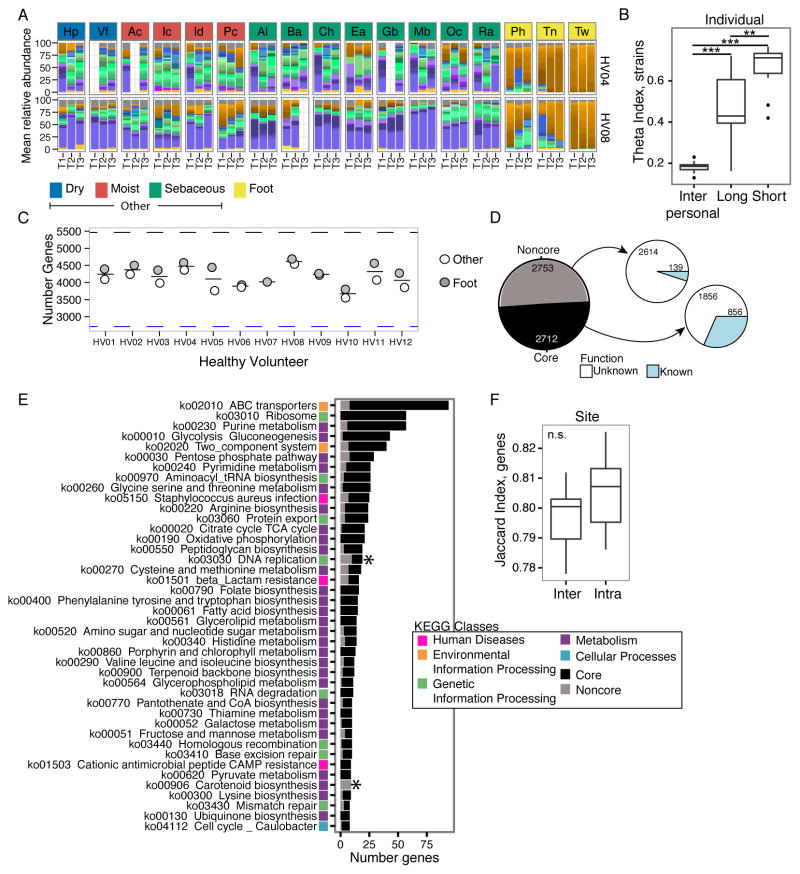
To determine if our observations in P. acnes extended to other members of the skin community, we applied our analyses to evaluate the stability of S. epidermidis strains over time. Like P. acnes, S. epidermidis is a common skin commensal with well-documented sequence and gene content variation (Figure S6A). Multi-phyletic communities of S. epidermidis strains are stably maintained over time irrespective of body site (Figures 7A and S6B); strain similarity over the long- and short-term exceeded similarity between individuals (P-value< 0.001, Figure 7B), suggesting that new strains are rarely acquired from outside sources. Because S. epidermidis is maintained at an overall lower abundance on the skin than P. acnes (<10% versus 40%), we lacked sufficient depth for SNV analyses.
Figure 7. S. epidermidis Strains Remain Stable over Time and Communities do not Reach Gene Saturation.
(A) Relative abundance plots of S. epidermidis strains across body sites, color-coded by site characteristic. Full set of taxonomic classifications is shown in Figure S6B. Strain colors defined in Figure S6A. Due to coverage S. epidermidis, foot sites were combined as “foot” and all moist, dry, and sebaceous sites were combined as “other” (Figure S6E). Combined samples with <1 million S. epidermidis reads were excluded. (B) Boxplots of theta indices indicate the stability of S. epidermidis strains within an individual over long or short time compared to between individuals (Inter) (θ = 1 means identical). **P < 0.01; ***P < 0.001, Wilcoxon rank-sum test. (C) Number of genes present on foot and other body sites, with mean number shown as thin bar. Black dashed line indicates the 5,465 genes present in S. epidermidis pangenome. Blue dashed lines indicate the 2,712 genes present in all foot and other combined samples. (D) Pie chart indicates distribution of S. epidermidis genes between those present in all individuals “core” and those missing from some individuals “noncore”. Majority of core and noncore genes are pathway “unknown” when compared to a KEGG database. (E) Distribution of genes between core and noncore for the most prevalent KEGG pathways. Colors indicate the broader KEGG class of each pathway. Pathways indicated with * are functionally enriched in noncore based on Fisher exact test with FDR < 0.05. (F) S. epidermidis gene repertoire is not statistically more similar within a site (‘Intra’) than between (‘Inter’) sites, calculated with Jaccard theta. See also Figure S6.
The S. epidermidis pangenome, assembled from 61 reference genomes, is larger than P. acnes, containing 5,465 unique clusters and a noncore of 3,583 genes (Figure S6C) with 23 genes added from each additional genome (Figure S6D). To achieve sufficient coverage for pangenome analyses, S. epidermidis reads from three foot sites were pooled by individual to create ‘foot’, while non-foot body sites were pooled to yield a composite “other” sample (Figure S6E). This grouping was based on B clade S. epidermidis dominance on all foot sites (Figure S6B). Using samples with greater than 1 million S. epidermidis reads (Figure S6F), we found that individuals did not appear to reach gene saturation, and generally possessed from 65 to 85% (3,552 to 4,693 genes) of the available pangenome (Figure 7C). In addition to lower saturation of the pan-genome, S. epidermidis gene content varied 20% between individuals, in contrast to 5% for P. acnes. Accordingly, our newly defined S. epidermidis healthy ‘core’ is 2,712 genes. Unique genes could encode similar functions, but be unrecognized as homologs, which could increase the number of genes in the S. epidermidis pan-genome, leading to lower full representation. However, the differences in the functional/gene saturation may also be explained by the relatively narrow niche of the sebaceous gland where P. acnes primarily resides. S. epidermidis has a broader range, which could be reflected in the larger complement of genes needed for strains to persist in a niche.
To examine the functional diversity of the healthy noncore, we examined KEGG annotations for enrichment. Similar to P. acnes, the noncore was statistically enriched for pathway “None”, with only 139 annotations for 2,753 genes (Figure 7ED. Of annotated clusters, we identified DNA replication and carotenoid biosynthesis as enriched within the noncore (Figure 7E). Other enriched pathways demonstrated significant trends, including beta-lactam resistance, cysteine and methionine metabolism, bacterial secretion system, vancomycin resistance, and steroid hormone biosynthesis. Discovering multiple mechanisms of drug resistance in the noncore was unsurprising, given that individuals have varied histories of antibiotic usage, and intraspecies transfer of drug resistance is common. Finally, we looked for functional differences in the foot compared to non-foot sites and found overall gene content was not statistically significantly different (Figure 7F), despite the presence of 212 genes specific to foot and 58 genes to non-foot sites.
Discussion
Despite the continuous perturbation that human skin undergoes in daily life, healthy adults stably maintain their skin communities for up to two years, similar to the stability observed in the gut (Faith et al., 2013; Schloissnig et al., 2013). Homeostasis of skin microbial communities is largely maintained by fixation of abundant species, although a smaller number of low abundance species are also stably maintained and contribute to an individual’s unique microbial signature. We suspect that larger, longer-term studies will show a larger reservoir of transients entering and exiting the community, consistent with previous observations of individuals sharing and receiving microbes from the home and other individuals (Lax et al., 2014). Such stochastic drift likely increases over time, unless other constraints, like geographic restriction, lifestyle, or host immune surveillance narrow the transient pool.
The exception to low abundance transients was the class of DNA eukaryotic viruses, which displayed little sharedness across individuals and less site-specificity than bacterial and fungal communities, which are strongly shaped by the nutritional availability, pH, and moisture of skin microenvironments. Eukaryotic viruses occupy a different niche (human cells) and interact with different constraints within the complex skin ecosystem. Interestingly, even high-abundance viral species were observed transiently, at least within the detection limits of metagenomic sequencing. This further supports a hypothesis for differential regulation and selection for eukaryotic viruses. However, viral communities are unusually complex to characterize due to technical limitations in recovery and classification; further computational and experimental approaches will be valuable to refine viral dynamics in the skin.
We surmise that in the absence of major perturbations, dominant characteristics of skin microbial communities would remain stable indefinitely, a conclusion previously extrapolated for gut communities (Faith et al., 2013; Schloissnig et al., 2013). This stability extends beyond the species level into SNVs and strains, which can impart unique functional contributions to a niche or individual. Total functional content variation, however, differed depending on skin species—P. acnes, a dominant skin commensal, showed low content variation in comparison to S. epidermidis. However, integration of metagenomic, or ‘coding’ potential with transcriptional or metabolomics profiles may better delineate community function, as SNVs and small variants can impact actual functional levels.
Future studies will define what and how extrinsic perturbations can alter the skin microbiota; these include antimicrobial treatment (e.g., Naik et al., 2012), probiotics, prebiotics, long-term environmental relocations, or diet (Kang et al., 2015). Intrinsic conditions, like immunosuppression, illness, or the occurrence of disease, have also been shown to cause major shifts in skin communities (Kong et al., 2012; Oh et al., 2013). In future disease studies, sequence data can generate hypotheses about which strains contribute to the disease and which are bystanders in the greater microbial consortia. Subsequently, valuable functional information can be gained from culturing and sequencing of primary isolates associated with metagenomic datasets. Functional assaying of individual and mixed strain groups in vitro and in animal models will be particularly relevant for determining the causality of diseases. Such studies are the prelude to prebiotic, probiotic and transplantation approaches of skin microbes in the context of disease amelioration and prevention.
Experimental Procedures
Subject Recruitment, Sampling, Processing, Sequencing and Classification
Expanding our previous metagenomic survey, we re-sampled 12 healthy volunteers from our original study (Oh et al., 2014) approved by the Institutional Review Board of the National Human Genome Research Institute (http://www.clinicaltrials.gov/ct2/show/NCT00605878). Longitudinal sampling occurred at 10–30 months (‘long’), and 5–10 weeks (‘short’) (Figure S1B). Recruitment criteria, sampling procedure, sample processing, sequencing, taxonomic classification and strain tracking were as described previously, except for a new Refseq viral genome database as of 06.2015.
Identification of SNVs in the P. acnes core
For high coverage back and manubrium samples (Figure S4C), metagenomic reads were mapped against the P. acnes core genome and the alignment file was processed with GATK’s IndelRealigner (McKenna et al., 2010), samtools (Li, 2011), and bcftools to identify possible variants with criteria previously described (Lieberman et al., 2014).
Pangenome analyses of dominant skin species
As illustrated in Figure S5A, P. acnes and S. epidermidis nucleotide-coding sequences were clustered into non-redundant orthologs with usearch (Edgar, 2010) and validated with single copy marker genes (Greenblum et al., 2015). A gene was considered present when 40% of its length was covered with reads (Zhu et al., 2015).
Statistics
All statistical analyses were performed in R. Data are represented as mean ± standard error of the mean unless otherwise indicated. Spearman correlations of non-zero values were used for all correlations. Supervised random forest models were implemented with the package random Forest (Liaw and Wiener, 2002). For all boxplots, center lines represent the median and edges the first and third quartiles. The nonparametric Wilcoxon rank-sum test was used to determine statistically significant differences between populations. Statistical significance was ascribed to an alpha level of the adjusted P-values ≤ 0.05. Similarity between samples was assessed using the Yue–Clayton theta or Jaccard similarity.
Data deposition
Metagenomic read data are available from SRA under bioproject 46333. Scripts and supplemental data available at github https://github.com/julia0h/longitudinal.git.
Supplementary Material
Acknowledgments
Amynah Pradhan, Sharon Osgood, Clay Deming, Cynthia Ng, Brian Schmidt, Pamela Thomas and Sean Conlan provided underlying efforts; Segre lab and Mark Udey engaged in helpful discussions. Work was supported by NHGRI and NCI Intramural Research Programs, and Chanel/CE.R.I.E.S. research award to J.A.S. Study utilized the high-performance computational capabilities of the NIH Biowulf Linux cluster. Sequencing was funded by grants from National Institutes of Health (4UH3AR057504-02). IGS Analysis Engine at University of Maryland School of Medicine provided structural and functional annotation of genomes.
Footnotes
Author contribution
J.O., A.L.B., H.H.K., J.A.S. designed study and drafted manuscript. H.H.K. collected patient samples. Sequencing was carried out by NISC. J.O., A.L.B., M.P. analyzed sequence data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alekseyenko AV, Perez-Perez GI, De Souza A, Strober B, Gao Z, Bihan M, Li K, Methe BA, Blaser MJ. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. Moving pictures of the human microbiome. Genome biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Mijares LA, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, et al. NISC Comp Seq Program. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome biology. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Colombel JF, Gordon JI. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:633–640. doi: 10.1073/pnas.1418781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, et al. Program NISC Comp Seq Program. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, Leff JW, Vazquez-Baeza Y, Gonzalez A, Knight R, et al. Temporal variability is a personalized feature of the human microbiome. Genome biology. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan JM, Huttenhower C. Identifying personal microbiomes using metagenomic codes Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, et al. Relating the metatranscriptome and metagenome of the human gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2329–2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, et al. Temporal dynamics of the human vaginal microbiota. Science translational medicine. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen K, Orvis J, Daugherty S, Creasy HH, Angiuoli S, White O, Wortman J, Mahurkar A, Giglio MG. The IGS Standard Operating Procedure for Automated Prokaryotic Annotation. Stand Genomic Sci. 2011;4:244–251. doi: 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome research. 2015;25:1514–1520. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, et al. NISC Comp Seq Program. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nature reviews Microbiology. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, Minot S, Bushman FD, Grice EA. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio. 2015;6:e01578–01515. doi: 10.1128/mBio.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Shi B, Erfe MC, Craft N, Li H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Science translational medicine. 2015;7:293ra103. doi: 10.1126/scitranslmed.aab2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, et al. Program NISC Comp Seq Program. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome research. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by random Forest. R News. 2002;2:18–22. [Google Scholar]
- Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nature genetics. 2014;46:82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yan R, Zhong Q, Ngo S, Bangayan NJ, Nguyen L, Lui T, Liu M, Erfe MC, Craft N, et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. The ISME journal. 2015;9:2116. doi: 10.1038/ismej.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZS. Power law analysis of the human microbiome. Mol Ecol. 2015;24:5428–45. doi: 10.1111/mec.13394. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA Program NISC Comp Seq Program. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Freeman AF, Park M, Sokolic R, Candotti F, Holland SM, Segre JA, Kong HH Program NISC Comp Seq Program. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome research. 2013;23:2103–2114. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nature reviews Microbiology. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- Scharschmidt TC, Fischbach MA. What Lives On Our Skin: Ecology, Genomics and Therapeutic Opportunities Of the Skin Microbiome. Drug discovery today Disease mechanisms. 2013;10 doi: 10.1016/j.ddmec.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, Li H. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio. 2013;4:e00003–00013. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JC, Clayton MK. A similarity measure based on species proportions. Comm Statist Theory Methods. 2005;34:2123–2131. [Google Scholar]
- Zhu A, Sunagawa S, Mende DR, Bork P. Inter-individual differences in the gene content of human gut bacterial species. Genome biology. 2015;16:82. doi: 10.1186/s13059-015-0646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.