Abstract
The lung is constantly exposed to environmental atmospheric cues. How it senses and responds to these cues is poorly defined. Here, we show that Roundabout receptor (Robo) genes are expressed in pulmonary neuroendocrine cells (PNECs), a rare, innervated epithelial population. Robo inactivation in mouse lung results in an inability of PNECs to cluster into sensory organoids and triggers increased neuropeptide production upon exposure to air. Excess neuropeptides lead to an increase in immune infiltrates, which in turn remodel the matrix and irreversibly simplify the alveoli. We demonstrate in vivo that PNECs act as precise airway sensors that elicit immune responses via neuropeptides. These findings suggest that the PNEC and neuropeptide abnormalities documented in a wide array of pulmonary diseases may profoundly affect symptoms and progression.
In humans, approximately 5 to 8 liters of air passes in and out of the lung per minute when resting. The air can vary in oxygen and CO2 concentration, may carry allergens, and confers different extents of mechanical stretch of the airway and gas-exchange surfaces. These signals are sensed, relayed, and processed into physiological outputs such as the control of pulmonary blood pressure, immune responses, and breathing rhythm, but the mechanism is unclear. Pulmonary neuroendocrine cells (PNECs) are found in a wide array of organisms from fish to mammals (1). In the mammalian lung, PNECs are the only innervated airway epithelial cells and represent less than 1% of the total lung epithelial cell population (2). Although in vitro evidence has implicated PNECs in oxygen sensing, bronchial and vascular smooth muscle tonus, and immune responses (1, 3), these roles have not been demonstrated in vivo. A recent study showed that genetic ablation of PNECs in the adult did not compromise homeostasis or airway repair, leaving in question the in vivo importance of these cells (4). PNEC pathologies, in particular an increase in PNEC number, have been documented in a large array of lung diseases, including asthma, bronchopulmonary dysplasia, cystic fibrosis, chronic obstructive pulmonary disease, congenital diaphragmatic hernia, neuroendocrine hyperplasia of infancy, sudden infant death syndrome, and pulmonary hypertension (5–8). In each case, it remains unclear whether the PNEC increase is a cause for or the consequence of symptoms.
In mouse lung, most PNECs reside in clusters of ~3 to 20 cells called neuroepithelial bodies (NEBs) (3, 9). Both solitary and clustered PNECs contain dense core vesicles, filled with bioactive neuropeptides such as calcitonin gene-related peptide (CGRP) or amines such as serotonin (1). These are released in response to stimuli, such as changes in oxygen level. Neuropeptides and amines have been implicated in some of the same processes as PNECs (10–12), raising the possibility that they may mediate PNEC function. However, a causal link has not been demonstrated in vivo.
We initiated the current study to uncover the mechanisms underlying congenital diaphragmatic hernia (CDH), a birth defect associated with considerable lung dysfunction, including heightened immune response and pulmonary hypertension (13). In a genetic mouse model of CDH, we uncovered a defect of failed PNEC clustering. This is followed by a sequence of events: an increase in PNEC neuropeptides, an increase in immune infiltrates, and remodeling of lung structure. These findings offer an in vivo demonstration of PNEC function. Because changes in PNEC number and associated neuropeptides have been documented in many lung diseases, our results have wide implications beyond CDH.
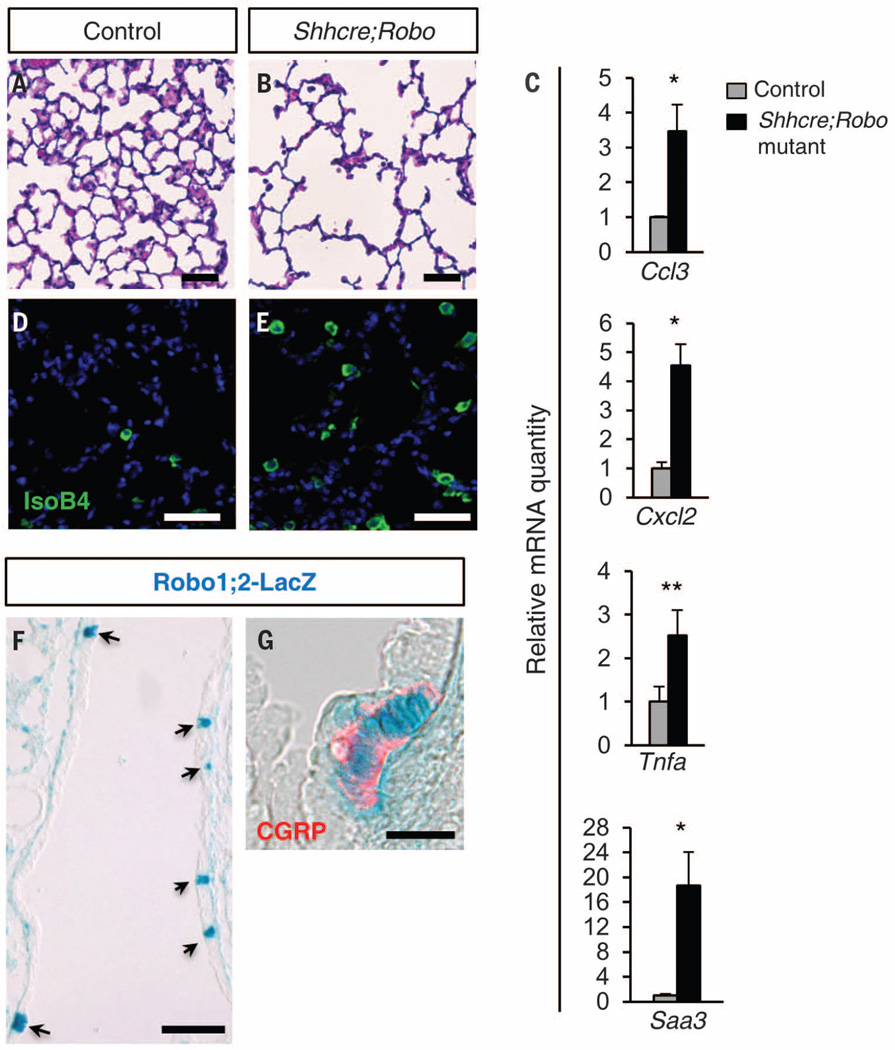
In humans, mutations in roundabout receptor (ROBO) genes have been associated with CDH (13, 14). To study the lung defects associated with CDH, we inactivated both Robo1 and Robo2 in endoderm-derived epithelium, including the lung, using Shhcre (hereafter Shhcre;Robo mutant) in mice (15, 16). Although these mutants survive, they exhibit reduced gas-exchange surface area starting at postnatal day (P) 15 (Fig. 1, A and B, and fig. S1). We performed microarray followed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) at P7, before reduction of gas-exchange surface. Fifteen of the top 20 differentially expressed genes have been implicated in immune responses, and all are significantly increased, including Ccl3, Cxcl2, Tnfa, and Saa3 (Fig. 1C). Consistent with this signature, we observed elevated numbers of immune cells, including neutrophils, eosinophils, macrophages, and T cells (Fig. 1, D and E, and fig. S2). Furthermore, there is an increase in the proportion of M2 and a decrease in the proportion of M1 macrophages (fig. S3). These findings indicate that Shhcre;Robo mutants show heightened immune sensitivity, mimicking a common CDH comorbidity (13).
Fig. 1. Robo mutants exhibit alveolar simplification and heightened immune response.
(A and B) Hematoxylin and eosin (H&E) staining of alveolar region at P22. (C) qRT-PCR of P7 lungs. (D and E) IsoB4 labeling of macrophages at P22. (F and G) X-Gal–stained heterozygous Robo1lacZ/+;Robo2lacZ/+ lungs. (F) P0, arrows indicate expression in PNECs. (G) E15.5, with anti-CGRP immunostaining (red). *P < 0.05; **P < 0.001.
Although Robo is expressed in the alveolar region of the lung mesenchyme (fig. S4), its expression in the epithelium is restricted to rare cells along the airway (Fig. 1F). Colabeling with CGRP antibody revealed that Robo-expressing epithelial cells are PNECs (Fig. 1G). To confirm that Robo genes are required within PNECs for function, we inactivated Robo using Ascl1creERT2 (17), a knock-in cre driver that confers PNEC-specific activity in the lung epithelium (fig. S5). We found that Ascl1creERT2;Robo mutants exhibited both alveolar simplification and macrophage increase, recapitulating the Shhcre;Robo phenotypes (fig. S6). These findings together demonstrate that Robo is required specifically in PNECs for restricting immune cell number and preventing alveolar simplification.
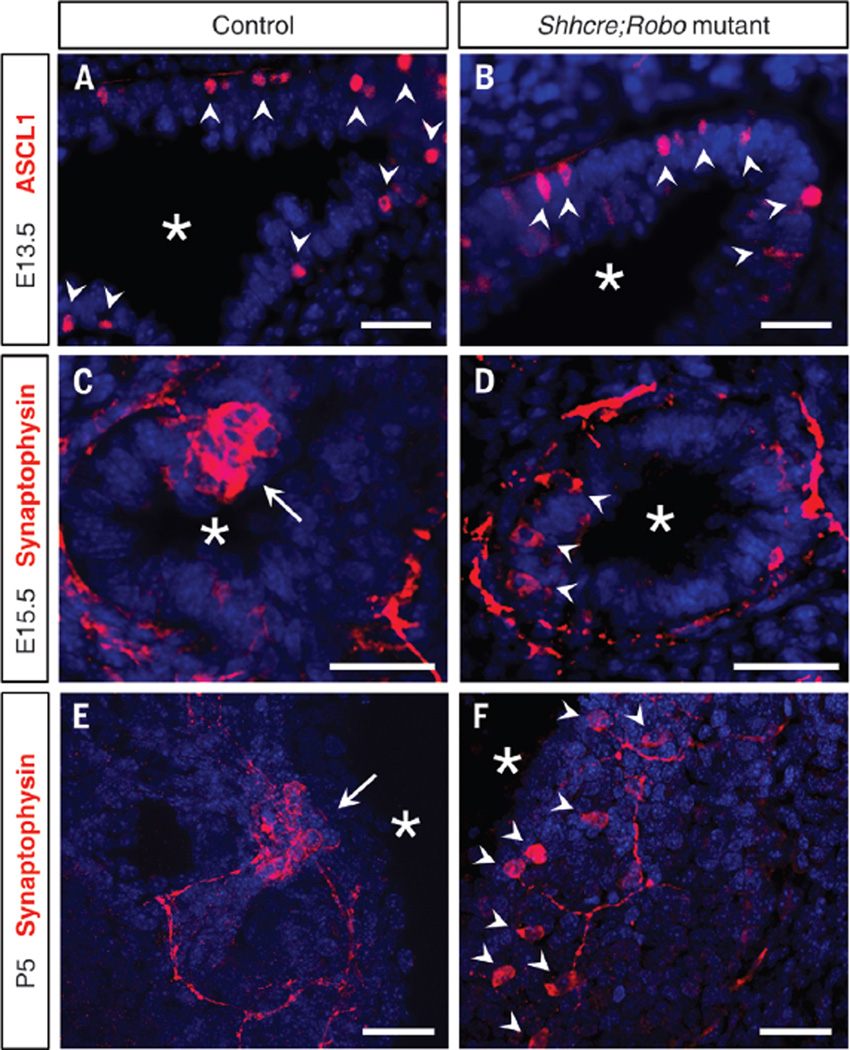
At embryonic day (E) 13.5, newly specified PNECs were solitary cells in both control and Shhcre;Robo mutant lungs (Fig. 2, A and B). By E15.5, a majority of PNECs had aggregated into NEBs in the control. However, PNECs were not clustered in Shhcre;Robo mutants (Fig. 2, C and D). This highly penetrant phenotype persisted in postnatal lungs (Fig. 2, E and F, and fig. S7). Total PNEC cell number appears unaffected, as supported by normal expression of Ascl1 and other PNEC markers (fig. S8). Unclustered cells in the mutant lose their wedge shape and are more rounded (figs. S7 and S9). Furthermore, in contrast to controls, where solitary PNECs are not innervated (9), ~33.3% (31 of 93 cells) of unclustered PNECs are innervated in the mutant (Fig. 2F and fig. S9), suggesting that PNEC innervation is not dependent on cluster formation or on Robo function in PNECs.
Fig. 2. Robo1;2 are required for PNEC clustering.
Immunostained vibratome lung slices. (A and B) ASCL1 immunostaining labels nascent PNECs. (C to F) Synaptophysin immunostaining labels differentiated PNECs and their associated nerves. Arrowheads indicate solitary PNECs, arrows indicate clustered PNECs in NEBs, and asterisks indicate airway lumen. Scale bars, 30 µm.
Ascl1creERT2;Robo mutants also exhibit PNEC unclustering (fig. S10). This phenotype manifested even when Robo inactivation was induced postnatally, which is subsequent to NEB formation. Together, our results establish that Robo is required for PNEC assembly and maintenance in NEBs.
Robo can function either dependently or independently of its ligand, Slit (18). Analysis of Slit mutants show that, whereas PNEC clustering is not affected in any of the single mutants, it is reduced in Slit1;3 mutants (fig. S11, A to D). This result suggests that Robo function in this process is likely ligand dependent.
Slit and Robo function primarily to mediate cellular repulsion and rarely attraction (19). To determine whether Slit acts as a repulsive or attractive signal for Robo-expressing PNECs, we first determined where Slit genes are expressed. Combined Slit1;2-GFP (green fluorescent protein) reporter revealed expression in only about 1 to 3 PNECs within large NEBs, raising the possibility that the Slit1/2-expressing cells may be the nucleating cells of the cluster (fig. S11E). This also indicates PNEC subspecialization within a cluster. Slit3 expression is restricted to the vascular smooth-muscle-cell layer surrounding arteries, which runs alongside the main bronchi where most NEBs are found (fig. S11, F and G). Together, the close proximity of Slit-expressing cells to Robo-expressing PNECs raised the possibility that Slit ligands may provide an attractive cue for PNECs.
To test this, flow cytometry sorted GAD1-GFP+ PNECs were seeded in the top chamber of a Boyden cell migration culture insert. When Slit protein was added with the cells in the top chamber, ~52% fewer (P = 8.5 × 10−4) PNECs migrated to the bottom (fig. S12, J to I). Conversely, when Slit protein was added to the bottom chamber, 18% more (P = 7.5 × 10−5) PNECs migrated to the bottom (fig. S11, L to N). These results suggest that Slit-Robo drive PNEC clustering into NEBs, likely through cellular attraction.
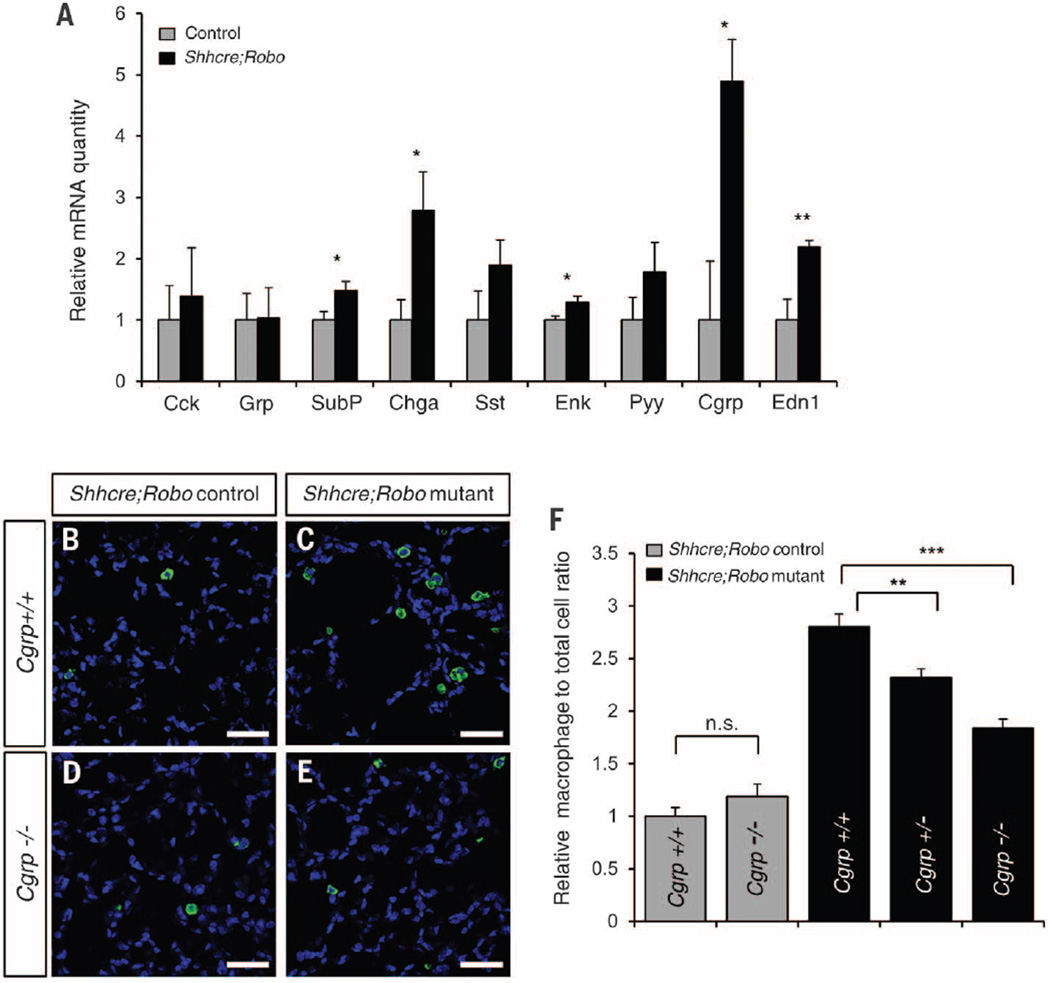
To test a possible link between PNECs and immune response, we assayed the expression of neuropeptides produced by PNECs (1). Of the nine neuropeptide genes assayed, five were significantly up-regulated in Shhcre;Robo mutants (Fig. 3A). Staining with antibody against CGRP revealed that, although its expression remains in PNECs in the mutant, the staining intensity is increased and it is no longer restricted to the basal side of these cells (figs. S7 and S9). We also note that, while unclustering occurred by E15.5, neuropeptide up-regulation is only observed after birth, presumably upon exposure to air (fig. S12).
Fig. 3. Increase in neuropeptide level contributes to increased macrophage infiltration.
(A) qRT-PCR of PNEC peptide transcript levels in P5 lungs. *P < 0.05, **P < 0.001. (B to E) IsoB4 labeling of macrophages at P22. Scale bars, 40 µm. (F) Quantification presented as the relative percentage of macrophage to total cell ratio normalized to Shhcre; Robo+/−;Cgrp+/+. **P < 0.01; ***P < 0.0001; n.s., not significantly different (P ≥ 0.05).
To determine whether the increase in neuropeptides contributes to the immune response, we focused on CGRP because its transcript shows the largest increase among all assayed (Fig. 3A). We countered this increase by breeding a mutant allele of Cgrp into the Shhcre;Robo background (20). In Robo controls, loss of Cgrp did not alter macrophage numbers (Fig. 3, B, D, and F). However, in Shhcre;Robo mutants, loss of Cgrp significantly decreased macrophage numbers in a dose-dependent manner (Fig. 3, C, E, and F). We also found that loss of Cgrp partially reversed the alveolar simplification phenotype (fig. S13). We note that neither macrophage increase nor alveolar simplification were entirely prevented, suggesting that the increase in other neuropeptides may add to downstream outcomes. Together, these results provide in vivo genetic demonstration that neuropeptides mediate PNEC function.
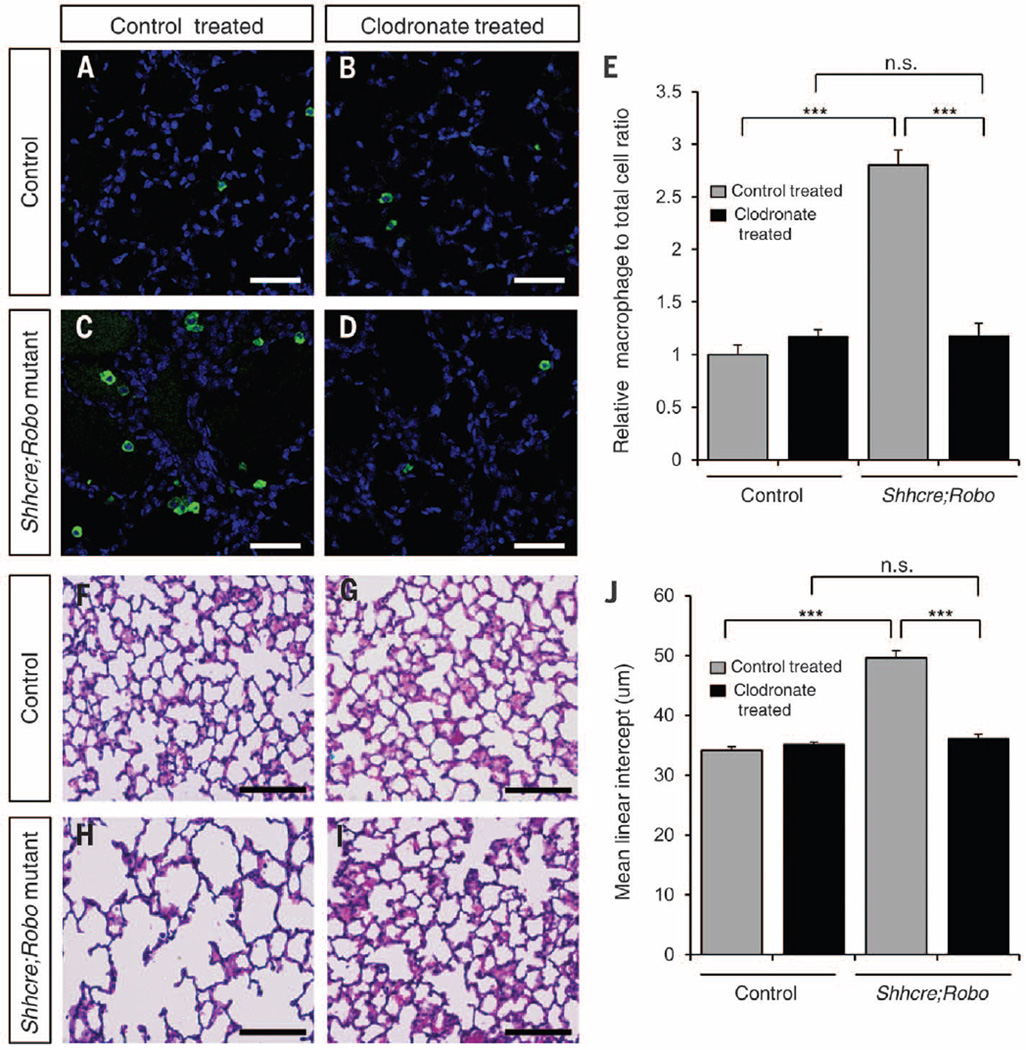
As normal alveologenesis initiates at P4 (21), the late appearance of alveolar simplification at P15 suggests that disruption of alveologenesis may not be a primary cause. Although no change in cell death was observed by P10, there was a clear reduction of elastin (fig. S14), which is a possible trigger of simplification (22). Immune cells such as macrophages express matrix metalloproteinases that degrade elastin (23). Furthermore, the increase in macrophages is observed before simplification (figs. S1 and S2), raising the possibility of a causal relationship. To test this, we treated Shhcre;Robo and control lungs with clodronate, a hydrophilic drug that depletes macrophages (24). Treatment starting at P5, before the immune cell increase, effectively restricted alveolar macrophage numbers to baseline level in Shhcre;Robo mutants (Fig. 4, A to E). This attenuated the decrease in elastin and entirely prevented simplification (Fig. 4, F to J, and fig. S15). Together these data offer in vivo demonstration that increased immune infiltrates are responsible for alveolar simplification and that both are downstream consequences of PNEC dysfunction.
Fig. 4. Macrophage reduction by clodronate treatment attenuates alveolar simplification.
(A to D) IsoB4 labeling of macrophages at P22. Scale bar, 50 µm. (E) Macrophage quantification as the relative percentage of macrophage to total cell ratio normalized to control mice with liposome control treatment. (F to I) H&E staining of alveolar region at P22. Scale bar, 100 µm. (J) Quantification of mean linear intercept (MLI). ***P < 0.0001; n.s., not significantly different (P ≥ 0.05).
In this study, we present in vivo genetic evidence demonstrating that PNECs, despite their rarity, have profound impact on postnatal lung function. Although the PNEC defect is already evident at E15.5 in the Shhcre;Robo mutant, the physiological outcomes, beginning with up-regulation of neuropeptides, initiates after birth. This suggests that the effect of PNEC is dependent on lung exposure to air. Thus, our findings delineate a mode of signal transduction in which PNECs are sensitive rheostats on the airway wall that translate environmental cues non–cell-autonomously into immune responses.
Our results establish Robo1,2 as a set of genes that control PNEC clustering into NEBs. It also presents Slit and Robo as players in selective cell sorting in the epithelium of amammalian organ. Inactivation of Robo after NEB formation also led to unclustering, suggesting that the clusters are actively maintained. Although Slit-Robo are largely known to mediate cellular repulsion, our data indicate that they drive PNEC clustering through cellular attraction. Robo inactivation led to altered innervation and loss of basal-biased localization of neuropeptides. These changes may underlie the altered downstream impact of PNECs.
An increase in PNEC number has been documented in a large array of lung-associated diseases, ranging from rare disorders such as CDH to common conditions such as asthma (5–8). We note that the Robo mutant PNEC phenotype is distinct from increased PNEC number. However, both are associated with increased neuropeptides, which we show to be potent effectors of PNEC function. Our findings thereby predict that rather than being a passive readout of the disease, the documented PNEC pathologies and neuropeptide increasesmay serve as active contributors to symptoms in a large array of respiratory diseases.
Supplementary Material
Acknowledgments
We thank X. Ai, T, Gomez, and E. Chapman for discussion; N. Hernandez-Santos for immune analysis; L. Ma, M. Tessier-Lavigne, J. Johnson, L. Wadiche, M. Zylka, and Mutant Mouse Regional Resource Center for mouse strains; and A. Lashua for technical support. This work was supported by American Heart Association predoctoral fellowship 14PRE20490146 and NIH predoctoral training grant T32 GM007133 (to K.B.), NIAID postdoctoral fellowship 5T32AI007635 (to L.N.), and NHLBI RO1 HL113870, HL097134, HL122406, University of Wisconsin Romnes Faculty Fellowship, and Wisconsin Partnership Program grant 2897 (to X.S.).
Footnotes
Supporting data and methods are presented in the supplementary materials.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/351/6274/707/suppl/DC1
Materials and Methods
Figs. S1 to 15
References (25–28)
REFERENCES AND NOTES
- 1.Cutz E, Pan J, Yeger H, Domnik NJ, Fisher JT. Semin. Cell Dev. Biol. 2013;24:40–50. doi: 10.1016/j.semcdb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Boers JE, den Brok JL, Koudstaal J, Arends JW, Thunnissen FB. Am. J. Respir. Crit. Care Med. 1996;154:758–763. doi: 10.1164/ajrccm.154.3.8810616. [DOI] [PubMed] [Google Scholar]
- 3.Linnoila RI. Lab. Invest. 2006;86:425–444. doi: 10.1038/labinvest.3700412. [DOI] [PubMed] [Google Scholar]
- 4.Song H, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutz E, Yeger H, Pan J. Pediatr. Dev. Pathol. 2007;10:419–435. doi: 10.2350/07-04-0267.1. [DOI] [PubMed] [Google Scholar]
- 6.Perrin DG, McDonald TJ, Cutz E. Pediatr. Pathol. 1991;11:431–447. doi: 10.3109/15513819109064779. [DOI] [PubMed] [Google Scholar]
- 7.Sunday ME, Shan L, Subramaniam M. Endocr. Pathol. 2004;15:91–106. doi: 10.1385/ep:15:2:091. [DOI] [PubMed] [Google Scholar]
- 8.Gu X, et al. Am. J. Respir. Cell Mol. Biol. 2014;50:637–646. doi: 10.1165/rcmb.2013-0199OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo CS, Krasnow MA. Cell. 2015;163:394–405. doi: 10.1016/j.cell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tjen-A-Looi S, Ekman R, Lippton H, Cary J, Keith I. Am. J. Physiol. 1992;263:H681–H690. doi: 10.1152/ajpheart.1992.263.3.H681. [DOI] [PubMed] [Google Scholar]
- 11.Dunzendorfer S, Meierhofer C, Wiedermann CJ. J Leukoc. Biol. 1998;64:828–834. doi: 10.1002/jlb.64.6.828. [DOI] [PubMed] [Google Scholar]
- 12.Moore BD, et al. Respiration. 2012;83:529–542. doi: 10.1159/000336835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantarci S, Donahoe PK. Am. J. Med. Genet. C. Semin. Med. Genet. 2007;145C:217–226. doi: 10.1002/ajmg.c.30132. [DOI] [PubMed] [Google Scholar]
- 14.Longoni M, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12450–12455. doi: 10.1073/pnas.1412509111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domyan ET, et al. Dev. Cell. 2013;24:52–63. doi: 10.1016/j.devcel.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. PLOS ONE. 2011;6:e18472. doi: 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hivert B, Liu Z, Chuang CY, Doherty P, Sundaresan V. Mol. Cell. Neurosci. 2002;21:534–545. doi: 10.1006/mcne.2002.1193. [DOI] [PubMed] [Google Scholar]
- 19.Englund C, Steneberg P, Falileeva L, Xylourgidis N, Samakovlis C. Development. 2002;129:4941–4951. doi: 10.1242/dev.129.21.4941. [DOI] [PubMed] [Google Scholar]
- 20.McCoy ES, Taylor-Blake B, Zylka MJ. PLOS ONE. 2012;7:e36355. doi: 10.1371/journal.pone.0036355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mund SI, Stampanoni M, Schittny JC. Dev. Dyn. 2008;237:2108–2116. doi: 10.1002/dvdy.21633. [DOI] [PubMed] [Google Scholar]
- 22.Shifren A, Durmowicz AG, Knutsen RH, Hirano E, Mecham RP. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L778–L787. doi: 10.1152/ajplung.00352.2006. [DOI] [PubMed] [Google Scholar]
- 23.Wallace AM, et al. COPD. 2008;5:13–23. doi: 10.1080/15412550701817789. [DOI] [PubMed] [Google Scholar]
- 24.van Rooijen N, Hendrikx E. Methods Mol. Biol. 2010;605:189–203. doi: 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.