Abstract
N-methyl-D-aspartate receptors (NMDARs) are major targets of both acute and chronic alcohol, as well as regulators of plasticity in a number of brain regions. Aberrant plasticity may contribute to the treatment resistance and high relapse rates observed in alcoholics. Recent work suggests that chronic alcohol treatment preferentially modulates both the expression and subcellular localization of NMDARs containing the GluN2B subunit. Signaling through synaptic and extrasynaptic GluN2B-NMDARs has already been implicated in the pathophysiology of various other neurological disorders. NMDARs interact with a large number of proteins at the glutamate synapse, and a better understanding of how alcohol modulates this proteome is needed. We employed a discovery-based proteomic approach in subcellular fractions of hippocampal tissue from chronic intermittent alcohol (CIE) exposed C57Bl/6j mice to gain insight into alcohol-induced changes in GluN2B signaling complexes. Protein enrichment analyses revealed changes in the association of postsynaptic proteins, including scaffolding, glutamate receptor, and PDZ-domain binding proteins with GluN2B. In particular, GluN2B interaction with metabotropic glutamate (mGlu)1/5 receptor-dependent long-term depression (LTD) associated proteins such as Arc and Homer 1 was increased, while GluA2 was decreased. Accordingly, we found a lack of mGlu1/5 induced-LTD while α1-adrenergic receptor-induced LTD remained intact in hippocampal CA1 following CIE. These data suggest that CIE specifically disrupts mGlu1/5-LTD, representing a possible connection between NMDAR and mGlu receptor signaling. These studies not only demonstrate a new way in which alcohol can modulate plasticity in the hippocampus but emphasize the utility of this discovery-based proteomic approach to generate new hypotheses regarding alcohol-related mechanisms.
Keywords: Alcohol Addiction, NMDA Receptor, Proteomics
Introduction
Alcoholism is a complex and multifaceted disease with a high persistence over the lifespan due in large part to high rates of relapse. These high relapse rates indicate both that current treatment options are largely ineffective and that alcohol use imparts long-lasting neural changes that persist even after discontinued use. NMDARs are likely candidates for these enduring changes given their necessity in various forms of neural plasticity. Indeed, acute (Dildy and Leslie, 1989; Hoffman et al., 1989; Lovinger et al., 1989) and chronic (Follesa and Ticku, 1996; Kalluri et al., 1998; Kumari, 2001; Nagy et al., 2003; Pawlak et al., 2005; Sheela Rani and Ticku, 2006) alcohol exposures are known modulators of NMDARs and disrupt NMDAR-plasticity in a number of regions (Blitzer et al., 1990; Givens, 1995; Grover and Frye, 1996; Hendricson et al., 2002; Izumi et al., 2005; Schummers and Browning, 2001; Weitlauf et al., 2004). Therefore, understanding mechanisms by which alcohol exerts its effects on NMDARs will be critical in developing new therapeutic interventions to intercede in the aberrant plasticity imparted by chronic alcohol use.
NMDARs are composed of two obligatory GluN1 subunits and either two GluN2 or GluN3 subunits. The GluN2 subunits are predominately expressed in the adult forebrain, with lower levels of GluN3 subunit expression. Of these GluN2 subunits, GluN2A and GluN2B have the highest expression and their incorporation into the receptor complex regulates many NMDAR properties (Cull-Candy et al., 2001; Traynelis et al., 2010). Several studies have explored the potential subunit selective effects of acute alcohol treatments and find mixed results depending on experimental parameters: recombinant receptors, brain region assessed, the phosphorylation state of subunits, and age [reviewed in (Wills and Winder, 2013)].
In contrast to the acute inhibitory effects of alcohol on NMDARs, withdrawal from chronic alcohol treatments have been frequently reported to enhance their function (Follesa and Ticku, 1996; Kalluri et al., 1998; Kumari, 2001; Nagy et al., 2003; Pawlak et al., 2005; Sheela Rani and Ticku, 2006). This enhancement is preferential to the GluN2B subunit, which shows a general increase in expression and function in numerous brain regions after chronic alcohol exposure (Blevins et al., 1997; Blevins et al., 1995; Follesa and Ticku, 1995; Hardy et al., 1999; Hendricson et al., 2007; Henniger et al., 2003; Hu et al., 1996; Kalluri et al., 1998; Kash et al., 2009; Matsumoto et al., 2001; Nagy et al., 2003; Narita et al., 2000; Qiang et al., 2007; Wills et al., 2012), however, GluN2A increases have also been seen in a few studies (Follesa and Ticku, 1996; Kalluri et al., 1998). Chronic alcohol use and withdrawal also alters the location of these NMDARs. For example, chronic alcohol use increases synaptic clustering of NMDARs (Carpenter-Hyland et al., 2004; Hendricson et al., 2007; Qiang et al., 2007), while withdrawal re-locates GluN2B-NMDARs to extrasynaptic sites (Clapp et al., 2010; Wills et al., 2012). The localization of these GluN2B-NMDARs can differentially promote either LTP or LTD (Barria and Malinow, 2005; Bartlett et al., 2007; Brigman et al., 2010; Liu et al., 2004; Massey et al., 2004) presumably as a result of divergent GluN2B signaling at either synaptic or extrasynaptic locations (Newpher and Ehlers, 2009). Numerous recent studies have implicated extrasynaptic GluN2B-NMDAR transmission in the pathologies of a number of neurological disorders (Gladding and Raymond, 2011; Hardingham and Bading, 2010). Elucidating the mechanisms responsible for the effects of alcohol on subcellular localization of GluN2B-NMDARs and corresponding changes in their signaling cascades will be critical for developing treatments to rectify aberrant plasticity produced during alcoholism.
The current studies were designed to identify novel alcohol-related changes in GluN2B signaling. While many studies have demonstrated that synaptic and extrasynaptic NMDARs engage distinct signaling pathways, it is unknown what signaling is involved in alcohol-induced re-localization of GluN2B-NMDARs and how this might affect the subsequent function of these receptors. To elucidate these mechanisms, we used a proteomic approach to obtain an unbiased global view of proteins associated with GluN2B-NMDARs and the impact of withdrawal from chronic intermittent ethanol (CIE-W) exposure on these interactions in the hippocampus. CIE-W has been shown to be a good model of dependence as it increases voluntary ethanol consumption and produces increase in anxiety-like behavior (Becker and Lopez, 2004; Kash et al., 2009). Glutamatergic synapses in the hippocampus are very well characterized and modulated by alcohol. In this study we targeted proteins specifically associated with immunoprecipitated GluN2B-NMDARs and separately evaluated synaptic and non-synaptic GluN2B-NMDAR protein pools. We were able to identify GluN2B-associated proteins that are sensitive to chronic alcohol exposure in synaptic and non-synaptic locations. The association of post-synaptic density (PSD) scaffolding proteins was particularly sensitive to alcohol with a consistently decreased association with GluN2B, suggesting that chronic alcohol dissociates GluN2B-NMDARs from the PSD. A second constellation of proteins that were significantly changed by chronic alcohol have all been linked to mGlu1/5-LTD. Electrophysiology experiments further confirmed that mGlu1/5-LTD was selectively disrupted after chronic alcohol treatment.
Materials and Methods
Animals
Male C57BL/6J (6–8 wk of age; Jackson Laboratories) and GluN2B KO (C57BL/6J background; described in (Wills et al., 2012)) were housed in groups of two to five in a temperature- and humidity-controlled animal facilities are maintained on a 12:12 -h light:dark cycle (lights on 0600–1800 hours). Food and water were available ad libitum. All procedures were approved by the Animal Care and Use Committee at Vanderbilt.
Treatment/Tissue Collection
In these studies, adult male C57Bl/6J mice underwent the chronic intermittent ethanol and withdrawal (CIE-W) procedure. Mice were given a daily injection of either pyrazole (Air control, 1mmol/kg) or pyrazole + ethanol (ethanol group, 1mmol/kg + 0.8 g/kg, respectively) to impair the metabolism of ethanol. Thirty minutes after the injection mice were placed in their home cages into a chamber filled with volatilized ethanol (20.3 ± 0.2 mg/L) or volatilized water (Air group). Airflow through the chambers was maintained at 5.5 L/min, and volatilization was maintained at 1.5 L/min. After 16 h of exposure, mice were removed from the chambers and returned to standard animal housing. Ethanol chamber exposure occurred from 1600-0800 the following day. Using these parameters, we can reliably obtain blood ethanol concentrations in the range of 150–185 mg/dL. CIE-W was comprised of two, 4 day cycles of 16 hours in ethanol vapor chambers and 8 hours out of vapor chambers (Figure 1A). Five hours following the final vapor chamber exposure, 0.35 mm tissue punches were collected from 500 μm slices within the hippocampus (8 mice/sample; Figure 1B) containing portions of CA1, CA3, and the dentate gyrus. Tissue punches were also obtained from the hippocampus of conditional GluN2B KO mice (Wills et al., 2012) with an ~80% reduction in levels of GluN2B (Badanich et al., 2011). GluN2BKO mice were not exposed to vapor chambers. For western blot analysis, a separate cohort of mice was exposed to ethanol or air as described above. Five hours following the final vapor chamber exposure, whole hippocampi were dissected for subcellular fractionation, GluN2B IP, and then western blot analysis.
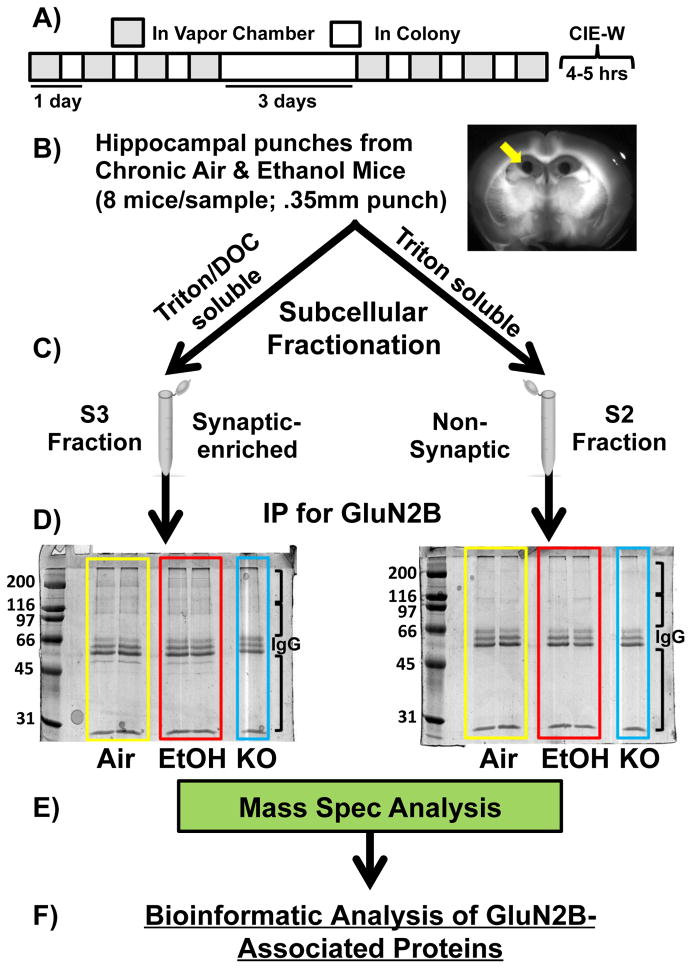
Figure 1. Schematic of Proteomic Analysis.
(A) Chronic Intermittent Ethanol and Withdrawal (CIE-W): Two 4 day cycles of 16 hours in ethanol vapor chambers and 8 hours out of vapor chambers. Brains collected five hours after the final vapor exposure. (B) 0.35mm tissue punches were collected from the hippocampus (8 mice/sample). (B) Subcellular Fractionation. Synaptic-enriched/S3 [triton- & deoxycholate (DOC)-soluble] and non-synaptic/S2 (triton-soluble) fractions were generated. (C) GluN2B Immunoprecipitation (IP). Coomassie stain of blots for GluN2B IPs in chronic air and ethanol mice (4 samples/treatment; 8 mice/sample) and GluN2B knockout mice (1 sample; 8 mice/sample) for S2 and S3 fractions. (D) Mass spectrometry. Sample lanes were subdivided into three separate samples (black brackets on blots) to maximize protein coverage in the mass spectrometer (MS). (E) Bioinformatic analysis of GluN2B-assoicated proteins.
Subcellular Fractionation & GluN2B Immunoprecipitations (IPs)
Synaptic (triton & deoxycholate (DOC)-soluble) and non-synaptic (triton-soluble) fractions were generated from all of these tissue samples (Baucum et al., 2013; Gustin et al., 2011) (Figure 1C). Tissue was homogenized in homogenization buffer (150 mM KCl, 50 mM Tris–HCl pH 7.5, 1 mM DTT, 0.2 mM PMSF, 1 mM benzamidine, 1 μM pepstatin, 10 μg/ml leupeptin, and 1 μM microcystin-LR) using Kontes glass tissue grinders at 4°C. Total homogenate was rocked for 30 min at 4°C and spun down at 100,000 x g for 1 h yielding an S1 fraction (soluble cytosolic protein pool) and a P1 pellet (insoluble fraction). The cytosolic fractions (S1) were not used in further analysis. P1 was resuspended in homogenization buffer containing 1% (v/v) Triton X-100 using a Kontes, rounded tip cone pestle and rocked for 30 min at 4°C. The homogenate was then spun down at 16,000 x g for 10 min at 4°C yielding an S2 fraction (membrane-associated protein pool) and a P2 pellet (Triton-insoluble fraction). The P2 was sonicated at 4°C in Homogenization buffer containing 1% (v/v) Triton X-100 and 1% (w/v) sodium deoxycholate and rocked for 30 min at 4°C. Precleared samples (300 μl) were incubated at 4°C for 1 h with mouse antisera to GluN2B (7.5 μl; antibody from BD Transduction Laboratories), protein G magnetic beads (Pierce Biotechnology) were added, and samples were incubated overnight at 4 °C. Immunoprecipitates were washed 3 times with 1 ml of IP buffer and suspended in 2 x SDS sample buffer and separated by SDS-PAGE (Baucum et al., 2013; Baucum et al., 2010) (Figure 1D).
Mass Spectrometry
Sample lanes were subdivided into three separate samples (black brackets on blots; Figure 1) to maximize protein coverage in the mass spectrometer (MS). Co-IPs from the synaptic and non-synaptic fractions in Air, CIE-W, and KO mice were then analyzed via LC-MS/MS (Vanderbilt Proteomics Core; LTQ-Orbitrap; Figure 1E). MS/MS spectra of the peptides were obtained using data-dependent scanning, in which one full MS spectra was followed by sixteen MS/MS spectra, drawn from the most intense ions observed in each MS spectra. Peptiome 1.1.50 (Dasari et al., 2012) identified peptides corresponding to the MS/MS scans by comparing them to spectra from the NIST mouse ion trap library [or an equal number of shuffled decoy spectra (Lam et al., 2010)]. IDPicker 3 filtered away peptide-spectrum matches with a greater than 2% q-value, and then proteins were assembled with the requirement that each protein group was required to have at least two distinct peptides in evidence, with parsimony applied (Zhang et al., 2007). A protein false discovery rate of 4.93% resulted from the set of fifty-five LC-MS/MS experiments. Data were placed into a hierarchy that separated early and late experiments, with synaptic and non-synaptic samples separated within the folders, with each of these folders separated into “KO,” “EtOH,” and “Air” categories (only early experiments included “KO”).
Proteomic Statistics
The average spectral counts of synaptic Air and CIE-W samples were compared to those of KO samples. Only proteins with higher average spectral count in the experimental samples compared to KO samples were included in further analysis. This KO edit was included to remove non-specific protein interactions, however, an “all or not” approach was not employed due the incomplete GluN2B deletion in these mice (Badanich et al., 2011). This same KO filtering process was applied to non-synaptic groups. Fisher’s exact test was used to test if the spectral counts were different between experiment/control groups. A 2 by 2 contingency table is constructed for each protein. The 4 components in the table were A) spectral counts for this protein in the experimental group, B) spectral counts for this protein in the control group, C) spectral counts for all other proteins in the experimental group, and D) spectral counts for all other proteins in the control group. The p-values from duplicated MS runs in synaptic or non-synaptic fractions were combined by Fisher’s p-value combination method. All the p-values were adjusted by Benjamini-Hochberg multiple comparison adjustment.
Immunoblot Analysis
The proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Nitrocellulose membranes were blocked in 5% milk in TBST and incubated with the appropriate primary and secondary antibodies. For detection using the Odyssey system (LiCor Biosciences, Lincoln, NE), infrared-conjugated secondary antibodies (LiCor) were used. Densitometry was performed using Image J (National Institutes of Health, Bethesda, MD) on images linearly adjusted for brightness and contrast. Inputs were normalized to Ponceau-S staining while samples from GluN2B-IPs were unnormalized since there were no treatment differences in GluN2B. The following primary antibodies were used: Homer1 (Synaptic Systems; 1:2,000), PSD-95 (Affinity Bioreagents; 1:20,000), GluA2 (Millipore Anti-GluR2; 1:2,000), Shank 3 (UC Davis/NIH NeuroMab), Arc (Santa Cruz; 1:500), and mGluR5 (Millipore; 1:3,000).
Slice Physiology
Five hours following the final vapor chamber exposure, brains were quickly removed and placed in ice-cold sucrose artificial cerebrospinal fluid (ACSF): (in mM) 194 sucrose, 20 NaCl, 4.4 KCl, 2 CaCl2, 1 MgCl2, 1.2 NaH2PO4, 10.0 glucose, and 26.0 NaHCO3 saturated with 95% O2/5% CO2. Slices of 300 μm in thickness were prepared using a Tissue Slicer (Leica). Slices were then transferred to a submerged recording chamber where they were perfused with heated (28°C), oxygenated (95% O2–5% CO2) ‘normal’ ACSF (in mM: 124 NaCl, 4.4 KCl, 2 CaCl2, 1.2 MgSO4, 1 NaH2PO4, 10.0 glucose, and 26.0 NaHCO3, pH 7.2–7.4; 290–310 mOsm) or modified low magnesium ACSF (in mM: 124 NaCl, 4.4 KCl, 3.7 CaCl2, 1 NaH2PO4, 10 glucose, and 26 NaHCO3, pH 7.2–7.4; 290–310 mOsm) for NMDA receptor recordings at a rate of about 2 ml/min. After dissection, slices were transferred to an interface recording chamber where they were perfused with heated (~29°C), oxygenated (95% O2 – 5% CO2) ACSF at a rate of ~2 ml/min. Slices were allowed to equilibrate in ACSF for at least 1.5 hr before experiments began. A bipolar stainless steel stimulating electrode and a borosilicate glass recording electrode filled with ACSF were placed in the CA1 region of the hippocampus to elicit and record an extracellular field response in CA3 region. Baseline responses of excitatory field potentials were recorded for 20 min (at 0.05 Hz) followed by application of DHPG (10 mins; 100 μM) and 50 min washout. In other experiments, 10 mins of stable fEPSPs were recorded prior to methoxamine application (10 mins; 40 μM) and 30 min washout.
Results
Discovery-Based Proteomic Screen
Our discovery-based proteomic approach was designed to explore the effect of withdrawal from chronic intermittent ethanol (CIE-W; Figure 1A) on GluN2B-NMDAR associated proteins in C57Bl6/J male mouse hippocampus. Regional specificity was obtained by collecting 0.35 mm tissue punches from the hippocampus (Figure 1B) in mice exposed to CIE-W and air controls (8 mice/treatment/sample). Since the literature supports NMDAR synaptic localization changes following chronic alcohol exposure, extracts of the punched tissue were fractionated to separately enrich synaptic and non-synaptic GluN2B-associated proteins (Baucum et al., 2013; Gustin et al., 2011) (Figure 1C). Fractionated samples were immunoprecipitated (IP) using an antibody against GluN2B (Baucum et al., 2013; Baucum et al., 2010; Baucum et al., 2012; Brown et al., 2008) to isolate GluN2B-associated proteins. The specificity of the IPs was established by analyzing samples prepared in parallel from conditional GluN2B-KO mice that express ≤ 20% of wildtype GluN2B protein levels in hippocampal tissue (Badanich et al., 2011). Similar strategies have proven effective in other proteomic analyses (Baucum et al., 2010). Immune complexes were separated by SDS-PAGE and the gels were stained with Coomassie blue. Figure 1D shows a representative gel comparing GluN2B immune complexes from synaptic-enriched (S3) and non-synaptic (S2) fractions from CIE-W, air controls, and conditional GluN2B-KO mice. Numerous proteins were detected in representative samples from air and CIE-W exposed mice, which were much less abundant in samples from GluN2B-KO mice. Moreover, these relatively abundant GluN2B-associated proteins appeared to be distinct in the S3 and S2 fractions, with generally more proteins in the S3 fraction compared to the S2 fraction, as would be expected. Mass spectrometry (MS, LTQ-Orbitrap; Figure 1E) was performed on all samples (4 samples/treatment). Proteins detected in high abundance in KO tissue were removed from subsequent analyses (see Methods). From the MS, there were 829 unique proteins identified in all fractions/all samples/all treatments, of which 105 were removed due to above threshold detection in GluN2B-KO samples, leaving a total of 724 proteins. Significant treatment effects were determined with Fisher’s Exact Test on protein spectral counts. Of the proteins exhibiting significant treatment effects, 64 proteins were detected in the S3 fraction (Table 1) and 22 proteins were detected in the S2 fraction (Table 2).
Table 1.
Chronic alcohol and withdrawal-induced changes in GluN2B-associated proteins in S3 (synaptic) fraction.
| Accession | S3 Fraction Proteins | Air ΣSpectra | CIE ΣSpectra | Fisher pValue | Fisher FDR | Treatment Change |
|---|---|---|---|---|---|---|
| NP_001020422 | myelin basic protein | 51 | 142 | 0.000 | 0.000 | Increase |
| NP_034053* | 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (isoform 1 & 2) | 37 | 101 | 0.000 | 0.000 | Increase |
| NP_671705 | homer protein homolog 1 isoform L | 53 | 89 | 0.000 | 0.000 | Increase |
| NP_033059* | RNA binding motif protein, X-linked-like-1 | 28 | 61 | 0.000 | 0.000 | Increase |
| NP_033477 | tubulin beta-4A chain | 176 | 286 | 0.000 | 0.002 | Increase |
| NP_036145* | core histone macro-H2A.1 (isoform 1–4) | 44 | 82 | 0.000 | 0.004 | Increase |
| NP_666228 | tubulin beta-4B chain | 228 | 334 | 0.001 | 0.013 | Increase |
| NP_032717 | neurofilament medium polypeptide | 88 | 146 | 0.001 | 0.014 | Increase |
| NP_035785 | tubulin beta-5 chain | 237 | 339 | 0.002 | 0.016 | Increase |
| NP_033476 | tubulin beta-2A chain | 251 | 357 | 0.002 | 0.020 | Increase |
| NP_056602 | histone H1.4 | 183 | 205 | 0.006 | 0.047 | Increase |
| NP_570954 | isocitrate dehydrogenase 3, beta subunit | 4 | 17 | 0.008 | 0.058 | Increase |
| NP_766333 | elongation factor Tu, mitochondrial | 1 | 15 | 0.011 | 0.077 | Increase |
| NP_035040 | neurofilament light polypeptide | 95 | 139 | 0.011 | 0.080 | Increase |
| NP_783597 | histone H2B type 2-B | 59 | 77 | 0.012 | 0.082 | Increase |
| NP_663759 | histone H1.3 | 206 | 236 | 0.013 | 0.086 | Increase |
| NP_033124 | 40S ribosomal protein S8 | 6 | 18 | 0.013 | 0.088 | Increase |
| NP_061260* | activity-regulated cytoskeleton-associated protein (Arc) | 5 | 18 | 0.014 | 0.092 | Increase |
| NP_036183 | 60S ribosomal protein L8 | 4 | 16 | 0.018 | 0.112 | Increase |
| NP_032953 | dual specificity mitogen-activated protein kinase kinase 1 | 0 | 11 | 0.018 | 0.112 | Increase |
| NP_033907 | complement C1q subcomponent subunit B precursor | 10 | 21 | 0.019 | 0.112 | Increase |
| NP_082724 | centromere protein V | 24 | 37 | 0.019 | 0.113 | Increase |
| NP_038505 | V-type proton ATPase subunit d 1 | 4 | 15 | 0.025 | 0.137 | Increase |
| NP_998779 | septin-5 | 1 | 12 | 0.027 | 0.142 | Increase |
| NP_666212 | alpha-internexin | 13 | 34 | 0.028 | 0.151 | Increase |
| NP_038790 | 60S ribosomal protein L3 | 0 | 10 | 0.031 | 0.162 | Increase |
| NP_872591* | heterogeneous nuclear ribonucleoproteins A2/B1 (isoform 1&2) | 1 | 10 | 0.044 | 0.216 | Increase |
| NP_033408 | thy-1 membrane glycoprotein preproprotein (Thy1) | 6 | 16 | 0.046 | 0.219 | Increase |
| NP_038749 | 60S ribosomal protein L7a | 1 | 11 | 0.047 | 0.225 | Increase |
| NP_001182350* | histone H4 | 29 | 50 | 0.049 | 0.226 | Increase |
| NP_031890* | disks large homolog 4 isoform 2 (PSD-95) | 513 | 413 | 0.000 | 0.000 | Decrease |
| NP_659170 | sodium/potassium-transporting ATPase subunit alpha-3 | 461 | 307 | 0.000 | 0.000 | Decrease |
| NP_112442 | heat shock cognate 71 kDa protein | 261 | 198 | 0.000 | 0.000 | Decrease |
| NP_035937 | disks large homolog 2 isoform 1 (PSD-93) | 406 | 333 | 0.000 | 0.000 | Decrease |
| NP_659149 | sodium/potassium-transporting ATPase subunit alpha-1 | 329 | 223 | 0.000 | 0.001 | Decrease |
| NP_848492 | sodium/potassium-transporting ATPase subunit alpha-2 | 372 | 259 | 0.000 | 0.002 | Decrease |
| NP_038635* | myosin-11 (isoform 1 & 2) | 113 | 70 | 0.000 | 0.003 | Decrease |
| NP_766024 | calcium-binding mitochondrial carrier protein Aralar1 | 143 | 90 | 0.000 | 0.004 | Decrease |
| NP_058027* | disks large homolog 3 (isoform 1 & 2) (SAP-102) | 134 | 97 | 0.000 | 0.005 | Decrease |
| NP_542364 | aconitate hydratase, mitochondrial | 62 | 25 | 0.000 | 0.007 | Decrease |
| NP_032197 | glutamate receptor ionotropic, NMDA 2B | 325 | 254 | 0.000 | 0.007 | Decrease |
| NP_032195 | glutamate receptor ionotropic, NMDA 1 isoform 1 | 234 | 218 | 0.001 | 0.010 | Decrease |
| NP_035354 | glycogen phosphorylase, muscle form | 63 | 40 | 0.001 | 0.012 | Decrease |
| NP_031485 | AP-2 complex subunit alpha-2 | 87 | 51 | 0.001 | 0.014 | Decrease |
| NP_001171127 | glutamate receptor ionotropic, NMDA 1 isoform 2 | 209 | 195 | 0.002 | 0.016 | Decrease |
| NP_032766 | vesicle-fusing ATPase | 154 | 116 | 0.002 | 0.022 | Decrease |
| NP_001108136 | IQ motif and SEC7 domain-containing protein 2 (Iqsec2) | 204 | 156 | 0.003 | 0.025 | Decrease |
| NP_001030931* | AP-2 complex subunit beta (isoform a & b) | 124 | 79 | 0.004 | 0.034 | Decrease |
| NP_001070982 | excitatory amino acid transporter 2 (GLT-1) | 114 | 64 | 0.005 | 0.038 | Decrease |
| NP_032327* | heat shock-related 70 kDa protein 2 | 106 | 82 | 0.006 | 0.047 | Decrease |
| NP_001156647 | brain-enriched guanylate kinase-associated protein (Begain) | 70 | 53 | 0.006 | 0.047 | Decrease |
| NP_031484* | AP-2 complex subunit alpha-1 isoform b | 94 | 61 | 0.008 | 0.059 | Decrease |
| NP_032196 | glutamate receptor ionotropic, NMDA 2A | 128 | 95 | 0.011 | 0.080 | Decrease |
| NP_001139251* | kinesin-like protein KIF2A (isoform 1 & 2) | 18 | 6 | 0.012 | 0.084 | Decrease |
| NP_031534 | V-type proton ATPase catalytic subunit A | 76 | 48 | 0.019 | 0.112 | Decrease |
| NP_941020 | disks large-associated protein 3 (SAPAP 3) | 207 | 142 | 0.019 | 0.113 | Decrease |
| NP_034600* | heterochromatin protein 1-binding protein 3 (isoform 1 & 2) | 45 | 28 | 0.023 | 0.132 | Decrease |
| NP_001156960* | 6-phosphofructokinase, muscle type | 34 | 18 | 0.024 | 0.132 | Decrease |
| NP_038568 | glutamate receptor 2 isoform 2 (GluA2) | 49 | 29 | 0.044 | 0.216 | Decrease |
| NP_031486 | AP-3 complex subunit delta-1 | 21 | 9 | 0.044 | 0.216 | Decrease |
| NP_031954* | serine/threonine-protein kinase MARK2 (isoform 1–4) (PAR-1b) | 13 | 4 | 0.045 | 0.217 | Decrease |
| NP_722476 | glycogen phosphorylase, brain form | 35 | 23 | 0.046 | 0.219 | Decrease |
| NP_808482 | ERC protein 2 (CAST1) | 41 | 32 | 0.049 | 0.226 | Decrease |
| NP_067398 | SH3 and multiple ankyrin repeat domains protein 3 (Shank3) | 156 | 121 | 0.049 | 0.226 | Decrease |
The mouse IDPicker 3 database was used to determine accession numbers for identified proteins. The number is the first number given by the IDPicker and * denotes if there were multiple accession number from the analysis. The next column denotes the corresponding protein name that corresponds with this accession number. The spectral columns gives the total spectral counts from Air and CIE-treated mice. These are followed by p-values and false discovery rate (FDR) for the product of the individual p-values and FDR by treatment for each cohort. The final column represents the direction of the treatment change from the Air-treated group to the CIE-treated group.
Table 2.
Chronic alcohol and withdrawal-induced changes in GluN2B-associated proteins in S2 (non-synaptic) fraction.
| Accession | S2 Fraction Proteins | Air ΣSpectra | CIE ΣSpectra | Fisher pValue | Fisher FDR | Treatment Change |
|---|---|---|---|---|---|---|
| NP_034053* | 2′,3′-cyclic-nucleotide 3′-phosphodiesterase | 16 | 59 | 0.000 | 0.000 | Increase |
| NP_001020422 | myelin basic protein | 41 | 96 | 0.000 | 0.000 | Increase |
| NP_035010 | nucleolin | 0 | 15 | 0.000 | 0.024 | Increase |
| NP_035354 | glycogen phosphorylase, muscle form | 35 | 46 | 0.040 | 0.575 | Increase |
| NP_001170867 | ankyrin repeat and sterile alpha motif domain-containing protein 1B | 30 | 5 | 0.001 | 0.048 | Decrease |
| NP_067262 | spectrin beta chain, brain 2 | 37 | 11 | 0.002 | 0.085 | Decrease |
| NP_031600 | complement C1q subcomponent subunit C | 13 | 0 | 0.006 | 0.179 | Decrease |
| NP_071855 | myosin-9 | 37 | 12 | 0.006 | 0.180 | Decrease |
| NP_001070022* | spectrin alpha chain, non-erythrocytic 1 isoform 3 | 187 | 135 | 0.012 | 0.292 | Decrease |
| NP_001171138 | spectrin alpha chain, non-erythrocytic 1 isoform 2 | 187 | 135 | 0.012 | 0.292 | Decrease |
| NP_033739 | actin, cytoplasmic 2 | 215 | 146 | 0.012 | 0.292 | Decrease |
| NP_031419 | actin, cytoplasmic 1 | 210 | 147 | 0.012 | 0.292 | Decrease |
| NP_659149 | sodium/potassium-transporting ATPase subunit alpha-1 | 93 | 53 | 0.014 | 0.299 | Decrease |
| NP_848492 | sodium/potassium-transporting ATPase subunit alpha-2 | 109 | 64 | 0.015 | 0.299 | Decrease |
| NP_035937 | disks large homolog 2 (PSD 93) | 80 | 44 | 0.021 | 0.408 | Decrease |
| NP_780469 | myosin-10 | 53 | 25 | 0.025 | 0.445 | Decrease |
| NP_031485 | AP-2 complex subunit alpha-2 | 30 | 9 | 0.025 | 0.445 | Decrease |
| NP_001003908 | clathrin heavy chain 1 | 135 | 78 | 0.031 | 0.498 | Decrease |
| NP_032766 | vesicle-fusing ATPase | 21 | 4 | 0.031 | 0.498 | Decrease |
| NP_671705 | homer protein homolog 1 | 38 | 17 | 0.032 | 0.498 | Decrease |
| NP_001170842 | drebrin | 20 | 6 | 0.045 | 0.626 | Decrease |
| NP_659170 | sodium/potassium-transporting ATPase subunit alpha-3 | 142 | 95 | 0.048 | 0.654 | Decrease |
The mouse IDPicker 3 database was used to determine accession numbers for identified proteins. The number is the first number given by the IDPicker and * denotes if there were multiple accession number from the analysis. The next column denotes the corresponding protein name that corresponds with this accession number. The spectral columns gives the total spectral counts from Air and CIE-treated mice. These are followed by p-values and false discovery rate (FDR) for the product of the individual p-values and FDR by treatment for each cohort. The final column represents the direction of the treatment change from the Air-treated group to the CIE-treated group.
Pathway Analysis of Alcohol-induced Protein Changes
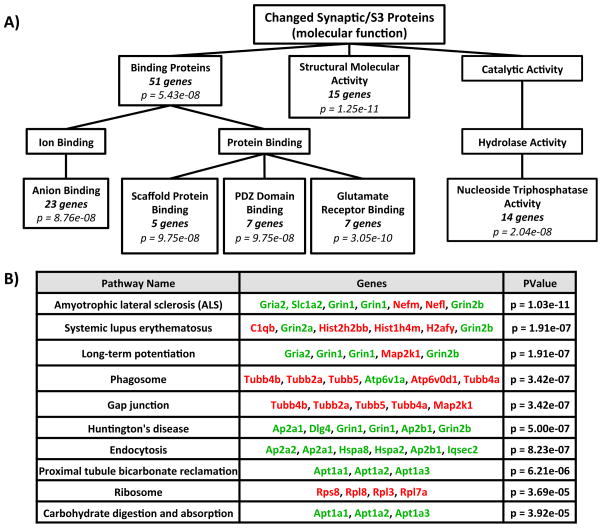
To gain an understanding of the signaling pathways that are significantly changed following CIE-W, we used several pathway enrichment programs, focusing on proteins in the S3 fraction over the S2 fraction because the larger sample size (64 versus 22 proteins) was better suited to detect affected pathways. The list of proteins that were significantly changed (either increased or decreased) in the S3 fraction by CIE-W was compared to the entire mouse genome. The enriched Gene Ontology (GO) Analysis divided many of the synaptically (S3) located proteins that were significantly altered by CIE-W into several broad categories (Figure 2A). These categories are: binding proteins (51 genes; p = 5.43e-08), structural activity proteins (15 genes; p = 1.25e-11), and catalytic activity (nucleoside-triphosphatase; 14 genes; p = 2.04e-08). In the binding protein category, these could be further subdivided into: anion binding (23 genes; p = 8.76e-08), scaffold protein binding (5 genes; p = 9.75e-08), PDZ-domain binding (5 genes; p = 9.75e-08), and glutamate receptors (7 genes; p = 3.05e-10). The second enrichment strategy used was the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway analysis, which assesses the enhancement of the significant changed S3 proteins against genes in known signaling cascades. The KEGG pathway analysis found 9 signaling pathways (see Figure 2B) that were significantly changed by CIE-W treatment: amyotrophic lateral sclerosis (ALS; 7 genes; p = 1.03e-11), systemic lupus erythematosus (6 genes; p = 1.91e-07), long-term potentiation (LTP; 5 genes; p = 1.91e-07), phagosome (6 genes; p = 3.42e-07), gap junction (5 genes; p = 3.42e-07), Huntington’s disease (6 genes; p = 5.00e-07), endocytosis (6 genes; p = 8.23e-07), proximal tubule bicarbonate reclamation (3 genes; p = 6.21e-06), ribosome (4 genes; p = 3.69e-05), and carbohydrate digestion and absorption (3 genes; p = 3.92e-05).
Figure 2. Pathway Analysis of GluN2B-associated Proteins Changed by Alcohol.
(A) Modified diagram of enriched Gene Ontology (GO) analysis through WebGestalt. Analysis was performed on significantly changed synaptic/S3 proteins by chronic ethanol (see Table 1) compared to the whole mouse genome. (B) Modified diagram of Kegg Pathway analysis through Webgestalt. Analysis was performed on significantly changed synaptic/S3 proteins by chronic ethanol (see Table 1) compared to the whole mouse genome. Proteins in red font were increased by CIE and those in green font were decreased by CIE. Note: Both diagrams were modified from their original version to highlight the main treatment effects.
Immunoblot Validation of Selected Alcohol-induced Protein Changes
To confirm the effects of CIE-W on GluN2B-associated proteins from the above proteomic screen, immunoblot analyses were also performed. A separate cohort of mice was exposed to CIE-W and hippocampal tissue was collected. Subcellular fractionations and GluN2B IPs were performed on this tissue in the same manner as in proteomic screens. Immunoblots for GluN2B, Homer 1, PSD-95, and GluA2 were performed on both synaptic (S3) fraction and non-synaptic (S2) fraction. There was no change in levels of GluN2B in either S3 (t(9) = 1.158; p = 0.28, N.S.; data not shown) or S2 (t(10) = 0.213; p = 0.84, N.S.; data not shown) fractions (inputs) or in the GluN2B-IPs from S3 (t(9) = 1.247; p = 0.24, N.S.; Figure 3A) or S2 (t(10) = 1.079; p = 0.31, N.S.; Figure 3B). Homer 1 was increased from CIE-W treated mice compared to Air controls in GluN2B-IPs from S3 fractions (t(9) = 2.293; p < 0.05; Figure 3C) with no change in GluN2B-IPs from S2 fractions (t(10) = 0.427; p = 0.68, N.S.; Figure 3D). For PSD-95, there was a significant decrease in the GluN2B-IP S3 fraction in CIE-W compared to Air controls (t(10) = 2.422; p < 0.05; Figure 3E), with a corresponding increase in PSD-95 levels in GluN2B-IP S2 fraction for CIE-W compared to Air controls (t(10) = 2.697; p < 0.05; Figure 3F). Finally, for GluA2, there was a significant decrease in CIE-W treated mice compared to Air-treated controls in the GluN2B-IP S3 fraction (t(10) = 2.842; p < 0.05; Figure 3G). In conjunction, there was an increase in GluA2 in the GluN2B S2 fraction in CIE-W treated mice compared to Air treated controls (t(10) = 2.461; p < 0.05; Figure 3H). These findings confirm the selected alcohol-related protein changes found in the proteomic screen and thus validate this approach in its ability to uncover treatment-specific protein changes. Further, these findings reflect alcohol-induced changes specifically in GluN2B subunit association with these proteins because CIE-W had no effect on the input levels for Homer 1 (S3: t(9) = 1.605; p = 0.14, N.S.; S2: t(10) = 0.6968; p = 0.50, N.S.), PSD-95 (S3: t(10) = 1.095; p = 0.30, N.S.; S2: t(10) = 0.0046; p = 0.99, N.S.), and GluA2 (S3: t(8) = 1.306; p = 0.28, N.S.; S2: t(10) = .2242; p = 0.83, N.S.; data not shown). In contrast, we were unable to corroborate the proteomic data for Shank 3 and Arc GluN2B-IP changes identified in the S3 fraction with immunoblot-based analyses (Shank 3: t(10) = 0.731, p = 0.48, N.S.; and Arc: t(10) = 0.681, p = 0.51, N.S.; data not shown). There was also no treatment change in either protein from the S2 fraction (Shank 3: t(10) = 0.3197, p = 0.76, N.S.; Arc: t(10) = 0.6026, p = 0.56, N.S.; data not shown).
Figure 3. Immuoblot Analysis of Alcohol-induced Changes in S3 and S2 Fractions.
Bar graphs and picture insets show immunoblot data for (A/B) GluN2B, (C/D) Homer1, (E/F) PSD-95, and (G/H) GluA2 from S3 and S2 fractions of GluN2B-immunoprecipitated hippocampal tissue. Immunoblot images for each protein are from the same blot with black boxes used to separate treatment and fraction and numbers are used to delineate molecular weight marker. Bar graphs represent optical density of bands from immunoblots; n = 5–6; * p < .05
Proteomic Screen Predicts Alcohol-induced Disruption of Group 1 mGlu-LTD
Several of the CIE-induced changes in GluN2B-associated proteins from the synaptic-enriched fraction are indicative of upregulated LTD-associated signaling [Table 1; (Cavarsan et al., 2012; Jakkamsetti et al., 2013; Loweth et al., 2013; Potschka et al., 2002; Tappe and Kuner, 2006; Wolf and Tseng, 2012)]. Specifically, we find increased Arc and MEK 1, which can promote group 1 mGlu-LTD, as well as, increased Homer 1 (a mGlu scaffold protein). Further there is a decrease in the GluA2 subunit the AMPA receptor, which could be reflective of LTD.
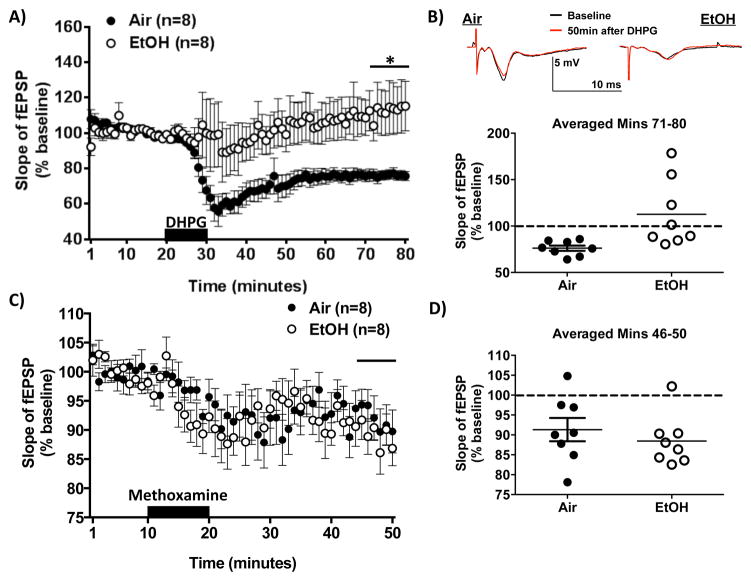
A single report has indicated that acute, high concentration ethanol can block LTD in the immature hippocampus induced by the broad spectrum agonist ACPD (Overstreet et al., 1997), but no studies have evaluated the effects of chronic ethanol exposure in the mature hippocampus. To do this, we measured excitatory field potentials (fEPSPs) evoked by Schaffer collateral stimulation in area CA1 (Sch-CA1) of the hippocampus in slices from either CIE-W or Air controls during exposure to the Group 1 mGlu agonist, DHPG (10 mins). DHPG produced a rapid and long lasting depression of fEPSPs in slices from Air controls but failed to have a significant effect on fEPSPs in slices from CIE-W mice (t(7.661) = 2.753; p < 0.05; Figure 4A/B).
Figure 4. DHPG and Methoxamine-induced LTD following CIE-W.
(A) Hippocampal slope of excitatory field potentials before/after DHPG (10 mins; 100 μM). (B) Top - Representative traces of excitatory field potentials before/after DHPG application in air and ethanol treated mice. Bottom -Summary of 40–50 mins post drug; n = 8; * p < .05. (C) Hippocampal slope of excitatory field potentials before/after Methoxamine (10 mins; 40 μM). (D) Summary of 25–30 mins post drug; n = 8.
We next performed immunoblot analyses for mGlu5 in hippocampus, and found no significant changes in levels of mGlu5 protein in S1 (cytoplasmic; t(10) = 0.4384; p = 0.61, N.S.), S2 (t(10) = 2.162; p = 0.06, N.S.), and S3 (t(10) = 0.0167; p = 0.99, N.S.) fractions or of immunoprecipitated GluN2B S2 (t(22) = 0.3069; p = 0.91, N.S.) or S3 (t(10) = 0.9736 p = 0.35, N.S.) fractions.
α1-adrenergic Receptor-induced LTD Remains Intact Following Chronic Alcohol
We next wanted to determine if this disruption in LTD was specific to Group 1 mGlu-LTD or if other forms of LTD were also altered by alcohol. Since NMDAR-dependent low frequency stimulation (LFS)-LTD can only be induced in the immature hippocampus (Kemp et al., 2000), it was not feasible to evaluate the effects of CIE-W on this form of plasticity. Therefore, we focused on another form of Gq GPCR linked-LTD induced by activation of α1 adrenergic receptors using an α1 adrenergic receptor agonist, Methoxamine (Scheiderer et al., 2004). In the hippocampus, it has been demonstrated that other Gq-coupled GPCRs induced-LTDs share a common mechanisms ((Scheiderer et al., 2004; Scheiderer et al., 2008). To do this, we measured fEPSPs in the Sch-CA1 region of the hippocampus in slices from either CIE-W or Air controls during exposure to Methoxamine (10 mins; 40μM). Methoxamine produced a long lasting depression of fEPSPs in slices from both CIE-W and Air- treated mice (t(14) = 0.7759; p = 0.45, N.S.; Figure 4C/D).
Discussion
We used a discovery-based proteomic approach to assess the impact of chronic alcohol treatment (CIE-W) on GluN2B-associated proteins in synaptic and non-synaptic fractions, identifying a unique subset of CIE-W regulated interactions in different subcellular compartments. This type of proteomic approach has been previously utilized with the AMPAR and NMDAR to identify both known and novel signaling complexes (Husi et al., 2000; Kang et al., 2012). While our methods were focused on enriching the GluN2B-NMDAR proteome, it was encouraging that the proteins that were identified overlapped nicely with the previously identified NMDAR proteome (Husi et al., 2000). An indication into how these proteins might be mechanistically related can be seen in the protein enrichment strategies that we employed. For example, GO enrichment analysis found that many proteins could be classified into PDZ-domain proteins, scaffolding proteins, glutamate receptor binding proteins, and structural activity proteins; which are all important in receptor localization and glutamate synapse plasticity. Further, pathway enrichment analysis also implicated several pathways of interest: LTP (a form of plasticity known to be disrupted by alcohol), endocytosis (mechanism responsible for AMPAR internalization), and Huntington’s disease [a disease with a pathophysiology related to extrasynaptic GluN2B-NMDA receptor transmission; (Gladding and Raymond, 2011)]. The changes in these proteins are capable of providing important clues into the molecular mechanisms responsible for alcohol-induced effects on GluN2B trafficking and how these could relate to altered plasticity.
In addition to mechanisms identified by these pathway enrichment strategies, we also surveyed the literature for potentially affected signaling cascades indicated by the protein changes found in these studies. In doing this, we identified several CIE-sensitive proteins linked to mGlu-LTD. Since there was no previous evidence that this form of LTD was modulated by alcohol in the mature hippocampus, we set out to test this mechanism. Our studies here showed that chronic alcohol treatment (CIE-W) was able to profoundly disrupt Group 1 mGlu-LTD, but not α1-adrenergic receptor induced-LTD.
Disruption of GluN2B and Scaffolding Proteins at the PSD
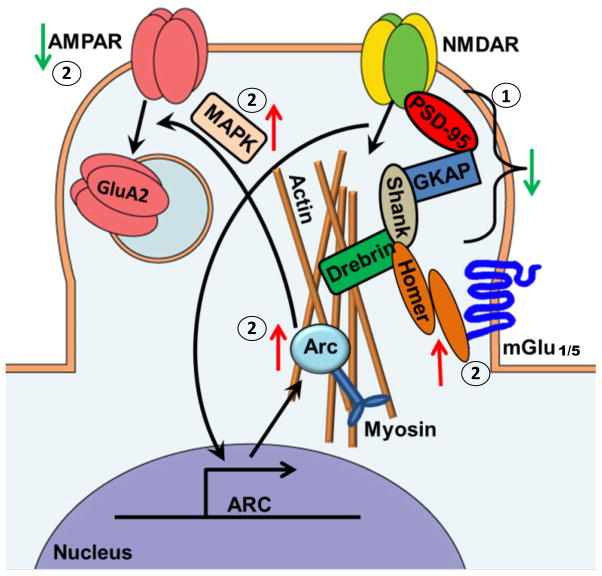
The GO enrichment analysis illustrated that PDZ domain and scaffold proteins were significantly changed by alcohol exposure. The model in Figure 5 illustrates many of the scaffolding proteins located at glutamatergic synapses. NMDARs are bound within the synaptic cytoarchitecture by PSD-95. PSD-95 binds to GKAPs, which acts as a link between NMDAR-PSD and Shank-Homer (Luo et al., 2012). Dynamic regulation of GKAPs was recently shown to be critical for homeostatic synaptic scaling (Shin et al., 2012). In these studies, alcohol exposure produces a decrease in GluN2B association with PSD proteins (PSD-95, PSD-93, SAP-102), GKAP (SAPAP 3), and Shank 3 in the synaptic-, and PSD-93 and Drebrin in the non-synaptic fraction. This decrease in GluN2B association with the PSD scaffolding proteins further supports prior work in hippocampal cultures demonstrating the relocalization of these NMDARs after chronic ethanol (Clapp et al., 2010). Relocalization is also supported by the decrease in NMDAR subunit (GluN1, GluN2B, GluN2A) association following CIE-W in the synaptic fractions. In other work, genetic knockouts of PSD-95 have been shown to alter various alcohol-related behaviors and thus illustrate the importance of the PSD complex in the regulation of alcohol’s effects (Camp et al., 2011).
Figure 5. Schematic of GluN2B-associated Alcohol-Induced Protein Changes in the Synaptic (S3) Fraction of Hippocampal Glutamatergic Synapses.
Arrows indicate the direction of alcohol-induced change. Up arrows indicate proteins that were significantly increased by chronic alcohol while down arrows indicate proteins that were significantly decreased by chronic alcohol. 1) Illustrates the destabilization of many key PSD scaffolding proteins following CIE. 2) Illustrates the CIE-induced changes in numerous proteins involved in the mGlu1/5-LTD signaling cascade.
In hippocampal primary cultures, chronic alcohol produces a synaptic clustering of NMDARs, especially GluN2B-NMDARs (Carpenter-Hyland and Chandler, 2006; Carpenter-Hyland et al., 2004). Other work confirmed this GluN2B synaptic enhancement and illustrated that during alcohol withdrawal GluN2B moved to extrasynaptic locations through lateral diffusion (Clapp et al., 2010). Further, our previous work in the bed nucleus of the stria terminalis (BNST) also showed that CIE-W seems to enhance transmission at extrasynaptic GluN2B-NMDARs (Wills et al., 2012). Transmission through these extrasynaptic GluN2B-NMDARs is normally thought to preferentially promote LTD versus LTP (Massey et al., 2004; Newpher and Ehlers, 2009), although evidence in the BNST suggests a potentially enhancing role on transmission (Wills et al., 2012). Extrasynaptic GluN2B transmission is involved in various excitotoxic processes and implicated in a number of pathological conditions [e.g. Huntingtons disease, ischemia, Parkinson’s disease, Alzheimer’s disease (Gladding and Raymond, 2011; Hardingham and Bading, 2010)]. It is interesting to note that Huntington’s disease was identified in our pathway enrichment analysis and its pathology involves extrasynaptic GluN2B. It is possible that common mechanisms may be responsible for the relocation of these receptors in both diseases.
Regulation mGlu-LTD by Alcohol
Several of the proteins altered by CIE were found to be involved in mGlu-LTD. Our results found an increased association of synaptic GluN2B with Arc signaling proteins (Arc, MEK 1) and Homer 1 (a mGlu scaffolding protein) along with a decrease in GluA2 (AMPAR subunit). Arc is an immediate early gene that undergoes rapid transcription in response to stimuli and then translocates to the synapse to regulate local translation (Bramham et al., 2008). Arc is a dynamic regulator of various forms of plasticity in the hippocampus and is critically involved in the endocytosis of AMPARs that occurs during mGlu-LTD. Further, synaptic translation of Arc primes neurons for induction of group 1 mGlu-LTD (Jakkamsetti et al., 2013). Therefore, in the current work the increased synaptic association of Arc could be indicative of neurons primed for mGlu-LTD. It is important to note that the change in Arc was not validated with our western blot. Western approaches have weaknesses in their own right and since the current proteomic screen was so robust in its sample size and statistical analyses, it is potentially more reliable.
Homer proteins are scaffold proteins that serve as the linker between NMDAR-PSD-GKAP-Shank complexes and mGlu1/5. Thus these proteins serve as a convergence point of NMDAR and mGlu signaling. In our studies, we find that Homer 1 association with GluN2B is increased in the synaptic fraction and decreased in the non-synaptic fraction following CIE-W. Homer proteins are known regulators mGlu1/5 trafficking and signaling. mGlu1/5 are highly expressed in the hippocampus, where their activation leads to LTD (Huber et al., 2001; Inta et al., 2012). Drugs of abuse can induce mGlu-LTD in the accumbens through the internalization of GluA2-containing AMPARs (Loweth et al., 2013; Wolf and Tseng, 2012). mGlu5 signaling is known to play a role in many alcohol related behaviors (Adams et al., 2010; Backstrom et al., 2004; Besheer et al., 2008; Besheer et al., 2010; Besheer et al., 2006; Cowen et al., 2005; Hodge et al., 2006; Lominac et al., 2006; Sinclair et al., 2012). Recent work further demonstrates a role of group I mGlu-homer signaling on ethanol binge drinking (Cozzoli et al., 2013). Ethanol has also been shown to occlude GluN2B-dependent LTD in the nucleus accumbens (Jeanes et al., 2011) indicating a potential connection between GluN2B transmission and LTD mechanisms. Since the current work finds an increased synaptic association of GluN2B and Homer 1, it suggests that CIE-W is disrupting mGlu1/5-LTD in the hippocampus.
Indeed we find no evidence of DHPG-induced depression (LTD) in alcohol-treated mice, which was readily induced in control mice. This result suggests that chronic alcohol disrupts mGlu-LTD induction mechanisms or that LTD is induced in vivo and thus occludes further LTD by DHPG. Our proteomic data argues for the latter since increased Arc and decreased GluA2 are indicative of LTD induction. Further, there is no change in mGluR5 protein levels, which also suggests that induction mechanism seem to be intact. This disruption of group I mGlu-LTD was not generalizable to other forms of Gq GPCR-linked LTD since α1-adrenergic receptor induced-LTD was intact following CIE-W. This α1-adrenergic receptor induced-LTD at these hippocampal synapses has previously been shown to be activity and NMDAR-dependent and rely on Erk signaling (Scheiderer et al., 2004; Scheiderer et al., 2008). Thus the retention of this LTD following CIE-W suggests that both NMDAR-dependent induction mechanisms and Erk signaling remain intact. This separable disruption of Gq GPCR-linked-LTD has been observed in the BNST, where stress specifically alters α1-adrenergic receptor induced-LTD and leaves mGlu1/5-LTD intact. The cause for these disparate effects was due to distinct postsynaptic LTD maintenance mechanisms (McElligott et al., 2010). These studies not only demonstrate a new way in which alcohol can modulate plasticity in the mature hippocampus but emphasize the utility of this discovery-based proteomic approach to generate new hypotheses regarding alcohol-related mechanisms.
Conclusions
This study illuminates new mechanisms by which chronic alcohol and withdrawal effect plasticity in a region known to be critical to the memory impairing effects of alcohol, the hippocampus. The hippocampus is a highly studied region, which is highly involved in contextual associations and episodic memory. This context association plays a pivotal role in context-induced relapse to alcohol and alcohol regulation of memory in this region is responsible for “blackouts” [see Review (Zorumski et al., 2014)]. Additionally, chronic alcohol is known to induce neurotoxicity in hippocampus through activation of NMDA receptors (particularly GluN2B-containing). This neurotoxicity is thought to be involved in certain memory impairments and seizures (Harris et al., 2003; Lewis et al., 2012; Stepanyan et al., 2008). These behaviors are all critical to the etiology and treatment of alcohol use disorders and thus understanding their mechanisms is necessary. Further, these proteomic data have highlighted key proteins that may be responsible for the relocalization of GluN2B-NMDARs to extrasynaptic locations during alcohol withdrawal. This understanding of how signaling mechanisms change as these NMDARs relocate will provide critical information into new therapeutic targets to combat this pathological signaling. Furthermore, information gained from these studies could provide novel insights into numerous neuropsychiatric disorders, which involve disrupted GluN2B signaling(Gladding and Raymond, 2011).
Acknowledgments
D.L.T. and Y.Y.C. were supported by NIH/NCI U01 CA152647
R.C., A.J.B., and J.G.P. were supported by R01-MH063232, K01-NS073700, and F32MH100747
D.G.W., T.W., and K.L. were supported by R01-AA019455, K99-AA22651, and F31 AA021623-01A1 Generation of GluN2B KO mice were generated by E.D and supported by U01-AA013514
Footnotes
Author Contributions
Tiffany Wills: substantial contributions to conception and design, acquisition of data, and analysis and interpretation of data; drafting the article; final approval of the version to be published
Anthony J. Baucum: contributions to design, acquisition of data, and analysis and interpretation of data; revising it critically for important intellectual content;
Katherine M. Louderback: acquisition of data and analysis
Yaoyi Chen: analysis and interpretation of data
Johanna G. Pasek: acquisition of data and analysis
Eric Delpire: acquisition of data and editing of manuscript
David L. Tabb: analysis and interpretation of data and editing of manuscript
Roger J. Colbran: contributions to design; revising it critically for important intellectual content
Danny G. Winder: contributions to design; drafting the article or revising it critically for important intellectual content; final approval of the version to be published
References
- Adams CL, Short JL, Lawrence AJ. Cue-conditioned alcohol seeking in rats following abstinence: involvement of metabotropic glutamate 5 receptors. British journal of pharmacology. 2010;159:534–542. doi: 10.1111/j.1476-5381.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom P, Bachteler D, Koch S, Hyytia P, Spanagel R. mGluR5 antagonist MPEP reduces ethanol-seeking and relapse behavior. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2004;29:921–928. doi: 10.1038/sj.npp.1300381. [DOI] [PubMed] [Google Scholar]
- Badanich KA, Doremus-Fitzwater TL, Mulholland PJ, Randall PK, Delpire E, Becker HC. NR2B Deficient Mice are More Sensitive to the Locomotor Stimulant and Depressant Effects of Ethanol. Genes Brain Behav. 2011 doi: 10.1111/j.1601-183X.2011.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Bartlett TE, Bannister NJ, Collett VJ, Dargan SL, Massey PV, Bortolotto ZA, Fitzjohn SM, Bashir ZI, Collingridge GL, Lodge D. Differential roles of NR2A and NR2B-containing NMDA receptors in LTP and LTD in the CA1 region of two-week old rat hippocampus. Neuropharmacology. 2007;52:60–70. doi: 10.1016/j.neuropharm.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Baucum AJ, 2nd, Brown AM, Colbran RJ. Differential association of postsynaptic signaling protein complexes in striatum and hippocampus. J Neurochem. 2013;124:490–501. doi: 10.1111/jnc.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ, 2nd, Jalan-Sakrikar N, Jiao Y, Gustin RM, Carmody LC, Tabb DL, Ham AJ, Colbran RJ. Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Molecular & cellular proteomics : MCP. 2010;9:1243–1259. doi: 10.1074/mcp.M900387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ, 2nd, Strack S, Colbran RJ. Age-dependent targeting of protein phosphatase 1 to Ca2+/calmodulin-dependent protein kinase II by spinophilin in mouse striatum. PloS one. 2012;7:e31554. doi: 10.1371/journal.pone.0031554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28:1829–1838. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- Besheer J, Faccidomo S, Grondin JJ, Hodge CW. Regulation of motivation to self-administer ethanol by mGluR5 in alcohol-preferring (P) rats. Alcoholism, clinical and experimental research. 2008;32:209–221. doi: 10.1111/j.1530-0277.2007.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besheer J, Grondin JJ, Cannady R, Sharko AC, Faccidomo S, Hodge CW. Metabotropic glutamate receptor 5 activity in the nucleus accumbens is required for the maintenance of ethanol self-administration in a rat genetic model of high alcohol intake. Biological psychiatry. 2010;67:812–822. doi: 10.1016/j.biopsych.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besheer J, Stevenson RA, Hodge CW. mGlu5 receptors are involved in the discriminative stimulus effects of self-administered ethanol in rats. European journal of pharmacology. 2006;551:71–75. doi: 10.1016/j.ejphar.2006.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Mirshahi T, Chandler LJ, Woodward JJ. Effects of acute and chronic ethanol exposure on heteromeric N-methyl-D-aspartate receptors expressed in HEK 293 cells. J Neurochem. 1997;69:2345–2354. doi: 10.1046/j.1471-4159.1997.69062345.x. [DOI] [PubMed] [Google Scholar]
- Blevins T, Mirshahi T, Woodward JJ. Increased agonist and antagonist sensitivity of N-methyl-D-aspartate stimulated calcium flux in cultured neurons following chronic ethanol exposure. Neuroscience letters. 1995;200:214–218. doi: 10.1016/0304-3940(95)12086-j. [DOI] [PubMed] [Google Scholar]
- Blitzer RD, Gil O, Landau EM. Long-term potentiation in rat hippocampus is inhibited by low concentrations of ethanol. Brain research. 1990;537:203–208. doi: 10.1016/0006-8993(90)90359-j. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigman JL, Wright T, Talani G, Prasad-Mulcare S, Jinde S, Seabold GK, Mathur P, Davis MI, Bock R, Gustin RM, Colbran RJ, Alvarez VA, Nakazawa K, Delpire E, Lovinger DM, Holmes A. Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning. J Neurosci. 2010;30:4590–4600. doi: 10.1523/JNEUROSCI.0640-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, Baucum AJ, Bass MA, Colbran RJ. Association of protein phosphatase 1 gamma 1 with spinophilin suppresses phosphatase activity in a Parkinson disease model. J Biol Chem. 2008;283:14286–14294. doi: 10.1074/jbc.M801377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp MC, Feyder M, Ihne J, Palachick B, Hurd B, Karlsson RM, Noronha B, Chen YC, Coba MP, Grant SG, Holmes A. A novel role for PSD-95 in mediating ethanol intoxication, drinking and place preference. Addiction biology. 2011;16:428–439. doi: 10.1111/j.1369-1600.2010.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter-Hyland EP, Chandler LJ. Homeostatic plasticity during alcohol exposure promotes enlargement of dendritic spines. The European journal of neuroscience. 2006;24:3496–3506. doi: 10.1111/j.1460-9568.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Carpenter-Hyland EP, Woodward JJ, Chandler LJ. Chronic ethanol induces synaptic but not extrasynaptic targeting of NMDA receptors. J Neurosci. 2004;24:7859–7868. doi: 10.1523/JNEUROSCI.1902-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarsan CF, Tescarollo F, Tesone-Coelho C, Morais RL, Motta FL, Blanco MM, Mello LE. Pilocarpine-induced status epilepticus increases Homer1a and changes mGluR5 expression. Epilepsy research. 2012;101:253–260. doi: 10.1016/j.eplepsyres.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Clapp P, Gibson ES, Dell’acqua ML, Hoffman PL. Phosphorylation regulates removal of synaptic N-methyl-D-aspartate receptors after withdrawal from chronic ethanol exposure. J Pharmacol Exp Ther. 2010;332:720–729. doi: 10.1124/jpet.109.158741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen MS, Djouma E, Lawrence AJ. The metabotropic glutamate 5 receptor antagonist 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]-pyridine reduces ethanol self-administration in multiple strains of alcohol-preferring rats and regulates olfactory glutamatergic systems. The Journal of pharmacology and experimental therapeutics. 2005;315:590–600. doi: 10.1124/jpet.105.090449. [DOI] [PubMed] [Google Scholar]
- Cozzoli DK, Courson J, Wroten MG, Greentree DI, Lum EN, Campbell RR, Thompson AB, Maliniak D, Worley PF, Jonquieres G, Klugmann M, Finn DA, Szumlinski KK. Binge Alcohol Drinking by Mice Requires Intact Group1 Metabotropic Glutamate Receptor Signaling Within the Central Nucleus of the Amygdale. Neuropsychopharmacology. 2013 doi: 10.1038/npp.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzoli DK, Goulding SP, Zhang PW, Xiao B, Hu JH, Ary AW, Obara I, Rahn A, Abou-Ziab H, Tyrrel B, Marini C, Yoneyama N, Metten P, Snelling C, Dehoff MH, Crabbe JC, Finn DA, Klugmann M, Worley PF, Szumlinski KK. Binge drinking upregulates accumbens mGluR5-Homer2-PI3K signaling: functional implications for alcoholism. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8655–8668. doi: 10.1523/JNEUROSCI.5900-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Dasari S, Chambers MC, Martinez MA, Carpenter KL, Ham AJ, Vega-Montoto LJ, Tabb DL. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. Journal of proteome research. 2012;11:1686–1695. doi: 10.1021/pr200874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildy JE, Leslie SW. Ethanol inhibits NMDA-induced increases in free intracellular Ca2+ in dissociated brain cells. Brain research. 1989;499:383–387. doi: 10.1016/0006-8993(89)90789-0. [DOI] [PubMed] [Google Scholar]
- Follesa P, Ticku MK. Chronic ethanol treatment differentially regulates NMDA receptor subunit mRNA expression in rat brain. Brain research Molecular brain research. 1995;29:99–106. doi: 10.1016/0169-328x(94)00235-7. [DOI] [PubMed] [Google Scholar]
- Follesa P, Ticku MK. NMDA receptor upregulation: molecular studies in cultured mouse cortical neurons after chronic antagonist exposure. J Neurosci. 1996;16:2172–2178. doi: 10.1523/JNEUROSCI.16-07-02172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens B. Low doses of ethanol impair spatial working memory and reduce hippocampal theta activity. Alcohol Clin Exp Res. 1995;19:763–767. doi: 10.1111/j.1530-0277.1995.tb01580.x. [DOI] [PubMed] [Google Scholar]
- Gladding CM, Raymond LA. Mechanisms underlying NMDA receptor synaptic/extrasynaptic distribution and function. Mol Cell Neurosci. 2011 doi: 10.1016/j.mcn.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Grover CA, Frye GD. Ethanol effects on synaptic neurotransmission and tetanus-induced synaptic plasticity in hippocampal slices of chronic in vivo lead-exposed adult rats. Brain research. 1996;734:61–71. [PubMed] [Google Scholar]
- Gustin RM, Shonesy BC, Robinson SL, Rentz TJ, Baucum AJ, 2nd, Jalan-Sakrikar N, Winder DG, Stanwood GD, Colbran RJ. Loss of Thr286 phosphorylation disrupts synaptic CaMKIIalpha targeting, NMDAR activity and behavior in pre-adolescent mice. Mol Cell Neurosci. 2011;47:286–292. doi: 10.1016/j.mcn.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy PA, Chen W, Wilce PA. Chronic ethanol exposure and withdrawal influence NMDA receptor subunit and splice variant mRNA expression in the rat cerebral cortex. Brain research. 1999;819:33–39. doi: 10.1016/s0006-8993(98)01340-7. [DOI] [PubMed] [Google Scholar]
- Harris BR, Gibson DA, Prendergast MA, Blanchard JA, Holley RC, Hart SR, Scotland RL, Foster TC, Pedigo NW, Littleton JM. The neurotoxicity induced by ethanol withdrawal in mature organotypic hippocampal slices might involve cross-talk between metabotropic glutamate type 5 receptors and N-methyl-D-aspartate receptors. Alcohol Clin Exp Res. 2003;27:1724–1735. doi: 10.1097/01.ALC.0000093601.33119.E3. [DOI] [PubMed] [Google Scholar]
- Hendricson AW, Maldve RE, Salinas AG, Theile JW, Zhang TA, Diaz LM, Morrisett RA. Aberrant synaptic activation of N-methyl-D-aspartate receptors underlies ethanol withdrawal hyperexcitability. J Pharmacol Exp Ther. 2007;321:60–72. doi: 10.1124/jpet.106.111419. [DOI] [PubMed] [Google Scholar]
- Hendricson AW, Miao CL, Lippmann MJ, Morrisett RA. Ifenprodil and ethanol enhance NMDA receptor-dependent long-term depression. J Pharmacol Exp Ther. 2002;301:938–944. doi: 10.1124/jpet.301.3.938. [DOI] [PubMed] [Google Scholar]
- Henniger MS, Wotjak CT, Holter SM. Long-term voluntary ethanol drinking increases expression of NMDA receptor 2B subunits in rat frontal cortex. European journal of pharmacology. 2003;470:33–36. doi: 10.1016/s0014-2999(03)01787-4. [DOI] [PubMed] [Google Scholar]
- Hodge CW, Miles MF, Sharko AC, Stevenson RA, Hillmann JR, Lepoutre V, Besheer J, Schroeder JP. The mGluR5 antagonist MPEP selectively inhibits the onset and maintenance of ethanol self-administration in C57BL/6J mice. Psychopharmacology. 2006;183:429–438. doi: 10.1007/s00213-005-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PL, Rabe CS, Moses F, Tabakoff B. N-methyl-D-aspartate receptors and ethanol: inhibition of calcium flux and cyclic GMP production. J Neurochem. 1989;52:1937–1940. doi: 10.1111/j.1471-4159.1989.tb07280.x. [DOI] [PubMed] [Google Scholar]
- Hu XJ, Follesa P, Ticku MK. Chronic ethanol treatment produces a selective upregulation of the NMDA receptor subunit gene expression in mammalian cultured cortical neurons. Brain Res Mol Brain Res. 1996;36:211–218. doi: 10.1016/0169-328x(95)00223-f. [DOI] [PubMed] [Google Scholar]
- Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis--dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nature neuroscience. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Inta D, Filipovic D, Lima-Ojeda JM, Dormann C, Pfeiffer N, Gasparini F, Gass P. The mGlu5 receptor antagonist MPEP activates specific stress-related brain regions and lacks neurotoxic effects of the NMDA receptor antagonist MK-801: significance for the use as anxiolytic/antidepressant drug. Neuropharmacology. 2012;62:2034–2039. doi: 10.1016/j.neuropharm.2011.12.035. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Nagashima K, Murayama K, Zorumski CF. Acute effects of ethanol on hippocampal long-term potentiation and long-term depression are mediated by different mechanisms. Neuroscience. 2005;136:509–517. doi: 10.1016/j.neuroscience.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Jakkamsetti V, Tsai NP, Gross C, Molinaro G, Collins KA, Nicoletti F, Wang KH, Osten P, Bassell GJ, Gibson JR, Huber KM. Experience-Induced Arc/Arg3.1 Primes CA1 Pyramidal Neurons for Metabotropic Glutamate Receptor-Dependent Long-Term Synaptic Depression. Neuron. 2013;80:72–79. doi: 10.1016/j.neuron.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes ZM, Buske TR, Morrisett RA. In vivo chronic intermittent ethanol exposure reverses the polarity of synaptic plasticity in the nucleus accumbens shell. J Pharmacol Exp Ther. 2011;336:155–164. doi: 10.1124/jpet.110.171009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri HS, Mehta AK, Ticku MK. Up-regulation of NMDA receptor subunits in rat brain following chronic ethanol treatment. Brain research Molecular brain research. 1998;58:221–224. doi: 10.1016/s0169-328x(98)00112-0. [DOI] [PubMed] [Google Scholar]
- Kang MG, Nuriya M, Guo Y, Martindale KD, Lee DZ, Huganir RL. Proteomic analysis of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor complexes. J Biol Chem. 2012;287:28632–28645. doi: 10.1074/jbc.M111.336644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash TL, Baucum AJ, 2nd, Conrad KL, Colbran RJ, Winder DG. Alcohol exposure alters NMDAR function in the bed nucleus of the stria terminalis. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:2420–2429. doi: 10.1038/npp.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp N, McQueen J, Faulkes S, Bashir ZI. Different forms of LTD in the CA1 region of the hippocampus: role of age and stimulus protocol. Eur J Neurosci. 2000;12:360–366. doi: 10.1046/j.1460-9568.2000.00903.x. [DOI] [PubMed] [Google Scholar]
- Kumari M. Differential effects of chronic ethanol treatment on N-methyl-D-aspartate R1 splice variants in fetal cortical neurons. J Biol Chem. 2001;276:29764–29771. doi: 10.1074/jbc.M100317200. [DOI] [PubMed] [Google Scholar]
- Lam H, Deutsch EW, Aebersold R. Artificial decoy spectral libraries for false discovery rate estimation in spectral library searching in proteomics. Journal of proteome research. 2010;9:605–610. doi: 10.1021/pr900947u. [DOI] [PubMed] [Google Scholar]
- Lewis B, Wellmann KA, Kehrberg AM, Carter ML, Baldwin T, Cohen M, Barron S. Behavioral deficits and cellular damage following developmental ethanol exposure in rats are attenuated by CP-101,606, an NMDAR antagonist with unique NR2B specificity. Pharmacology, biochemistry, and behavior. 2012;100:545–553. doi: 10.1016/j.pbb.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- Lominac KD, Kapasova Z, Hannun RA, Patterson C, Middaugh LD, Szumlinski KK. Behavioral and neurochemical interactions between Group 1 mGluR antagonists and ethanol: potential insight into their anti-addictive properties. Drug and alcohol dependence. 2006;85:142–156. doi: 10.1016/j.drugalcdep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, White G, Weight FF. Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science. 1989;243:1721–1724. doi: 10.1126/science.2467382. [DOI] [PubMed] [Google Scholar]
- Loweth JA, Tseng KY, Wolf ME. Adaptations in AMPA receptor transmission in the nucleus accumbens contributing to incubation of cocaine craving. Neuropharmacology. 2013 doi: 10.1016/j.neuropharm.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Li X, Fei Z, Poon W. Scaffold protein Homer 1: implications for neurological diseases. Neurochemistry international. 2012;61:731–738. doi: 10.1016/j.neuint.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Massey PV, Johnson BE, Moult PR, Auberson YP, Brown MW, Molnar E, Collingridge GL, Bashir ZI. Differential roles of NR2A and NR2B-containing NMDA receptors in cortical long-term potentiation and long-term depression. J Neurosci. 2004;24:7821–7828. doi: 10.1523/JNEUROSCI.1697-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Wilce PA, Buckley T, Dodd P, Puzke J, Spanagel R, Zieglgansberger W, Wolf G, Leng S, Rommelspacher H, Finckh U, Schmidt LG. Ethanol and gene expression in brain. Alcoholism, clinical and experimental research. 2001;25:82S–86S. doi: 10.1097/00000374-200105051-00015. [DOI] [PubMed] [Google Scholar]
- McElligott ZA, Klug JR, Nobis WP, Patel S, Grueter BA, Kash TL, Winder DG. Distinct forms of Gq-receptor-dependent plasticity of excitatory transmission in the BNST are differentially affected by stress. Proc Natl Acad Sci U S A. 2010;107:2271–2276. doi: 10.1073/pnas.0905568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J, Kolok S, Dezso P, Boros A, Szombathelyi Z. Differential alterations in the expression of NMDA receptor subunits following chronic ethanol treatment in primary cultures of rat cortical and hippocampal neurones. Neurochem Int. 2003;42:35–43. doi: 10.1016/s0197-0186(02)00062-1. [DOI] [PubMed] [Google Scholar]
- Narita M, Soma M, Mizoguchi H, Tseng LF, Suzuki T. Implications of the NR2B subunit-containing NMDA receptor localized in mouse limbic forebrain in ethanol dependence. Eur J Pharmacol. 2000;401:191–195. doi: 10.1016/s0014-2999(00)00428-3. [DOI] [PubMed] [Google Scholar]
- Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends in cell biology. 2009;19:218–227. doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Obara I, Bell RL, Goulding SP, Reyes CM, Larson LA, Ary AW, Truitt WA, Szumlinski KK. Differential effects of chronic ethanol consumption and withdrawal on homer/glutamate receptor expression in subregions of the accumbens and amygdala of P rats. Alcoholism, clinical and experimental research. 2009;33:1924–1934. doi: 10.1111/j.1530-0277.2009.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet LS, Pasternak JF, Colley PA, Slater NT, Trommer BL. Metabotropic glutamate receptor mediated long-term depression in developing hippocampus. Neuropharmacology. 1997;36:831–844. doi: 10.1016/s0028-3908(97)00031-2. [DOI] [PubMed] [Google Scholar]
- Pawlak R, Melchor JP, Matys T, Skrzypiec AE, Strickland S. Ethanol-withdrawal seizures are controlled by tissue plasminogen activator via modulation of NR2B-containing NMDA receptors. Proc Natl Acad Sci U S A. 2005;102:443–448. doi: 10.1073/pnas.0406454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potschka H, Krupp E, Ebert U, Gumbel C, Leichtlein C, Lorch B, Pickert A, Kramps S, Young K, Grune U, Keller A, Welschof M, Vogt G, Xiao B, Worley PF, Loscher W, Hiemisch H. Kindling-induced overexpression of Homer 1A and its functional implications for epileptogenesis. The European journal of neuroscience. 2002;16:2157–2165. doi: 10.1046/j.1460-9568.2002.02265.x. [DOI] [PubMed] [Google Scholar]
- Qiang M, Denny AD, Ticku MK. Chronic intermittent ethanol treatment selectively alters N-methyl-D-aspartate receptor subunit surface expression in cultured cortical neurons. Mol Pharmacol. 2007;72:95–102. doi: 10.1124/mol.106.033043. [DOI] [PubMed] [Google Scholar]
- Scheiderer CL, Dobrunz LE, McMahon LL. Novel form of long-term synaptic depression in rat hippocampus induced by activation of alpha 1 adrenergic receptors. Journal of neurophysiology. 2004;91:1071–1077. doi: 10.1152/jn.00420.2003. [DOI] [PubMed] [Google Scholar]
- Scheiderer CL, Smith CC, McCutchen E, McCoy PA, Thacker EE, Kolasa K, Dobrunz LE, McMahon LL. Coactivation of M(1) muscarinic and alpha1 adrenergic receptors stimulates extracellular signal-regulated protein kinase and induces long-term depression at CA3-CA1 synapses in rat hippocampus. J Neurosci. 2008;28:5350–5358. doi: 10.1523/JNEUROSCI.5058-06.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schummers J, Browning MD. Evidence for a role for GABA(A) and NMDA receptors in ethanol inhibition of long-term potentiation. Brain research Molecular brain research. 2001;94:9–14. doi: 10.1016/s0169-328x(01)00161-9. [DOI] [PubMed] [Google Scholar]
- Sheela Rani CS, Ticku MK. Comparison of chronic ethanol and chronic intermittent ethanol treatments on the expression of GABA(A) and NMDA receptor subunits. Alcohol. 2006;38:89–97. doi: 10.1016/j.alcohol.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M, Lee SH. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nature neuroscience. 2012;15:1655–1666. doi: 10.1038/nn.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair CM, Cleva RM, Hood LE, Olive MF, Gass JT. mGluR5 receptors in the basolateral amygdala and nucleus accumbens regulate cue-induced reinstatement of ethanol-seeking behavior. Pharmacology, biochemistry, and behavior. 2012;101:329–335. doi: 10.1016/j.pbb.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyan TD, Farook JM, Kowalski A, Kaplan E, Barron S, Littleton JM. Alcohol withdrawal-induced hippocampal neurotoxicity in vitro and seizures in vivo are both reduced by memantine. Alcohol Clin Exp Res. 2008;32:2128–2135. doi: 10.1111/j.1530-0277.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- Szumlinski KK, Lominac KD, Oleson EB, Walker JK, Mason A, Dehoff MH, Klugmann M, Cagle S, Welt K, During M, Worley PF, Middaugh LD, Kalivas PW. Homer2 is necessary for EtOH-induced neuroplasticity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:7054–7061. doi: 10.1523/JNEUROSCI.1529-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappe A, Kuner R. Regulation of motor performance and striatal function by synaptic scaffolding proteins of the Homer1 family. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:774–779. doi: 10.1073/pnas.0505900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:4541–4551. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitlauf C, Egli RE, Grueter BA, Winder DG. High-frequency stimulation induces ethanol-sensitive long-term potentiation at glutamatergic synapses in the dorsolateral bed nucleus of the stria terminalis. J Neurosci. 2004;24:5741–5747. doi: 10.1523/JNEUROSCI.1181-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills TA, Klug JR, Silberman Y, Baucum AJ, Weitlauf C, Colbran RJ, Delpire E, Winder DG. GluN2B subunit deletion reveals key role in acute and chronic ethanol sensitivity of glutamate synapses in bed nucleus of the stria terminalis. Proc Natl Acad Sci U S A. 2012;109:E278–287. doi: 10.1073/pnas.1113820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills TA, Winder DG. Ethanol effects on N-methyl-D-aspartate receptors in the bed nucleus of the stria terminalis. Cold Spring Harbor perspectives in medicine. 2013;3:a012161. doi: 10.1101/cshperspect.a012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ME, Tseng KY. Calcium-permeable AMPA receptors in the VTA and nucleus accumbens after cocaine exposure: when, how, and why? Frontiers in molecular neuroscience. 2012;5:72. doi: 10.3389/fnmol.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. Journal of proteome research. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorumski CF, Mennerick S, Izumi Y. Acute and chronic effects of ethanol on learning-related synaptic plasticity. Alcohol. 2014;48:1–17. doi: 10.1016/j.alcohol.2013.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]