Abstract
Insect mitochondrial genomes (mitogenomes) contain a conserved set of 37 genes for an extensive diversity of lineages. Previously reported dictyopteran mitogenomes share this conserved mitochondrial gene arrangement, although surprisingly little is known about the mitogenome of Mantodea. We sequenced eight mantodean mitogenomes including the first representatives of two families: Hymenopodidae and Liturgusidae. Only two of these genomes retain the typical insect gene arrangement. In three Liturgusidae species, the trnM genes have translocated. Four species of mantis (Creobroter gemmata, Mantis religiosa, Statilia sp., and Theopompa sp.-HN) have multiple identical tandem duplication of trnR, and Statilia sp. additionally includes five extra duplicate trnW. These extra trnR and trnW in Statilia sp. are erratically arranged and form another novel gene order. Interestingly, the extra trnW is converted from trnR by the process of point mutation at anticodon, which is the first case of tRNA reassignment for an insect. Furthermore, no significant differences were observed amongst mantodean mitogenomes with variable copies of tRNA according to comparative analysis of codon usage. Combined with phylogenetic analysis, the characteristics of tRNA only possess limited phylogenetic information in this research. Nevertheless, these features of gene rearrangement, duplication, and reassignment provide valuable information toward understanding mitogenome evolution in insects.
The metazoan mitochondrial genome (mitogenome) is an ideal model system for comparative and evolutionary genomic research. The typical mitogenome of metazoans encodes a conserved set of 37 genes for 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes1, with genome-level characters, such as genome size, gene content, and gene order, display high diversity in some lineages2,3. Gene rearrangements are observed frequently in some groups, while gene duplication and loss are distributed sporadically in limited lineages such as Bivalvia, Cephalopod, and Afrobatrachia4,5,6. These remaining duplicate genes and pseudogenes represent important data for exploring the evolutionary history and mechanisms of gene rearrangement and recruitment. For the arrangement of mitochondrial genes, the variation in relative positions of PCGs and rRNA genes are more limited compared with that of tRNA genes across organisms within a phylum7. The tRNA genes with characteristics of diverse changes in relative position, gene content, and secondary structure, are considered as an important tool in studying the evolution of mitogenome, in particular to the rearrangement mechanism8,9,10. Additionally, its variation is usually linked to evolutionary relationships in a wide range of lineages at different taxonomic levels suggesting these features of tRNA could be utilized as useful phylogenetic markers11.
The extensive gene rearrangements (including PCGs and RNA) of insect mitogenomes have been detected in several lineages within the Diptera (Trichoceridae, Cecidomyiidae), Hemiptera (Enicocephalidae), Hymenoptera, Thysanoptera, Psocoptera and Phthiraptera12,13,14,15,16,17,18, while most of investigated mitogenomes share the same gene order with the hypothesized ancestral pancrustacean mitogenome arrangement19 or possess rare tRNA rearrangement. Previously reported dictyopteran mitogenomes consistently display the typical ancestral gene order and content, however only two species are praying mantises and the rest are cockroaches and termites. Members of the Mantodea, a separate lineage within the Dictyoptera, have evolved many unique morphological and behavioural features as the ambush and pursuit predators20,21,22. A better understanding of the diversity of mitogenome evolution in this enigmatic order underlines the need for exploring more taxa with the diverse praying mantis.
Herein, we report eight new mitogenomes from Mantodea and describe their general characteristics. Two new gene rearrangements and reassignment of tRNA genes are described, and evolutionary mechanisms for the gene rearrangements and duplication are discussed. Further, we examine the relationship between tRNA gene duplication and codon usage, and investigate whether these tRNA features vary with phylogeny.
Results
General features of Mantodea mitogenomes
Seven complete and one nearly complete praying mantis mitogenomes, including the first representatives from Hymenopodidae and Liturgusidae, were generated. The complete mitogenomes are typical circular DNA, and most of the common characteristics of insect mitogenomes are in the Mantodea mitogenomes (Supplementary Table S1–8). Standard collection of 37 mitochondrial genes including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and an A + T-rich region were detected in these mitogenomes, whereas the additional tRNAs were found in four species (Creobroter gemmata, Mantis religiosa, Statilia sp., and Theopompa sp.-HN). The lengths of these new mitogenomes range from 15,531 bp in Tenodera sinensis (Mantidae) to about 17,370 bp in Theopompa sp.-HN (Liturgusidae). Although the length variation of A + T-rich region is the main factor for difference in mitogenome size, some extra tRNA gene contents and non-coding regions also contribute to genome length variation. Most of new and published mantodean mitogenomes retain typical insect gene arrangement without considering the multiple gene copies in the original position. However, some gene orders of tRNAs in Statilia and Liturgusidae species depart from the common arrangement (Fig. 1).
Figure 1. The gene map of the mitochondrial genomes for eight praying mantises.
Gene and genome sizes are not to scale. All genes without underline are transcribed in the direction form left to right, and with underline are transcribed in the direction form right to left. Genes labeled an asterisk are pseudogenes.
All analyzed mantodean mitogenomes show base composition biases (Table 1). The range of mantodean A + T content is from 70.1% in Humbertiella nada to 77.8% in Anaxarcha zhengi, with a mean of 74.2%. Overall, the A + T contents of Liturgusidae species are lower than that of other analyzed species. The distribution of A + T content changed based on various gene or region. For Liturgusidae species, PCGs show the lowest A + T content, while the highest A + T content presents in different region among these three species. The remaining six mitogenomes are under the similar distribution trend in that rRNA and A + T-rich regions possess higher A + T content than PCGs and tRNA, the exception being Tamolanica tamolana.
Table 1. Base composition of mantodean mitochondrial genomes.
| Family | Species | Whole genome | Protein-coding genes | rRNA genes | tRNA genes | A + T-rich region |
|---|---|---|---|---|---|---|
| Hymenopodidae | Anaxarcha zhengi | 77.8 | 76.9 | 80.2 | 77.4 | 79.3 |
| Creobroter gemmata | 76.0 | 75.4 | 77.4 | 75.2 | 79.9 | |
| Mantidae | Tenodera sinensis | 75.5 | 74.7 | 77.8 | 75.5 | 78.9 |
| Tamolanica tamolana | 75.3 | 74.7 | 77.0 | 76.5 | 74.7 | |
| Mantis religiosa | 76.7 | 76.1 | 77.9 | 75.8 | 81.1 | |
| Statilia sp. | 75.3 | 74.7 | 77.6 | 75.4 | 76.2 | |
| Liturgusidae | Humbertiella nada | 70.1 | 68.8 | 73.1 | 73.1 | 73.7 |
| Theopompa sp.-YN | 70.7 | 69.8 | 74.4 | 72.7 | 70.8 | |
| Theopompa sp.-HN | 71.7 | 70.6 | 74.4 | 75.5 | 71.2 |
In mantodean mitogenomes, all inferred start codons are canonical start codons ATN except for ATP6 and COI. ATP6 of C. gemmata uses GTG as the start codon which is accepted conventional start codon for invertebrate mitogenome23. COI begins with another accepted start condon TTG in M. religiosa, C. gemmata, Statilia sp. and A. zhengi. For three Liturgusidae species, the COI start codon was inferred as non-canonical start codon CTG. Although several candidate common start condons were found around this region, the alignment of amino acid sequences with other mantis species suggests that the triplet CTG could be the start codon for these COI. Stop codons are more conserved than start codons in mantodean mitogenomes. Most PCGs invariably end with the typical stop codon TAA or truncated codon TA/T, while limited PCGs (Theopompa sp.-YN COI and T. tamolana ND3) exhibit TAG as stop codon. Comparative analyses of the relative synonymous codon usage of nine mantodean mitogenomes show the similar pattern for codon usage bias (Supplementary Table S9). In particular, the codon usage for Arginine and Tryptophan encoded with multiple tRNA copies is also analogous among their relative species (Table 2).
Table 2. Codon usage of codons for Arginine and Tryptophan.
| Species | Codons for Arginine |
Codons for Tryptophan |
||||||
|---|---|---|---|---|---|---|---|---|
| CGA | CGU | CGC | CGG | Total | UGA | UGG | Total | |
| Anaxarcha zhengi | 30 | 23 | 1 | 2 | 56 | 98 | 4 | 102 |
| Creobroter gemmata | 31 | 17 | 1 | 8 | 57 | 97 | 7 | 104 |
| Tenodera sinensis | 36 | 19 | 1 | 2 | 58 | 96 | 8 | 104 |
| Tamolanica tamolana | 32 | 21 | 2 | 2 | 57 | 101 | 1 | 102 |
| Mantis religiosa | 31 | 24 | 0 | 2 | 57 | 98 | 6 | 104 |
| Statilia sp. | 31 | 18 | 6 | 2 | 57 | 93 | 11 | 104 |
| Humbertiella nada | 29 | 16 | 4 | 9 | 58 | 94 | 14 | 108 |
| Theopompa sp.-YN | 29 | 15 | 2 | 9 | 55 | 88 | 17 | 105 |
| Theopompa sp.-HN | 30 | 17 | 2 | 7 | 56 | 91 | 19 | 110 |
| Average of nine species | 31.0 | 18.9 | 2.1 | 4.8 | 56.8 | 95.1 | 9.7 | 104.8 |
Non-coding regions
In general, insect mitogenomes are highly compact with few short non-coding fragments except for the A + T-rich region. Most of sequenced mantodean mitogenomes share this feature; however, three Liturgusidae species contain 129–138 bp intergenic spacers between trnM and trnI (this part was not amplified in Theopompa sp.-HN) and 59–68 bp intergenic spacers between trnQ and ND2. Perhaps the origin of these inserts involved gene rearrangement of this tRNA cluster. Indeed, we detected the remnant of trnM from the inserts between trnQ and ND2 (Fig. 2A). But in the former inserts, none of homologous fragments of mitochondrial genes or regions were found. In Theopompa sp.-YN and C. gemmata, 69 bp and 68 bp intergenic spacers were found between trnA and trnR, and trnR and trnN, respectively. And these intergenic spacer sequences display high sequence similarity with trnR (Fig. 2B). The T. tamolana mitogenome also contains a relatively large non-coding sequence (297 bp) between trnM and ND2 including the tandem duplication unit of the A + T-rich region. In addition, the intergenic spacer between trnS2 and ND1 was found in all sequenced mantodean mitogenomes, ranging from 17 bp to 37 bp. Alignments of all these intergenic spacer sequences showed a 7 bp highly conserved motif (WTACTTA) which could be considered as the binding site of the transcription termination factor (DmTTF).
Figure 2.
(A) Alignment of trnM and pseudogene trnM sequences in Humbertiella nada, and Theopompa spp. (B) Alignment of trnR and pseudogene trnR sequences in Creobroter gemmata and Theopompa sp.-YN. (C) Alignment of trnW2 in Statilia sp. and trnR in nine praying mantises. Genes with an asterisk are pseudogenes.
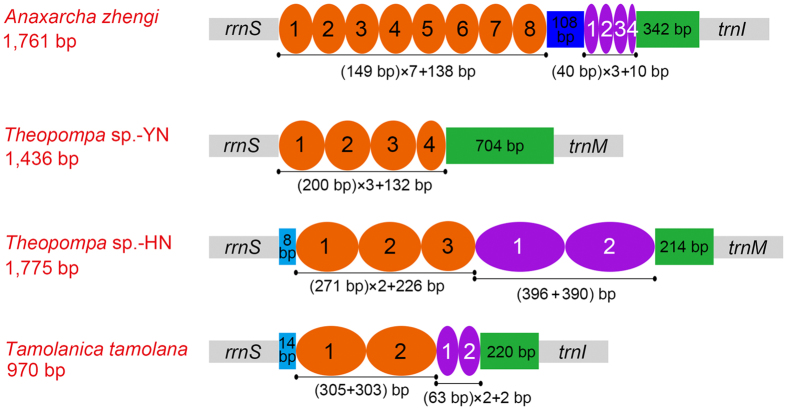
The A + T-rich region known as the control region for insect mitogenome is the largest non-coding region in all mantodean mitogenomes. The entire A + T-rich regions have distinguished differences in size, which range from 639 bp in M. religiosa to 1,775 bp in Theopompa sp.-HN. Four of comparatively larger sequences contain larger tandem repeats (Fig. 3) which are main factor for variation in the length. In Theopompa sp.-YN, only one tandem repeat unit with 200 bp extends three identical copies with a partial fourth. While the rest three species (A. zhengi, T. tamolana and Theopompa sp.-HN) include two different types of tandem repeats. Furthermore, plenty of microsatellite-like elements present in mantodean A + T-rich regions.
Figure 3. Organization of the A + T-rich region of Anaxarcha zhengi, Tamolanica tamolana, Theopompa sp.-YN and Theopompa sp.-HN mitochondrial genomes.
Oval with different color indicates tandem repeat sequence. Colored box shows the non-repeat region.
tRNA duplication, reassignment and rearrangements
Mitochondrial tRNA of Mantodea exhibits high evolutionary diversity including duplication, reassignment and rearrangement. Although the extra gene copy is uncommon in animal mitogenome, four of eight praying mantis mitogenomes contain multiple identical tRNA: two trnR in M. religiosa, three trnR in C. gemmata, eight trnR in Theopompa sp.-HN, and six trnR and five trnW2 (excluding trnW1) in Statilia sp. The trnW1 and trnW2 of Statilia sp. hold the same anticodon but show the low sequence similarity. The obvious sequence similarity of trnW2 and tnrR (Fig. 2C) indicates the duplicate trnR underwent a gene recruitment process, namely trnR converted to trnW by point mutation at the third anticodon position from triplet “TCG” to “TCA”. These uncommon changes contribute to the variable gene content and order in Mantodea mitogenomes. In three Liturgusidae species, trnM has moved into the position downstream of the A + T-rich region. The translocation of trnM poses a new gene order of this tRNA cluster (trnM-trnI-trnQ). Another novel gene order is present in tRNA cluster between ND3 and ND5. Duplicate trnR and trnW2 alternately occur between trnA and trnN and generate a unique gene order [ND3-trnA-(trnR)2-trnW-(trnR)2-(trnW)2-(trnR-trnW)2-trnN-trnS-trnE-trnF-ND5] for this tRNA cluster.
Phylogenetic analyses
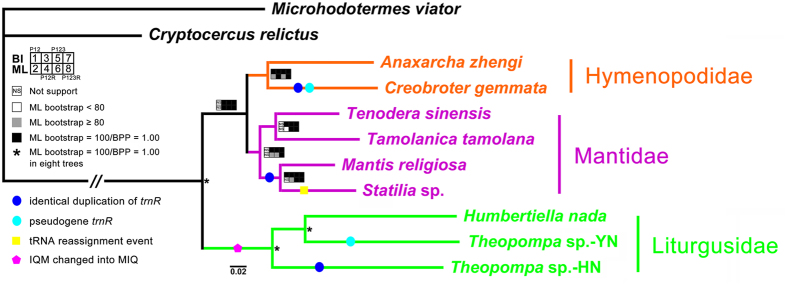
The phylogenetic relationships inferred from maximum likelihood (ML) and Bayesian inference (BI) analyses using three datasets (PCG123, PCG123R, and PCG12R) share similar topologies (Fig. 4). Our analyses based on datasets with all mitochondrial genes show a higher branch support values than PCGs dataset. Not surprisingly, Liturgusidae diverged basally from the rest of the species24, and all three families (Liturgusidae, Hymenopodidae, and Mantidae) form a monophyly respectively with strong support. Consistent with previous results, both Mantini species Statilia sp. and M. religiosa share a closer relationship within the Mantidae, and the other two species T. tamolana (Paramantini) and T. sinensis (Polyspilotini) cluster together as a sister group to the Mantini. While within the clade of Liturgusidae, Theopompa is unexpectedly identified as a non-monophyly group with topology ((Humbertiella nada + Theopompa sp.-YN) + Theopompa sp.-HN). The results of PCG12 dataset show different topologies for Mantidae but with low support values (ML bootstrap <50).
Figure 4. Phylogenetic relationship among nine praying mantises inferred from mitogenome data using BI and ML analyses.
Asterisk indicate Bayesian posterior probabilities (BPP) = 1.00 and ML bootstrap = 100 in all Phylogenetic analysis. Squares at the nodes correspond to BPP for 1 (PCG12), 3 (PCG12R), 5 (PCG123), and 7 (PCG123R) and ML bootstrap support values in percentages for 2 (PCG12), 4 (PCG12R), 6 (PCG123) and 8 (PCG123R). NS: not support; white square: ML bootstrap values <80; gray square: ML bootstrap values ≥80; black square: ML bootstrap values = 100 and BPP = 1.00.
Discussion
Mantodean mitogenomes show numbers of common features observed in other insects such as AT-biased base composition and codon usage. Compared within sequenced dictyopteran mitogenomes, the overall A + T contents of mantodean mitogenomes (70.1–77.8%) are comparable to those of Blattodea (72.0–76.0%)25,26 but relatively higher than those of Isoptera (63.4–71.4%)27. The length of mantodean coding region without duplicate genes is similar to that of other dictyopterans. The intergenic spacers are frequently found in mantodean mitogenomes contained pseudogenes or gene rearrangements, while most dictyopterans with ancestral gene arrangment present more compact mitogenome architecture. The A + T-rich region of mantodean mitogenomes also reflects the general characteristics for dictyopterans including tandem repeat sequences, microsatellite-like elements and poly-T structure. Although several fragments with relatively high similarity were found among close relative species, the conserved blocks could not be identified through all dictyopterans.
Gene duplication generally together with rearrangement occurs in the metazoan mitogenome, and the redundant gene copy might be rapidly deleted or pseudogenized with mutations under strong selective pressure23. Although the extra gene copy is uncommon in the metazoan mitogenome, five of eight praying mantis mitogenomes possess the duplication of tRNA. Slipped-strand mispairing could give rise to these repeated trnR during mitogenome replication28. The identical trnR gene sequences demonstrate that all of them are functional trnR and the duplicate is a very recent event. In fact, Theopompa sp.-YN and C. gemmata harbor another pseudogene trnR between trnA and trnR, and trnR and trnN respectively (Fig. 1). The pseudogene trnR of C. gemmata has a truncated and mutated 3′ end (acceptor stem secondary structure is absent), while mutations distribute through the degenerated trnR in Theopompa sp.-YN (Fig. 2B). The physical positions of two pseudogene trnR indicate that degeneration can occur in original and new copy of trnR, and both of two trnR for Theopompa sp.-YN may be functional genes before degeneration.
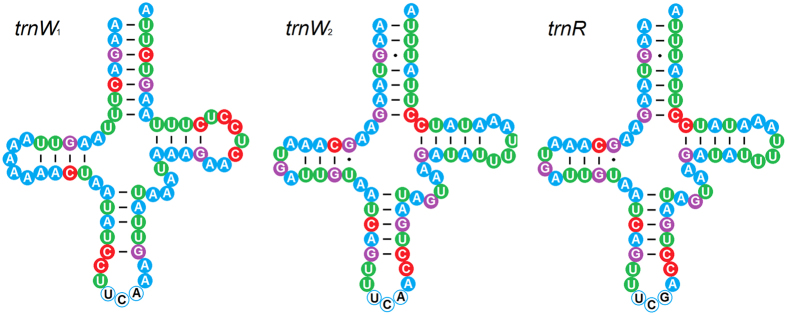
Gene duplication usually provides important source of material for formation of evolutionarily related genes with a specific function29. As the products of gene duplication, some copies function in the original role, while the others could serve a new function. In Statilia sp., two non-homologous sequences encode Tryptophan tRNA genes with same anticodon. The trnW2 is reassigned from trnR by point mutation at anticodon. Although two functional trnW is uncommon in animal mitogenomes, the stable secondary structure identical with full functional trnR implies potential function for trnW2 (Fig. 5). Remolding tRNA with only an anticodon point mutation may replace the role of a second disabled tRNA30. However, a single point mutation at the anticodon without any post-transcriptional modifications can not change the original advanced structure and identity elements which influence the recognition and interaction between tRNA and aminoacyl-tRNA synthetase. The first base pair A1-U72 (T72 at the DNA level) in acceptor stem and G73 are confirmed as the identity elements on trnW for bacterial tryptophanyl-tRNA synthetase31,32,33,34,35, however, unexpectedly both trnW1 and trnW2 harbor the same nucleotides for identity elements. Consequently, we cannot rule out the possibility that trnW2 participates in interactions, in particular to the coexistence with clear functional trnW1. Gene recruitment is an important mechanism for tRNA evolution, while the direct evidence of recent gene recruitment are detected in limited organisms such as Amoebidium parasiticum (Ichthyophonida), Axinella corrugata (Demospongiae), Crassostrea sp. and Pinctada maxima (Bivalvia), and Tropiocolotes tripolitanus (Gekkonidae)36,37,38,39. Most of mitochondrial tRNA recruitments happen in regular circular mitogenomes, the exception being some organism with linear chromosomes, where these recruitment features of Statilia sp. and T. tripolitanus provide evidences for verifying that the higher frequency of tRNA gene duplication would facilitate the process of gene recruitment.
Figure 5. Inferred secondary structure of trnR and two trnW genes for Statilia sp.
All available mitogenome organizations of Mantodea are shown in Fig. 1. Only two match the typical ancestral pancrustacean gene order when considering the multiple gene copies of trnR. The new gene orders of three Liturgusidae species and Statilia sp. are firstly reported in Dictyoptera which indicate Mantodea could no longer be treated as a lineage with low gene rearrangements. For the gene rearrangement of three Liturgusidae species, tandem duplication-random loss (TDRL) model is a plausible mechanism which has been proposed to explain the similar rearrangements in other insects40,41. We assume that the original gene order of this region is trnI-trnQ-trnM. The tandem duplication of trnI-trnQ-trnM was followed by random deletion of trnI-trnQ in first set and trnM in seconded set (Fig. 6A). In fact, the pseudogene remnant of trnM was confidently identified in intergenic spacer between trnQ and ND2 (Fig. 2A). Although mutations indicate trnM pseudogene could not fold into stable secondary structure (also including mutations of anticodon), the high level of sequence similarity for alignments of functional and pseudogenes trnM from these species respectively suggests that each of them is homologues.
Figure 6. Putative mechanism of mitochondrial gene rearrangements occurring in Humbertiella nada, Theopompa spp. and Statilia sp.
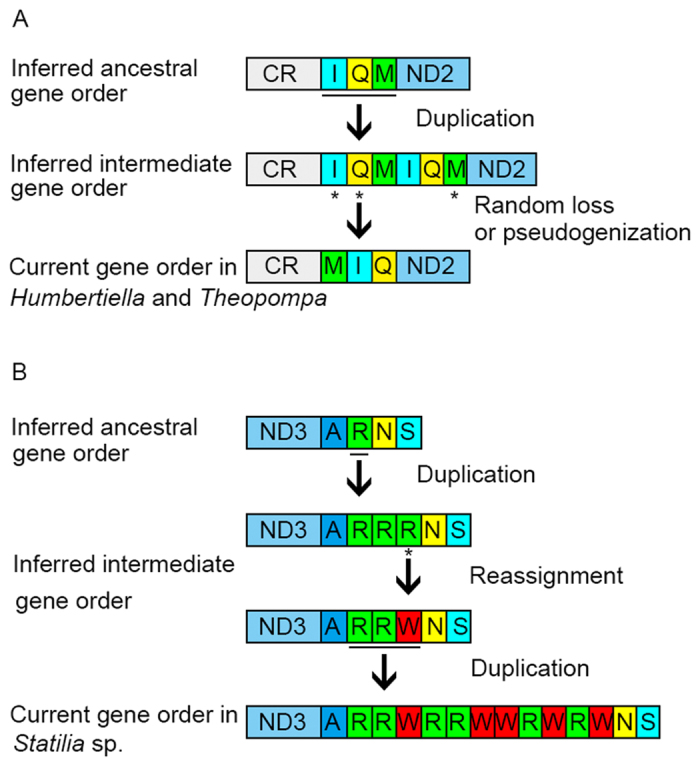
Even though various gene orders of the tRNA cluster between ND3 and ND5 were found in a number of insects42, the new tRNA gene arrangement in Statilia sp. [trnA-(trnR)2-trnW-(trnR)2-(trnW)2-(trnR-trnW)2-trnN] is novel for all sequenced pancrustacea. The unusual and extra trnR and trnW were generated by tRNA duplication and reassignment. Actually, trnR and trnW constantly alternated with different copy number indicating that the present gene order was derived from large duplication (might include deletion and/or other process) after reassignment for specific duplicate tRNA (Fig. 6B). We prefer this mechanism considering that reassignment is a rare event within the insect mitogenome, whereas gene duplication occurs more frequently in mitogenome. Moreover the mechanism mentioned above possesses more parsimonious scenario than multiple trnR duplication followed by random reassignment for several trnR.
Codon usage and tRNA content suffer the co-adaptation evolution in Escherichia coli, Saccharomyces cerevisiae and Caenorhabditis elegans genomes43,44,45 suggesting the strong correlation between the number of tRNA genes and codon usage. And all the isoaccepting tRNAs corresponding to preferential codons have the highest gene copy number. Recent studies proposed that change of tRNA gene content didn’t influence codon usage bias within vertebrate mitogenomes9,39. Among invertebrates, large duplications of tRNA gene is exceedingly rare, especially for Insecta. Large duplicate tRNA genes in Mantodea mitogenome provide the chance to investigate the relationship between tRNA gene number and codon usage. The frequencies of codon CGA corresponding trnR genes duplication in Statilia sp., M. religiosa, C. gemmata, Theopompa sp.-HN do not change compared to those of other mantises (Table 2). The use of overall Arginine codons is also stable for all analytical mantodean mitogenomes. In addition, a single trnW always decode two codon (UGA and UGG) for mitochondrial protein synthesis in invertebrate mitogenome. However, one trnW1 and five trnW2 appear in Statilia sp. mitogenome. Despite with five extra trnW2, the codon use UGA in Statilia sp. does not significantly differ from other mantises, and the relative frequency of UGA versus UGG is similar to other mantises. Moreover statistical findings in Table 2 do not show evidence for total Tryptophan codons use change (UGA + UGG). All evidence is indicative of increased tRNA gene copy does not change codon usage bias.
Recently, several molecular studies rejected the monophyly for several families of Mantodea24,46. Combining our results with previously reported phylogenetic relationships within Mantodea24, the common trait of ‘trnM-trnI-trnQ’ for these three species may derive from the ancestor of the Asian bark mantis rather than Liturgusidae, and this feature may also be found in other closely related species and genera. But the novel tRNA gene order in Statilia sp. underwent relatively recent multiple changes, implying this gene rearrangement characteristic may be less conserved and just an independent event for specific taxa. Gene order be regularly regarded as useful marker for phylogenetic analysis especially at higher taxonomic levels, but this characteristic of Mantodea is not appropriate for analysis at least in family-level. Another rare tRNA reassignment feature only happened in Statilia sp. mitogenome but not in other species with dupilicate trnR, although the high level of tRNA gene duplication may facilitate the process of gene recruitment. In animal mitogenomes, gene duplications are recognized as systematic or individual evolutionary event for different lineage. With respect to our findings that the duplication of tRNA occurs in five of nine mantis, including all three families, we cannot immediately confirm this duplication reflects phylogenetic information based on limited samples. But this feature might be helpful for studying lower level relationships.
In summary, we determined eight new mitogenomes of Mantodea including the first representatives from Hymenopodidae and Liturgusidae. These mitogenomes show several unusual features of genomic organization including gene rearrangement, gene duplication and gene reassignment. Comparative analysis of mitogenome sequences involving the changed gene order indicate that tandem duplication-random loss and duplication-reassignment-duplication are the persuasive mechanism for gene rearrangements in Asian bark mantis and Statilia sp. respectively. Abundant duplicate tRNA genes do not influence codon usage suggesting the absence of correlation between tRNA gene number and codon usage. In addition, tRNA reassignment by point mutation of anticodon generates an additional trnW2 in Statilia sp., which is really rare in Insecta. And all these various variations of tRNA were considered as potential markers for investigation of lower level relationships. With evolutionary diversity of tRNA, this enigmatic lineage, Mantodea, could expand the understanding of evolution for mitochondrial tRNA and gene order with more samples.
Methods
Mitochondrial Genome Sequencing
Eight new praying mantises mitochondrial genomes were obtained from a single specimen respectively (Supplementary Table S10) with overlapping PCR fragments and primer walking. DNA was extracted using the TIANamp Micro DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol. Universal primers47 and specific primers used to amplify mitochondrial genomes are listed in Supplementary Table S11. The overlapping fragments were amplified using FastPfu Fly DNA Polymerase (TransGen Biotech, Beijing, China) with the following cycling conditions: an initial denaturation for 1 min at 93 °C, followed by 35 cycles of 15 sec at 92 °C, 1 min at 45–57 °C, 2–7 min at 72 °C, and final extension of 10 min at 72 °C. After purification with PCR Purification Kit (Sangon Biotech, Shanghai, China), all PCR products were sequenced directly with the PCR primers and internal primers generated by primer walking.
Genome annotation and sequence analyses
Contiguous sequence fragments were assembled using Staden Package v1.7.048. Protein coding genes (PCGs) and ribosomal RNA (rRNA) genes were identified based on homologous regions of other insects using the Clustal X49,50. Transfer RNAs (tRNA) and their potential cloverleaf structures were identified by tRNAscan-SE 1.2151. The tRNAs, which were not detected by tRNA scan-SE v1.21, were identified by comparing the sequence to Tamolanica tamolana (GenBank accession number DQ241797). Tandem Repeat Finder v4.07 was used to identify tandem repeats in non-coding regions52. The base composition and codon usage were calculated with MEGA v5.153. All these new mitogenomes of Mantodea have been deposited at GenBank under the accessions KU201313–KU201320.
Phylogenetic analyses
To infer the phylogenetic relationships among sequenced mantises (Microhodotermes viator with GenBank accession number NC_018122 and Cryptocercus relictus with GenBank accession number NC_018132 as outgroup54), Four datasets (PCG123: 13 PCGs including all codon positions; PCG123R: two rRNAs, 22 tRNAs and 13PCGs including all codon positions; PCG12: 13 PCGs without third codon positions; PCG12R: two rRNAs, 22 tRNAs and 13PCGs without third codon positions) were used to draw the maximum likelihood (ML) and Bayesian inference (BI) phylogeny with the partitioning scheme of rRNA, tRNA and each codon for PCGs (merged codon positions across genes formed three independent codon subsets) to PCG123R and PCG12R, and codon-based partitions to PCG123 and PCG1255. We extracted and translated all 13 PCGs using the invertebrate mitochondrial genetic code. Then, the inferred amino acid sequences, rRNA and tRNA genes were individually aligned with Clustal X and ambiguously aligned regions were removed with Gblocks56. The best-fit model (GTR + Γ + I) for each partition was selected using Akaike information criterion in jModelTest57. For ML analyses implemented in RAxML ver.7.2.858, bootstrap analysis was performed with 1,000 replicates. For BI analyses implemented in MrBayes ver.3.1.259, two sets of four chains were allowed to run simultaneously for 1,000,000 generations. Each set was sampled every 100 generations with a burn-in of 25%.
Additional Information
How to cite this article: Ye, F. et al. Mitochondrial genomes of praying mantises (Dictyoptera, Mantodea): rearrangement, duplication, and reassignment of tRNA genes. Sci. Rep. 6, 25634; doi: 10.1038/srep25634 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Frank Wieland (Pfalzmuseum für Naturkunde-POLLICHIA-Museum) for helping to identify bark mantis samples, Dr. Qiang Xie (Nankai university) for giving useful suggestions of phylogenetic analysis and Dr. David Cone (Saint Mary’s University) for editorial comments on an early draft of the manuscript. This study was supported by the National Nature Science Foundation of China (31372158).
Footnotes
Author Contributions F.Y. conceived and designed the study and collected samples. F.Y., X.L. and W.Z. performed the experiments. F.Y. analyzed the data and wrote the manuscript. P.Y. elaborated the design of this study and revised the manuscript. All authors have read and approve the final version.
References
- Boore J. L. Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 21, 439–446 (2006). [DOI] [PubMed] [Google Scholar]
- Cameron S. L. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 59, 95–117 (2014). [DOI] [PubMed] [Google Scholar]
- Passamonti M., Ricci A., Milani L. & Ghiselli F. Mitochondrial genomes and Doubly Uniparental Inheritance: new insights from Musculista senhousia sex-linked mitochondrial DNAs (Bivalvia Mytilidae). BMC Genomics 12, 442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S., Fukuda N., Nakamura M., Aoyama T. & Oshima T. Long-term conservation of six duplicated structural genes in Cephalopod mitochondrial genomes. Mol. Biol. Evol. 21, 2034–2046 (2004). [DOI] [PubMed] [Google Scholar]
- Kurabayashi A. & Sumida M. Afrobatrachian mitochondrial genomes: genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genomics 14, 633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S., Thomas W. K., Whitfield K. M., Kumazawa Y. & Wilson A. C. Rearrangements of mitochondrial transfer RNA genes in marsupials. J. Mol. Evol. 33, 426–430 (1991). [DOI] [PubMed] [Google Scholar]
- Jühling F. et al. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40, 2833–2845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. et al. The evolution of mitochondrial genomes in modern frogs (Neobatrachia): nonadaptive evolution of mitochondrial genome reorganization. BMC Genomics 15, 691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. Evolution of the tRNA gene family in mitochondrial genomes of five Meretrix clams (Bivalvia, Veneridae). Gene 533, 439–446 (2014). [DOI] [PubMed] [Google Scholar]
- Littlewood D. T. J., Lockyer A. E., Webster B. L., Johnston D. A. & Le T. H. The complete mitochondrial genomes of Schistosoma haematobium and Schistosoma spindale and the evolutionary history of mitochondrial genome changes among parasitic flatworms. Mol. Phylogenet. Evol. 39, 452–467 (2006). [DOI] [PubMed] [Google Scholar]
- Beckenbach A. T. Mitochondrial genome sequences of Nematocera (lower Diptera): evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol. Evol. 4, 89–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLos One 7, e29419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao M., Gibson T. & Dowton M. Evolutionary dynamics of the mitochondrial genome in the Evaniomorpha (Hymenoptera)—a group with an intermediate rate of gene rearrangement. Genome Biol. Evol. 6, 1862–1874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R. & Barker S. C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 20, 362–370 (2003). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Mitochondrial genomes of two Barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLos One 8, e61685 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L., Johnson K. P. & Whiting M. F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 65, 589–604 (2007). [DOI] [PubMed] [Google Scholar]
- Atray I., Bentur J. S. & Nair S. The Asian rice gall midge (Orseolia oryzae) mitogenome has evolved novel gene boundaries and tandem repeats that distinguish its biotypes. PLos One 10, e0134625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C. E. The complete mitochondrial genome of the stomatopod crustacean Squilla mantis. BMC Genomics 6, 105 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson G. J. & Whiting M. F. Phylogeny of Mantodea based on molecular data: evolution of a charismatic predator. Syst. Entomol. 29, 359–370 (2004). [Google Scholar]
- Ehrmann R. Mantodea: Gottesanbeterinnen der Welt. (Natur und Tier, 2002). [Google Scholar]
- Loxton R. G. & Nicholls I. The functional morphology of the praying mantis forelimb (Dictyoptera: Mantodea). Zool. J. Linn. Soc. 66, 185–203 (1979). [Google Scholar]
- Wolstenholme D. R. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141, 173–216 (1992). [DOI] [PubMed] [Google Scholar]
- Svenson G. J. & Whiting M. F. Reconstructing the origins of praying mantises (Dictyoptera, Mantodea): the roles of Gondwanan vicariance and morphological convergence. Cladistics 25, 468–514 (2009). [DOI] [PubMed] [Google Scholar]
- Tang M. et al. Multiplex sequencing of pooled mitochondrial genomes–a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42, e166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Y., Xuan W. J., Zhao J. L., Zhu C. D. & Jiang G. F. The complete mitochondrial genome of the cockroach Eupolyphaga sinensis (Blattaria: Polyphagidae) and the phylogenetic relationships within the Dictyoptera. Mol. Biol. Rep. 37, 3509–3516 (2010). [DOI] [PubMed] [Google Scholar]
- Bourguignon T. et al. The evolutionary history of termites as inferred from 66 mitochondrial genomes. Mol. Biol. Evol. 32, 406–421 (2015). [DOI] [PubMed] [Google Scholar]
- Levinson G. & Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203–221 (1987). [DOI] [PubMed] [Google Scholar]
- Magadum S., Banerjee U., Murugan P., Gangapur D. & Ravikesavan R. Gene duplication as a major force in evolution. J. Genet. 92, 155–161 (2013). [DOI] [PubMed] [Google Scholar]
- Rawlings T. A., Collins T. M. & Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA 100, 15700–15705 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Asahara H., Tamura K. & Shimizu M. Identity determinants of E. coli tryptophan tRNA. Nucleic Acids Res. 19, 6379–6382 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q. et al. Recognition by tryptophanyl-tRNA synthetases of discriminator base on tRNATrp from three biological domains. J. Biol. Chem. 277, 14343–14349 (2002). [DOI] [PubMed] [Google Scholar]
- Xu F. et al. Species-specific differences in the operational RNA code for aminoacylation of tRNATrp. Nucleic Acids Res. 29, 4125–4133 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Jin Y. X. & Wang D. B. Mitochondrial tRNATrps imply a eubacterial origin. Acta Biochim. Biophys. Sin. (Shanghai) 32, 100–104 (2000). [PubMed] [Google Scholar]
- Chen L., Jin Y. X. & Wang D. B. A discriminator base in tDNATrp. Acta Biochim. Biophys. Sin. (Shanghai). 31, 466–468 (1999). [PubMed] [Google Scholar]
- Lavrov D. V. & Lang B. F. Transfer RNA gene recruitment in mitochondrial DNA. Trends Genet. 21, 129–133 (2005). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. New features of Asian Crassostrea oyster mitochondrial genomes: a novel alloacceptor tRNA gene recruitment and two novel ORFs. Gene 507, 112–118 (2012). [DOI] [PubMed] [Google Scholar]
- Wu X., Li X., Li L. & Yu Z. A unique tRNA gene family and a novel, highly expressed ORF in the mitochondrial genome of the silver-lip pearl oyster, Pinctada maxima (Bivalvia: Pteriidae). Gene 510, 22–31 (2012). [DOI] [PubMed] [Google Scholar]
- Kumazawa Y., Miura S., Yamada C. & Hashiguchi Y. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genomics 15, 930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In Computational biology series, Vol. 1 (eds Sankoff D. & Nadeau J.) 133–147 (Kluwer Academic Publishers, 2000). [Google Scholar]
- Cao Y. Q., Ma C., Chen J. Y. & Yang D. R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics 13, 276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S. & Hadrys H. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol. Phylogenet. Evol. 69, 393–403 (2013). [DOI] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16, 287–289 (2000). [DOI] [PubMed] [Google Scholar]
- Dong H., Nilsson L. & Kurland C. G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- Kanaya S., Yamada Y., Kudo Y. & Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species specific diversity of codon usage based on multivariate analysis. Gene 238, 143–155 (1999). [DOI] [PubMed] [Google Scholar]
- Legendre F. et al. Phylogeny of Dictyoptera: dating the origin of cockroaches, praying mantises and termites with molecular data and controlled fossil evidence. PLos One 10, e0130127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Buckley T. R., Frati F., Stewart J. B. & Beckenbach A. T. Incorporating molecular evolutioninto phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 37, 545–579 (2006). [Google Scholar]
- Staden R., Beal K. F. & Bonfield J. K. The Staden package, 1998. Methods Mol. Biol. 132, 115–130 (2000). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L., Barker S. C. & Whiting M. F. Mitochondrial genomics and the new insect order Mantophasmatodea. Mol. Phylogenet. Evol. 38, 274–279 (2006). [DOI] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S. L., Lo N., Bourguignon T., Svenson G. J. & Evans T. A. A mitochondrial genome phylogeny of termites (Blattodea: Termitoidae): robust support for interfamilial relationships and molecular synapomorphies define major clades. Mol. Phylogenet. Evol. 65, 163–173 (2012). [DOI] [PubMed] [Google Scholar]
- Cameron S. L., Lambkin C. L., Barker S. C. & Whiting M. F. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 32, 40–59 (2007). [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T. & Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 21, 456–463 (2005). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.