Abstract
The opportunistic pathogen Streptococcus pneumoniae is commonly carried asymptomatically in the human nasopharynx. Due to high rates of cocolonization with other pneumococcus strains, intraspecific competitive interactions partly determine the carriage duration of strains and thereby their potential to cause disease. These interactions may be mediated by bacteriocins, such as the type IIb bacteriocins encoded by the blp (bacteriocin-like peptide) locus. To understand blp diversity and evolution, we undertook a bioinformatic analysis of 4,418 pneumococcal genomes, including 168 newly sequenced genomes. We describe immense variation at all levels of genomic organization: Gene presence/absence, gene order, and allelic diversity. If we make the extreme and naive hypothesis that assumes all genes in this operon can assort randomly, this variation could lead to 1015 distinct bacteriocin-related phenotypes, each potentially representing a unique ecological strategy; however, we provide several explanations for why this extreme is not realized. Although rarefaction analysis indicates that the number of unique strategies is not saturated, even after sampling thousands of genomes, we show that the variation is neither unbounded nor random. We delimit three bacteriocin groups, which contain group-specific bacteriocins, immunity genes, and blp operon gene order, and argue that this organization places a constraint on realized ecological strategies. We additionally show that ecological strategy diversity is significantly constrained by pneumococcal phylogeny and clonal structure. By examining patterns of association between alleles within the blp operon, we show that bacteriocin genes, which were believed to function in pairs, can be found with a broad diversity of partner alleles and immunity genes; this overall lack of allelic fidelity likely contributes to the fluid structure of this operon. Our results clarify the diversity of antagonistic ecological strategies in the global pneumococcal population and highlight the potential role of blp bacteriocins in competition within the nasopharynx.
Keywords: bacteriocin, bioinformatics, ecological strategies, interference competition, antagonism
Introduction
Approximately 14 million cases of pneumococcal diseases—including pneumonia, sepsis, and meningitis—are caused by the Gram-positive bacterium Streptococcus pneumoniae each year, resulting in an estimated 800,000 deaths per year in children under 5 years old (O’Brien et al. 2009). Additionally, up to 88% of children under 5 years old are asymptomatically colonized by S. pneumoniae in the nasopharynx (Regev-Yochay et al. 2004; Wyllie et al. 2014), which serves as a reservoir out of which strains migrate and cause disease (Bogaert et al. 2001). Pneumococcal conjugate vaccines have reduced childhood deaths by over 75% in areas where they are administered (Black et al. 2000; Klugman et al. 2003; O’Brien et al. 2003; Cutts et al. 2005). This vaccine has also resulted in a shift from vaccine serotypes to nonvaccine serotypes in the commensal pneumococcal populations that reside in the nasopharynx (Tocheva et al. 2011; Spijkerman et al. 2012; Davis et al. 2013). Vaccine-induced replacement, together with more advanced methods of detection, has led to an increased appreciation that simultaneous cocolonization with multiple pneumococcal strains is very common (Sauver et al. 2000; García-Rodríguez and Fresnadillo Martínez 2002; Brugger et al. 2010; Wyllie et al. 2014). Competitive interactions among these strains within the nasopharynx can therefore influence clonal frequencies, colonization dynamics, and, in turn, the potential for different bacterial strains to cause disease.
One of the key drivers of competitive dynamics between coexisting pneumococcal strains is small-peptide bacteriocins that regulate intraspecific killing. Streptococcus pneumoniae has at least four bacteriocin systems: The competence-regulated CibAB bacteriocin responsible for fratricidal killing (Guiral et al. 2005), the Phr lantibiotic (Hoover et al. 2015), the recently discovered pneumocyclicin (Bogaardt et al. 2015), and the blp (bacteriocin-like peptides) operon (Dawid et al. 2007; Lux et al. 2007), which we focus on here. Regulation of the blp operon is coordinated by a typical Gram-positive quorum sensing two-component system (De Saizieu et al. 2000), with BlpH as a membrane-bound, histidine kinase receptor for the secreted quorum sensing signal peptide produced by blpC (De Saizieu et al. 2000; Reichmann and Hakenbeck 2000). When extracellular levels of BlpC surpass a threshold concentration, the peptide signal binds to BlpH (Pinchas et al. 2015), which activates the response regulator BlpR by phophorylation; this, in turn, increases production of the BlpC signal (De Saizieu et al. 2000) and activates putative bacteriocin genes blpD, blpE, blpI, blpJ, blpK, blpM, blpN, blpO, blpW, pncT, and pncW (Bogaardt et al. 2015). Before secretion, the N-terminal, double-glycine leader sequence of the translated signal and bacteriocins is cleaved, after which the mature peptide is exported by the ABC transporter system encoded by BlpA and BlpB (Håvarstein et al. 1995). blp bacteriocins are thought to be type IIb bacteriocins, which require equimolar production of two separate peptides to bind to the outside of a target cell to cause cell death (Nissen-Meyer et al. 2010). Notably, however, specific pairing has only been experimentally shown for a single pair of Blp bacteriocins, BlpM and BlpN (Dawid et al. 2007). Moreover, it remains unknown if pairs of two-peptide bacteriocins show absolute fidelity to one another, or if active bacteriocins can form between diverse peptide partners. To avoid bacteriocin-associated suicide as well as to defend against the bacteriocins of competing strains (Bogaardt et al. 2015), putative immunity genes blpF, blpG, blpL, blpP, blpX, blpY, blpZ, pncG, pncM, and pncP are also activated by the response regulator BlpR. Two additional genes, blpT and blpS, have an unknown role in the regulation of the blp operon (De Saizieu et al. 2000).
Although the general structure of the blp operon appears to be conserved among pneumococcal strains (Dawid et al. 2007; Lux et al. 2007), the specific composition of the operon is markedly variable (Bogaardt et al. 2015). First, the signal peptide pheromone of each strain, encoded by blpC, can vary. At least four distinct peptide types in the species have been described, each believed to bind most tightly to its specific cognate receptor (Pinchas et al. 2015), and here we describe the discovery of several additional potential signal types. Second, the set of bacteriocins carried by each strain can differ, indicating that there is variation in the chemical arsenal that each strain carries (Lux et al. 2007). Finally, strains vary in the number and type of immunity genes within this operon, implying between-strain differences in susceptibility (Lux et al. 2007). The various permutations of these components can be thought of as a vast ecological strategy set. Each strain from this set, with a unique combination of signal, killing, and immunity, thus expresses one ecological strategy that defines the fraction of potential competitors a given strain can kill, and the fraction to which it is susceptible. How many such strategies exist from among those that are possible? At present, the answer to this fundamental question remains unknown.
Using more than 4,000 publically available S. pneumoniae genomes together with 168 genomes sequenced for this study, we sought to answer this question using a bioinformatics approach. We ask the following specific questions: First, how diverse are blp signals, receptors, bacteriocins, and immunity genes, and what is the combinatorial complexity of this operon? Second, what subset of the combinatorial possibilities among these ecological strategies is actually observed? Is the realized strategy set biased to particular clonal complexes or combinations of alleles, implying that there are phylogenetic or functional constraints on operon structure? Finally, what is the correlational structure of signal, receptor, bacteriocin, and immunity genes? In brief, we identify unprecedented diversity in the pneumococcal blp operon with a combinatorial ecological strategy potential into the trillions. Yet despite this vast set of possibilities, the number of realized ecological strategies of signaling, bacteriocins, and immunity is significantly smaller. We discuss these results in the context of bacterial competitiveness and colonization dynamics within the human nasopharynx.
Materials and Methods
Genomes and Assemblies
We used S. pneumoniae genomic information from five publicly available data sets. This included the following: 297 fully assembled genomes from GenBank and the Sanger Institute FTP site, which included 121 assembled genomes from Georgia, United States (Chancey et al. 2015); 3,085 assembled contigs from Myanmar refugees (Maela data set; Chewapreecha et al. 2014); sequence reads for 616 carriage strains from Massachusetts (Croucher, Finkelstein, et al. 2013); sequence reads for 82 Complex 3 strains (Croucher, Mitchell, et al. 2013); and sequence reads for 242 PMEN-1 (Pneumococcal Molecular Epidemiology Network) strains (Croucher et al. 2011). Additionally, we extracted DNA and sequenced 142 carriage strains from The Netherlands (Hermans genome set; Bogaert et al. 2001) and 26 PMEN strains (McGee et al. 2001) using Hiseq Illumina sequencing (Leiden Genome Technology Center). These reads have been submitted to the European Nucleotide Archive as study PRJEB10892 and PRJEB10893 (Hermans genome set and PMEN strains respectively; accession numbers in supplementary table S1, Supplementary Material online).
For genome sets with sequence read data (the Massachusetts, Complex 3, PMEN-1, Hermans, and PMEN strains), we assembled sequence reads into contigs de nova using only unique sequence reads with no ambiguous bases and minimum Phred quality scores of 25, 35, and 45. Experimentally, we found that too many reads can interfere with the final quality of assembled genomes (data not shown), so we assembled genomes using a range of sequence reads that started at 1 million sequence reads, ended at the maximum number of sequence reads for each genome, and increased by intervals of 200,000 sequence reads. We selected the assembly with the highest N50 for each genome out of the assembly results. We used Velvet 1.2.10 (Zerbino and Birney 2008) and VelvetOptimiser 2.2.5 (Gladman and Seemann 2008) with hash values from 45 to 61 by intervals of 4 to assemble the genomes. All sets of reads were treated as unpaired, even in the presence of paired sequence reads, due to ambiguity in the distance between paired sequence reads.
The assembled contigs from the Maela genome set, as available from the Sanger FTP site, contained extensive evidence of perfectly duplicated blp regions. To overcome this artifact, we broke the assembled contigs from each of the Maela assembled genomes into fragments of 150 bp that overlapped by 25 bp, and then we reassembled the genomes as described above. Genomes thus assembled showed no evidence for the duplications in the original assemblies.
Algorithm for Identifying Homologs
We developed an iterative DNA reciprocal BLAST (Altschul et al. 1990) algorithm for finding alleles in genes of interest across draft, nonannotated genomes (supplementary fig. S1, Supplementary Material online). This algorithm used two databases: A genome database of all contigs from the assembled genomes, and a filter database initially consisting of the DNA sequence open reading frames (ORFs) of annotated and unknown genes in 25 well-annotated S. pneumoniae genomes (supplementary table S2, Supplementary Material online). All unique annotated sequences in the filter database for a query gene were used to create a BLAST queue. Each of these gene variants was BLASTed against the genome database in turn. Reported sequences with an e-value of less than 10.0 were then BLASTed back into the filter database. Although this was a lenient criterion, only protein-coding sequences with a top hit of the same query gene were reported, as the sequences were then reciprocal best-BLAST hits, and this always resulted in alleles that easily aligned. To check against the risk of excess leniency, we ran the same DNA reciprocal BLAST searches using a vastly reduced e-value of 10−20 for all putative blp bacteriocins. This analysis recovered identical alleles as with the higher e-value threshold. A recovered sequence was scored as an allele only if it satisfied the following criteria: 1) The contig on which it was found contained an in-frame stop codon before the sequence’s start codon or the contig on which it was found lacked an upstream, in-frame stop code, but the sequence was a full-length allele for the gene; 2) the sequence was followed by a stop codon; and 3) translation of the sequence was at least 20 residues long, with an overall length between 75% and 125% of the average allele for the given locus. Novel allele sequences were added to the filter database and to the BLAST queue. The BLAST queue was iteratively cycled through until no new reported sequences were found. This iterative process allowed the gene sequences to move further away in sequence space from the initial 25 well-annotated genomes while preserving the criterion of reciprocal best-BLAST hits. The algorithm was implemented using custom Python scripts.
Curating the Genome Set
To eliminate poorly assembled or misattributed genomes, we used the DNA reciprocal BLAST algorithm to search for three housekeeping genes that should be in every S. pneumoniae genome: groEL, gyrA, and rpoD. We eliminated genomes that lacked two or more of these housekeeping genes (strains 6938_7#19 and 6972_5#3) and genomes with non-Streptococcus variants of these genes (56 genomes; supplementary table S1, Supplementary Material online), as determined by top BLAST hits in the GenBank database. We also eliminated strain 484-93 because of its poor assembly and strain 06_01_003MEF_uid198409, as it contained 7,124 called genes (compared with a normal range of 1,800–2,100 called genes).
In order to estimate the whole-genome phylogeny of the strains, each genome was aligned to the R6_uid57859 genome using the following method. We first divided each assembled genome into 50 bp fragments that overlapped by 10 bp, and then we reassembled each genome with R6_uid57859 as a reference genome using Stampy 1.0.23 (Lunter and Goodson 2011) with a substitution rate of 0.01. For an alignment, we excluded sites with gaps or “N”s in more than 0.5% of genomes, resulting in 1,444,122 remaining sites. We aligned these sites with 43 Streptococcus sp. viridans genomes (supplementary table S3, Supplementary Material online) as an outgroup. We conducted 30 maximum-likelihood phylogenetic tree searches with ExaML 3.0 (Kozlov et al. 2015) using this alignment with 15 random, unique starting trees, and 15 unique parsimonious trees (as determined by RAxML 8.2.4; Stamatakis 2006) with the GTRCAT model of evolution and scored each resulting tree with the GTR + Gamma model of evolution under ExaML 3.0. We present the tree with the highest likelihood score from these searches (ln(likelihood) of −32145034.7). We created 100 random nonparametric bootstraps using RAxML 8.2.4 (Stamatakis 2006), and we searched for the best tree with a single ExaML search for each bootstrap (Kozlov et al. 2015) using a single starting parsimonious tree for each bootstrap (Stamatakis 2006). We collapsed branches with less than 75% bootstrap support. Genomes that clustered with the outgroup instead of with the remaining S. pneumoniae genomes were excluded from further analysis (12 genomes; supplementary table S1, Supplementary Material online). Together, this resulted in a final set of 4,418 S. pneumoniae genomes (supplementary table S1, Supplementary Material online). Of these, we considered 4,096 strains to be randomly sampled from global populations (by excluding the Complex 3 and PMEN-1 strains); percentages of gene presence are only for these randomly sampled genomes.
Locating Alleles within the Genome Set
We used the DNA reciprocal BLAST algorithm described above to locate alleles of the blp operon genes. As a starting set of blp operon genes, we searched for all blp- or pnc-annotated genes in the 25 well-annotated genomes (supplementary table S2, Supplementary Material online). To search for novel blp operon genes not present in these annotated genomes, we used the SEED server (Aziz et al. 2008) to find protein-encoding genes in the 4,418 genomes that 1) were located between two previously found blp operon genes, 2) were found less than 2,000 bp from a blp operon gene, and 3) did not have significant BLAST hits in GenBank for transposon-related genes. We then searched for any such blp operon genes using the DNA reciprocal BLAST algorithm. Three hundred seventy-one genomes contained blpT and pncP (which are the acknowledged ends of the blp operon; Bogaardt et al. 2015) on a single contig; additional blp operon genes that were not in any of these 371 randomly sampled genomes are calculated to occur at a frequency of 0.8% or lower in the global S. pneumoniae population (α = 0.05). We were unable to find blpW in the 4,418 genomes (Bogaardt et al. 2015).
In addition to blp operon genes, we additionally searched our genome set for comA, comB, and comC to directly compare these alleles with their paralogs blpA, blpB, and blpC, respectively. In order to assign strains to sequence types (ST) via the S. pneumoniae MLST database (September 24, 2015 data set; Jolley and Maiden 2010), we also identified the seven genes used for MLST sequence typing (aroE, gdh/gdhA, gki/glkA, recP, spi/lepB, xpt, and ddl/ddlA). We used eBURST v3 (Feil et al. 2004; Spratt et al. 2004) and the S. pneumoniae MLST database to assign ST to clonal complexes, where a clonal complex contained ST that shared at least six identical MLST alleles with at least one other ST within the clonal complex. Although 173 genomes (3.9%) could not be assigned to an ST due to missing sequence data, 47 of these genomes were unambiguously assigned to clonal complexes.
As evidence of the success of our DNA reciprocal BLAST algorithm, we found the seven MLST genes in 99.3–99.5% of genomes and comABC in 98.6–99.8% of genomes. This gave us confidence that we could detect genes at a discovery rate of at least 98.6%, which included incomplete genomes in our data set.
Finally, we briefly examined evidence for deteriorated and potentially active transposase sequences within the blp operon, which could possibly serve as foci for homologous recombination. We searched for transposase-like sequences by locating all annotated transposase genes in the 25 well-annotated genomes and using the DNA reciprocal BLAST algorithm. Importantly, this search did not require an ORF in the resulting sequences, as we were also interested in nonfunctional transposases. As a result, we did not iteratively search for new transposases after an initial search that used the starting transposase sequences.
The results of all searches were stored in a custom SQL database for easy querying.
Systemizing Variation
To assign putative functions for the blp operon genes, we BLASTed the amino acid sequence of all recovered alleles on both the UniProtKB/Swiss-Prot database and the nonredundant GenBank protein sequence database. We classified eight genes with no significant hits and shorter than 40 residues as probable untranslated ORFs (supplementary table S4, Supplementary Material online).
We identified extensive amino acid variation in blp operon genes across these thousands of genomes. To focus on larger patterns, we classified alleles within each locus into highly similar groups called “phylotypes”; while not designed as functionally distinct alleles, this grouping of alleles is more conservative than using amino acid variants. Phylotypes were determined differently for genes with or without presumed leader peptides. Quorum sensing signals and presumed bacteriocin peptides contain short leader peptides that are typically cleaved following a conserved double-glycine sequence before the mature bacteriocin is exported by ABC transporters. All unique sequences past the double-glycine were classified as a different phylotype for the quorum sensing signal blpC and putative bacteriocins blpD, blpE, blpI, blpI2, blpJ, blpK, blpM, blpN, blpO, blpQ, pncT, and pncW. Notably, we identified four presumed, distinct leader sequences in blp operon genes that were classified as putative bacteriocin by protein BLAST searches (supplementary table S5, Supplementary Material online). These presumed leader sequences had a Glu/Met residue preceding a Leu six residues before the double-glycine cleavage site (ELSNISGG, MLSEVYGG, and MLAXVEGG); this motif was also found in the leader sequence for BlpC (ELNQITGG), whereas ComC had an Asp-Leu motif six residues before the cleavage site (DLQKIKGG). In place of a double-glycine cleavage site, 13.9% of genomes had an Asp-Gly site specifically in PncW (MLAVRTEDG); as these residues were found in high frequency, it suggested the possibility of flexibility in the leader sequence cleavage site, assuming these sequences indeed permit mature peptide secretion. For genes without a leader sequence, we aligned protein variants for each locus and created a neighbor-joining tree using Geneious 7.1.5 (Kearse et al. 2012). The protein variants on these trees were impartially divided into phylotypes based on subclades using 3 rules: 1) Excluding branches with branch lengths over 3.5 standard deviations from the mean branch length; 2) excluding branches with branch lengths over 0.025; and 3) dividing clades so that the maximum intraclade distance was 0.05. For analysis, we created an arbitrary cut-off of 0.5% of the 4,096 randomly sampled genomes for “common” phylotypes to avoid focusing on singleton strains and their recently derived clone mates.
Statistical Analyses
Type IIb blp bacteriocins are predicted to work as a pair of peptides that interact at equimolar concentrations to induce lethality to target cells. Each active “pair” is believed to associate with an immunity gene that prevents self-toxicity. It is presently unknown if peptide pairs that create active bacteriocins are specific with respect to their patterns of association, or if peptides can assort more broadly to generate an active toxin. Equally, it remains unknown if immunity is specific to certain bacteriocins or if there is cross-immunity, where individual immunity proteins provide more generalized protection. We used the following procedure to test for significant coassociation of genes while accounting for phylogeny. For each phylotype, we scored each genome as either 1) containing the phylotype, 2) containing the same gene but a different phylotype, or 3) not containing the gene. In the last case, the genome was treated as “missing data” either from having incomplete information from draft genomes or from a complete genome not containing the gene. Then, for all pairs of phylotypes that co-occur in at least 0.5% of genomes and that belong to different genes, we used BayesTraits 2.0 (Pagel et al. 2004) to estimate the coassociation between phylotypes. BayesTraits calculated the maximum likelihood of the phylotypes’ patterns along the phylogenetic tree assuming that the phylotypes mutated their presence/absence independently or assuming that the phylotypes influenced each other. We used the whole-genome phylogenetic tree with a conservative bootstrap threshold of 75% in order to examine only well-supported clades. We tested for significance using a log likelihood test and post hoc corrected for multiple tests within each pair of genes using a Holm-Bonferroni correction (Holm 1979). All other statistical analyses were performed using RStudio 0.98.507 (RStudio Team 2015) and R 3.1.3 (R Core Team 2013).
Nucleotide diversity and dN/dS was measured from aligned alleles for each gene using DnaSP 5.10.01 (Librado and Rozas 2009). Analyses were performed on only amino acid coding regions. Aligned sites and whole sequences were removed if they contained indels found in less than 5% of the alleles.
Results
Diversity in the blp Operon
We investigated the diversity of the blp operon of 4,418 S. pneumoniae genomes across 5 publically available genome sets taken from around the world, together with 142 carriage strains isolated from healthy Dutch children (Hermans set, Bogaert et al. 2001) and 26 PMEN strains (McGee et al. 2001), which were sequenced for this study (fig. 1). Using a DNA reciprocal BLAST search (supplementary fig. S1, Supplementary Material online), we found 88,498 homologs of genes in the blp operon divided across 35 loci (table 1).
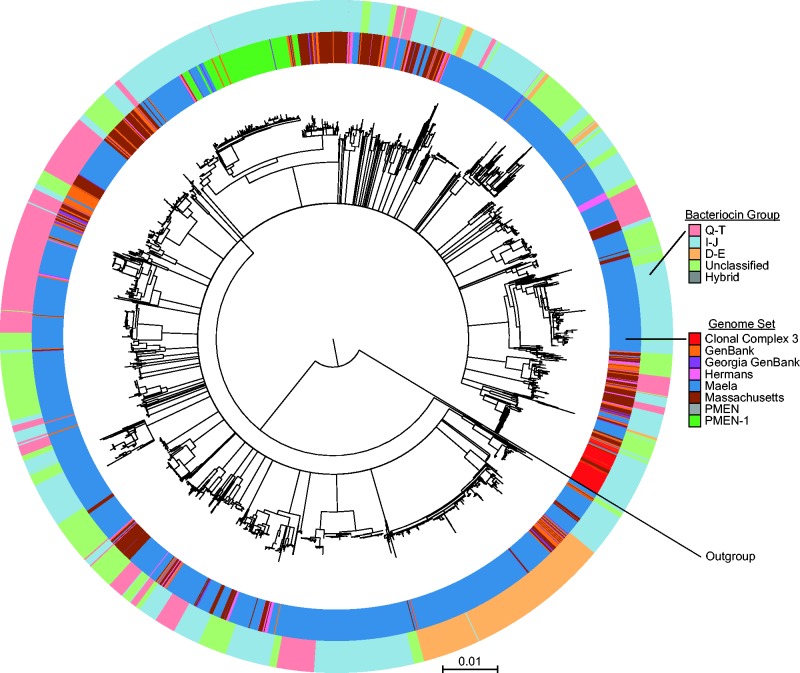
Fig. 1.—
Phylogenetic relationship between 4,418 Streptococcus pneumoniae genomes. We used 43 nonpneumoniae Streptococcus sp. as an outgroup. We collapsed clades with less than 75% nonparametric bootstrap support. The colored rings show the genome set (inner ring) and bacteriocin group (outer ring) of each genome.
Table 1.
Genes and Gene Frequencies of blp Operon and Related Genes
| Gene | Alternative Names | Functiona | Frequencyb,c | Amino Acid Variants | Phylotypes over 0.5% Frequencyb |
|---|---|---|---|---|---|
| blpA | spiCBA | ABC transporter | 0.249 | 272 | 23 |
| blpB | spiD | ABC transporter | 0.838 | 195 | 11 |
| blpC | spiP | QS signal | 0.991 | 29 | 9 |
| blpD | blpM, blpO | Bacteriocind | 0.115 | 4 | 1 |
| blpE | Bacteriocind | 0.116 | 2 | 1 | |
| blpF | Membrane proteind | 0.114 | 2 | 1 | |
| blpG | CAAX proteased | 0.111 | 3 | 1 | |
| blpH | spiH | QS receptor | 0.990 | 156 | 15 |
| blpI | pncA | Bacteriocind | 0.404 | 11 | 3 |
| blpI2 | Bacteriocind | 0.002 | 1 | 0 | |
| blpJ | pncD | Bacteriocin | 0.416 | 22 | 5 |
| blpK | pncE, thmA, blpU | Bacteriocin | 0.545 | 50 | 8 |
| blpL | pncH | Membrane proteind | 0.575 | 59 | 9 |
| blpM | pncI | Bacteriocin | 0.610 | 33 | 5 |
| blpN | pncJ, blpM, blpK | Bacteriocin | 0.757 | 26 | 6 |
| blpO | pncV, pncL | Bacteriocind | 0.605 | 24 | 4 |
| blpP | pncK | Membrane proteind | 0.864 | 18 | 2 |
| blpQ | pncR | Bacteriocind | 0.202 | 4 | 1 |
| blpR | spiR2 | Response regulator | 0.995 | 91 | 7 |
| blpS | spiR1 | Accessory protein | 0.938 | 70 | 9 |
| blpT | Unknown | 0.991 | 48 | 7 | |
| blpU1 | Unknown | 0.059 | 7 | 2 | |
| blpU2 | Unknown | 0.006 | 4 | 1 | |
| blpU3 | Unknown | 0.213 | 6 | 2 | |
| blpU4 | pncB | Unknown | 0.425 | 6 | 2 |
| blpU5 | Unknown | 0.422 | 7 | 1 | |
| blpV | Unknown | 0.116 | 2 | 1 | |
| blpX | pncN | Membrane proteind | 0.692 | 28 | 4 |
| blpY | pncO | CAAX proteased | 0.983 | 96 | 10 |
| blpZ | pncQ | Membrane proteind | 0.993 | 43 | 9 |
| comA | ABC transporter | 0.983 | 181 | 5 | |
| comB | ABC transporter | 0.992 | 121 | 2 | |
| pncG | Membrane proteind | 0.792 | 26 | 10 | |
| pncM | Membrane proteind | 0.633 | 18 | 4 | |
| pncP | SP0547 | CAAX proteased | 0.984 | 91 | 9 |
| pncT | Bacteriocind | 0.213 | 7 | 1 | |
| pncW | blpN, blpO | Bacteriocind | 0.302 | 20 | 4 |
aCAAX proteases and membrane proteins are both considered immunity genes.
bIn 4,096 randomly sampled genomes.
cFull length alleles only for blpA and blpB.
dPutative, based on sequence similarity.
Examining such a large number of genomes enabled us to identify six additional blp operon genes compared with previous studies (e.g., blpI2 and blpU1–blpU5; Lux et al. 2007; Bogaardt et al. 2015). We also exhaustively characterized within-locus allelic diversity for all identified blp operon genes (table 1). Overall, we found between 1 and 272 amino acid variants in these genes (average = 42.3 amino acid variants; table 1). With one exception, all blp operon genes were confined to the blp locus. blpK was present in a single copy in 47.6% of genomes and duplicated in 4.9% of genomes; this gene was located either within the blp locus (6.9% of strains), adjacent to the competence peptide transporter comAB elsewhere in the genome (11.2% of strains), or in an undetermined location (41.1% of strains). No other blp operon gene (table 1) was found more than 16,341 bp (the length of the largest fully assembled blp operon, in strain Taiwan19F_14_uid59119) outside of the canonical operon delimited by blpT or pncP (in a single exception to this finding, blpMNPO is directly adjacent to comAB in strain 6259_8#1).
Confusingly, three separate nomenclatures (i.e., blp, pnc, and spi) are currently used for genes within this operon (Bogaardt et al. 2015). Given the comprehensive nature of our DNA reciprocal BLAST search and to avoid confusion here and elsewhere, we adopted the most recent and inclusive nomenclature from Bogaardt et al. (2015). To their list, we add an additional putative bacteriocin gene blpI2, which occurs at low frequency (less than 0.5%) with no similar protein BLAST hit, and five hypothetical genes blpU1–blpU5 (tables 1 and 2). One blpM variant (distinct from blpMN; Bogaardt et al. 2015) in 1.1% of genomes encoded a hybrid bacteriocin containing BlpM and BlpN on a single reading frame, wherein BlpM was followed by a second leader sequence and BlpN without a stop codon between the two (table 2). It is possible that this structure would allow for production of two bacteriocin peptides from a single reading frame.
Table 2.
Mature Putative Bacteriocin Amino Acid Sequences
| Gene | Frequency in Randomly Sampled Genomesa | Amino Acid Sequence |
|---|---|---|
| blpD | 0.115 | TDWGTVGKGAVYGAGIGVAMCAVGGLLTGGSTWAMTAGCAWAGAKLGGSFTAIADNLWP |
| blpE | 0.116 | GLGGDVVVGALSGAFQAGQSCIAGGPQAYLICATGGAIVGGILAYGLRPPK |
| blpI | 0.371 | RGNLGSAIGGCIGAVLLAAATGPITGGAATLICVGSGIMSSL |
| 0.025 | ...............................T.......... | |
| 0.006 | .......................................P.. | |
| blpJ | 0.349 | YSSTDCQNALITGVTTGIITGGTGAGLATLGVAGLAGAFVGAHIGAIGGGLTCLGGMVGDKLGLSW |
| 0.027 | ...............................................R.................. | |
| 0.017 | ..F............................................................... | |
| 0.006 | ................................................S................. | |
| 0.005 | ...................................V............D................. | |
| blpK | 0.139 | GCNWGDFAKAGVGGGAARGLQLGIKTRTWQGAATGAVGGAILGGVAYAATCWW |
| 0.119 | ....................................A................ | |
| 0.099 | ................V...................A................ | |
| 0.067 | ................- - - - - - - - - - - - - - - - - - -............. | |
| 0.050 | ...............V....................A................ | |
| 0.044 | ................V...................A................ | |
| 0.034 | ..........................G.........A................ | |
| 0.029 | ..............A...........G.........A................ | |
| blpI2 | 0.002 | DKVGAGEVVQALGICTIGGAALGSVIPVVGTLAGGILGAQFCTAAWGAFRAS |
| blpM | 0.300 | KNNWQTNVLEGGGAAFGGWGLGTAICAASGVGAPFMGACGYIGAKFGVDLWAGVTGATGGF |
| 0.101 | ................................................A............ | |
| 0.096 | ................................................A.........S.. | |
| 0.092 | ........F...S...................................A............ | |
| 0.011 | ................................................A............QQKETC MNTYCNINETMLSEVYGG | |
| blpN | 0.374 | NS- - - - - - - - GGAAVVAALGCAAGGVKYGRLLGPWGAAIGGIGGAVVCGYLAYTATS |
| 0.149 | ..- - - - - - - - ...................KI.......................... | |
| 0.101 | KNNWQTNVLEGG .- - - -..F...................................... | |
| 0.057 | ..- - - - - - - - .......................L....................... | |
| 0.043 | GCNWGDFAKAGV ...................KI.......................... | |
| 0.011 | ..- - - - - - - - ...................KI........–-............... | |
| blpO | 0.547 | DIDWGRKISCAAGVAYGAIDGCATTV |
| 0.028 | ..........T............... | |
| 0.017 | ......................V... | |
| 0.007 | ......E................... | |
| blpQ | 0.200 | IFGVDDALFWAGLGYVAGSIVDTAIDDFTNQCRKNPHQWFCVRV |
| pncT | 0.207 | DDCFIGDIGCIGWGLLKSIGGMIKPAPYVPPVCIPKSSWNPAPPVPC |
| pncW | 0.132 | DVSDIYRGYANQRSPFASYPSILKNSGPFPVSGYCLRGYHDRGYIGAGFHLCGI |
| 0.126 | ............V...G...P..............P.................. | |
| 0.028 | ...G......Y.D...GP.................P...R.L............ | |
| 0.005 | ...G......Y.D...GP........D........P...R.L............ |
aFor genes present in over 0.5% of genomes, only variants found in at least 0.5% of genomes are shown.
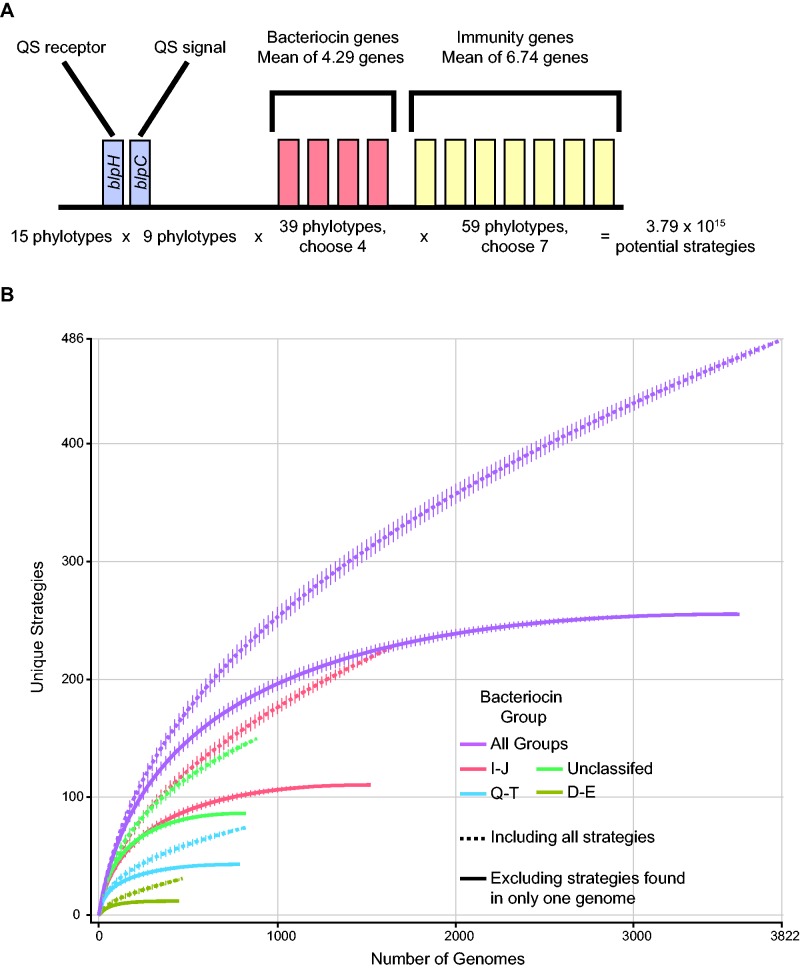
To simplify the analysis of allelic diversity (table 1), we grouped amino acid variants within individual genes into phylotypes (see Materials and Methods), focusing on the most common phylotypes occurring in at least 0.5% of the 4,096 genomes from randomly sampled strain collections (by excluding the Complex 3 and PMEN-1 genome sets; table 1). Even using this conservative measure to describe allelic diversity within genes, thousands of genomes were required to effectively sample global blp operon diversity, thus emphasizing the necessity of a large data set. We performed 10,000 bootstrap rarefactions on the 4,096 randomly sampled genomes, which revealed that variation in nonsingleton phylotypes was saturated at approximately 3,000 and 2,000 genomes for putative immunity and bacteriocin genes, respectively (supplementary fig. S2, Supplementary Material online). This gave us confidence that the vast majority of the total variation in global S. pneumoniae populations was included in our analyses.
To investigate the tempo of molecular evolution at this locus, we placed these genes into putative functional categories: Bacteriocin genes, immunity genes, blp regulatory genes, and blp genes with unknown function, as well as examined nine housekeeping genes outside of the blp operon (table 3). Although the mean rate of nucleotide substitution (π) was not significantly different between these categories (P > 0.054; Tukey’s HSD [Honestly Significant Difference] test), bacteriocins had a higher dN/dS ratio compared with housekeeping and immunity genes (P = 0.0066 and P = 0.040, respectively; Tukey’s HSD test).
Table 3.
Molecular Diversity of blp Operon and Selected Housekeeping Genes
| Genea | Category | Number of Sites | Number of Sequences | Nucleotide Diversity (π) | dN/dS |
|---|---|---|---|---|---|
| blpU3 | Unknown | 207 | 5 | 0.012 | 2.656 |
| blpI | Bacteriocin | 147 | 9 | 0.023 | 2.645 |
| blpJ | Bacteriocin | 226 | 18 | 0.012 | 1.814 |
| blpQ | Bacteriocin | 201 | 5 | 0.010 | 0.988 |
| blpO | Bacteriocin | 142 | 20 | 0.027 | 0.974 |
| blpC | Regulatory | 127 | 28 | 0.129 | 0.895 |
| pncW | Bacteriocin | 231 | 19 | 0.056 | 0.844 |
| blpU5 | Unknown | 147 | 8 | 0.012 | 0.712 |
| pncT | Bacteriocin | 192 | 7 | 0.009 | 0.569 |
| blpU1 | Unknown | 414 | 8 | 0.014 | 0.492 |
| pncG | Immunity | 84 | 13 | 0.066 | 0.476 |
| blpN | Bacteriocin | 126 | 19 | 0.030 | 0.462 |
| blpK | Bacteriocin | 213 | 38 | 0.040 | 0.395 |
| blpM | Bacteriocin | 219 | 24 | 0.020 | 0.360 |
| blpS | Regulatory | 327 | 74 | 0.027 | 0.352 |
| pncM | Immunity | 177 | 22 | 0.037 | 0.290 |
| blpX | Immunity | 357 | 23 | 0.023 | 0.242 |
| blpH | Regulatory | 1,308 | 157 | 0.086 | 0.206 |
| blpZ | Immunity | 204 | 35 | 0.055 | 0.199 |
| pncP | Immunity | 495 | 128 | 0.018 | 0.190 |
| blpT | Regulatory | 297 | 53 | 0.024 | 0.181 |
| blpL | Immunity | 222 | 68 | 0.068 | 0.153 |
| blpP | Immunity | 147 | 13 | 0.017 | 0.152 |
| aroE | Housekeeping gene | 852 | 121 | 0.017 | 0.151 |
| blpY | Immunity | 687 | 127 | 0.070 | 0.137 |
| blpb | Regulatory | 1,359 | 177 | 0.027 | 0.137 |
| gyrA | Housekeeping gene | 2,559 | 190 | 0.004 | 0.117 |
| blpU4 | Unknown | 114 | 5 | 0.014 | 0.092 |
| xpt | Housekeeping gene | 609 | 108 | 0.017 | 0.090 |
| blpAb | Regulatory | 2,151 | 74 | 0.015 | 0.083 |
| gdhA | Housekeeping gene | 1,344 | 161 | 0.018 | 0.071 |
| blpR | Regulatory | 666 | 114 | 0.049 | 0.064 |
| glkA | Housekeeping gene | 975 | 141 | 0.020 | 0.061 |
| ddlA | Housekeeping gene | 1,041 | 189 | 0.042 | 0.053 |
| rpoD | Housekeeping gene | 1,161 | 113 | 0.009 | 0.031 |
| lepB | Housekeeping gene | 654 | 101 | 0.020 | 0.028 |
| groEL | Housekeeping gene | 1,620 | 166 | 0.014 | 0.028 |
ablpD, blpE, blpF, blpG, blpI2, blpU2, and blpV were removed for having less than five unique sequences after trimming.
bFull-length blpA and blpB alleles only.
Diversity in Potential Ecological Strategies
There are four unambiguous functional classes of genes within the blp operon that could mediate interactions among strains. Bacteriocin and immunity genes define the range of killing and susceptibility, respectively. Additionally, the blp locus is regulated by the secretion of the BlpC peptide signal that binds to the quorum sensing receptor, BlpH. Because these signals are secreted, they can be potentially detected and bound by both the secreting cells as well as their competing neighbors. The diverse combinations of these four functional classes, together with the allelic diversity within them, define the potential ecological strategy set by which pneumococcal strains can interact and compete via blp bacteriocins. Each strain, by extension, expresses only a single strategy from among this total, and the sum of all such strategies in our data set represents the set of realized strategies.
To estimate the possible number of unique ecological strategies mediated by blp operon genes in S. pneumoniae, we calculated (using frequency data from our genome set) that an average genome contained a single peptide signal, a single peptide receptor, four bacteriocins, and seven immunity genes (fig. 2A). To conduct this analysis, we first examined an admittedly straw-man null hypothesis that assumed phylotypes are functionally distinct and that all genes and phylotypes can assort randomly, without influence of genetic linkage, phylogenetics, or functional constraints. By accounting for the phylotypic variation within these four gene classes (table 1), we calculated that approximately 3.79 × 1015 unique ecological strategies, or combinations of phylotypes, were possible (fig. 2A). Based on this estimate, we predicted that it would be necessary to sample an astronomical number of pneumococcal strains to fully characterize the realized diversity at the blp locus, and to some approximation this was true. As is evident in figure 2B, if we included the 47.5% of unique ecological strategies that appeared in single genomes, we estimated that on average, a new ecological strategy should be found for every 15.4 new strains sampled (assuming that the linear relationship at the end of the rarefaction curve—425 strategies until 486 strategies, r2 = 0.9996—is realized in perpetuity). In contrast, when we excluded singletons, the rarefaction curve saturated at 255 strategies (fig. 2B), providing strong evidence that our data set reflected the prevalent ecological strategies carried within the global pneumococcal population. It is notable that the curve that excluded singleton strategies reached its asymptote after 2,000 genomes, confirming again the necessity of such a large data set. Together, these results suggest there are a relatively small number of common ecological strategies from among all those that are possible, and that global S. pneumoniae populations contain a much higher diversity of strategies that occur at extremely low frequencies of less than 5.2 × 10−4 (as calculated by occurring in single strains out of 3,822 strains with complete strategies; fig. 2B). These data also clarify that the assumption of random assortment between blp operon genes and phylotypes is incorrect. Below, we explore different constraints on diversity that contribute to the more limited, although still vast, set of ecological strategies mediated by the blp operon, namely constraints imposed by overall operon organization and structure, physical or functional linkage, and phylogeny.
Fig. 2.—
Estimated and sampled ecological strategies in the blp operon. (A) We reduced the operon to an “average” operon consisting of a histidine kinase quorum sensing receptor (blpH), a quorum sensing signals (blpC), exactly four bacteriocin genes, and exactly seven immunity genes in order to estimate the number of potential ecological strategies. This average genome was based on frequency data from our genome set, as we detected: 99.1% of genomes containing a single blpC gene, with multiple co-occurring blpC genes not detected; 99.0% of genomes containing a single blpH gene, with multiple co-occurring blpH genes not detected; an average of 4.29 putative bacteriocins detected per genome; and an average of 6.74 putative immunity genes detected per genome. We included only phylotypes that were present in at least 0.5% of randomly sampled genomes. (B) Rarefaction of ecological strategies found in randomly sampled genomes. Only strains that contained blpC and blpH, and phylotypes that were present in at least 0.5% of randomly sampled genomes, were included. Error bars indicate standard deviation of 10,000 randomizations.
Constraints on Diversity
Operon Structure
In examining different explanations for the result that there were fewer ecological strategies than expected by gene combinatorics, an initial covariance analysis identified three sets of genes within the blp operon in which 1) all genes within each set were found together in genomes at a high frequency (>90%) and 2) genes from different sets were rarely (0.12%) found in the same genome. Based on these criteria, we were able to classify the 4,096 randomly sampled genomes into 3 discrete groups or as “unclassified” (fig. 3): Bacteriocin group Q–T, which included all genomes containing either blpQ, pncT, or blpU3 (875 genomes, 93.7% containing all 3 genes); group I–J, which included all genomes containing either blpI, blpJ, blpU4, or blpU5 (1,742 genomes, 92.0% containing all 4 genes); group D–E, which included all genomes containing either blpD, blpE, blpF, blpG, or blpV (475 genomes, 94.3% containing all 5 genes); and an unclassified group of genomes that contained none of these bacteriocin group-specific genes (999 genomes). Five genomes (0.12%, termed hybrid genomes) out of 4,096 contained genes for more than one of these bacteriocin groups. The groups had significantly different average number of blp bacteriocin and immunity genes (P ≤ 6.6 × 10−11 and P ≤ 0.0039 for all putative bacteriocin and immunity genes, respectively; P ≤ 1.3 × 10−5 and P ≤ 0.0038 excluding group-specific bacteriocin and immunity genes, respectively; Games–Howell test; supplementary fig. S3, Supplementary Material online), with the exception of an equivalent number of immunity genes in group I–J and in group D–E with group-specific genes (P = 0.84). As observed when we analyzed bacteriocin groups together, rarefaction analysis for each bacteriocin group separately again revealed that the number of unique ecological strategies within groups did not saturate with sampling until we removed ecological strategies found in single genomes (fig. 2B). In comparing the bacteriocin groups with the full-genome phylogeny, only the D–E group comprised of a single clade (fig. 1), while the other groups were broadly distributed across the phylogeny.
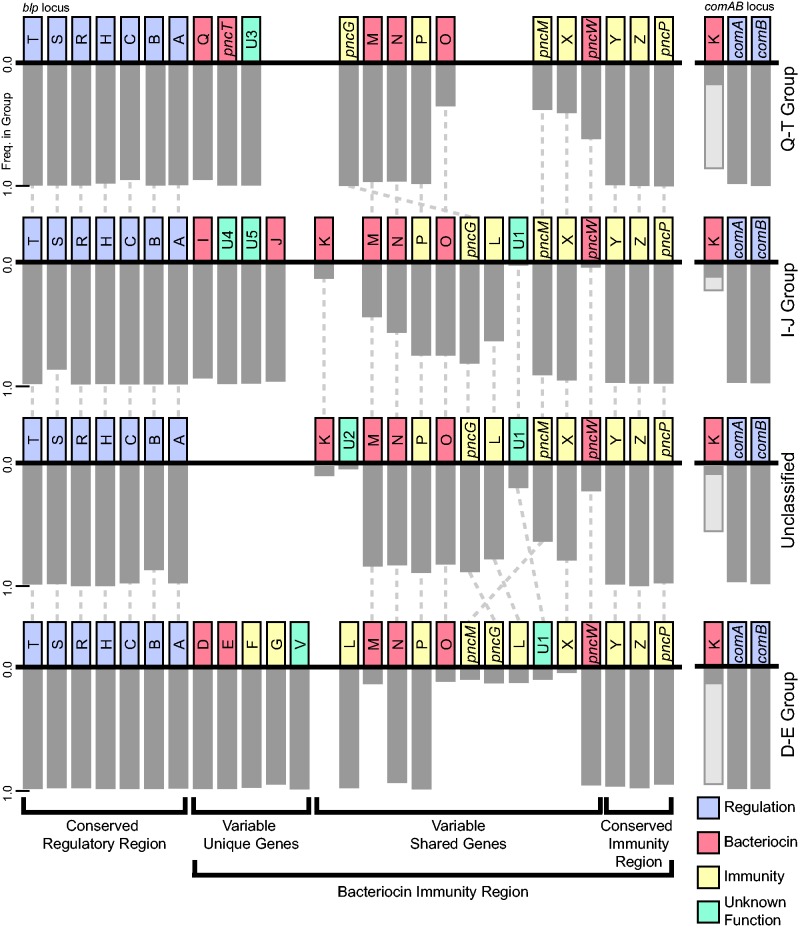
Fig. 3.—
Consensus blp operon gene structure across bacteriocin groups. This gene order was compatible with 99.5% of the Q–T group; 94.9% of the I–J group; 93.3% of unclassified genomes; and 99.4% of the D–E group. The frequency of the gene in the group is shown in gray bars; gene function is shown by the color of the gene. Lighter gray bars underneath blpK indicate the proportion of genomes with blpK in an unknown location. Genes occurring in less than 0.5% of genomes in the bacteriocin group are not shown.
Gene order was broadly conserved across all genomes, as previous noted (De Saizieu et al. 2000; Reichmann and Hakenbeck 2000; Bogaardt et al. 2015), as well as within bacteriocin groups. Figure 3 shows the consensus gene order for the bacteriocin groups, with each gene order compatible with over 93% of genomes in each group. The blp operon can be divided into four regions: The conserved regulatory region; the variable, bacteriocin group-specific genes; variable genes shared across bacteriocin groups; and the conserved immunity region. The last three of these regions is collectively known as the bacteriocin immunity region. In total, 91.4% of genomes carried all seven genes present in the conserved regulatory region, consisting of blpT, blpS, the response regulator blpR, the histidine kinase receptor blpH, the quorum sensing signal blpC, and ABC transporters blpB and blpA. Interestingly, only 23.5% of genomes had full-length, potentially functional, versions of blpB and blpA. However, all examined genomes placed blpBA sequences after blpC, and here we report the proportion of genomes with copies of blpB and blpA of any ORF length. The bacteriocin group-specific genes were always located together as a cluster after the conserved regulatory region (fig. 3). Following this region were the variable shared genes; while members of all bacteriocin groups contained these genes, the frequency varied widely across groups (e.g., the proportion of genomes with blpO ranged from 10.1% in group D–E to 75.9% in group I–J; fig. 3). Additionally, gene order within this region varied across bacteriocin groups, with pncG shifted in group Q–T and pncM shifted in group D–E. blpL was the only other gene besides blpK in which a significant number of genomes contained two copies (1.4% of genomes), each copy with a different phylotype; all except 3 of these 56 genomes were in group D–E. The conserved immunity region, which consists of blpY, blpZ, and pncP, flanked these variable shared genes; these genes were conserved in order and frequency across bacteriocin groups, with 94.9% of genomes containing all three genes.
The location of blpK was especially variable between bacteriocin groups. blpK was found in two places in group I–J and in unclassified genomes—at the beginning of the blp variable shared genes and in the comAB locus. The fragmented genome assemblies in these groups prevented determining the exact frequency of blpK at either of these loci. However, we found no genome with blpK in the blp operon for the other 2 bacteriocin groups—group Q–T and group D–E—out of 60 and 35 full genomes, respectively, with complete gene order data. In 95.9% of strains with a copy of blpK located in the comAB locus (440 out of 459 strains), we found a shared (93.9–100% identity) 65 bp sequence within 100 bp of blpK. This same 65 bp sequence was located in 89.0% of genomes (81.5–98.5% identity with the sequence in the comAB locus) between blpA and the first variable unique gene. The 65 bp sequences, which were identified in our DNA reciprocal BLAST search for transposase-like sequences, translated into the last 15 residues of an IS1381 transposase. Accordingly, this sequence could have or could still provide a homologous sequence for recombination or rearrangement between bacteriocin groups or between the blp and the comAB loci.
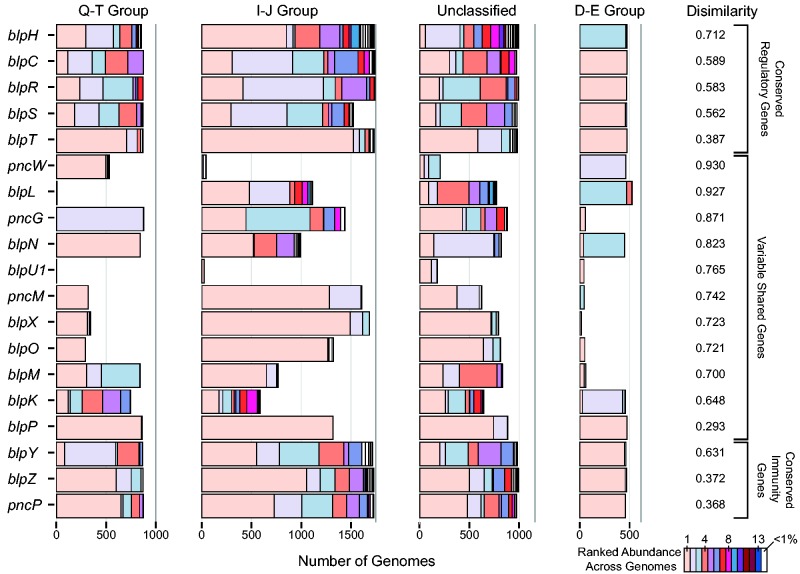
We next examined if phylotypes were biased to particular bacteriocin groups. In direct contrast to the bacteriocin group-specific genes, 81.2% of common phylotypes across all shared genes were not restricted to a single bacteriocin group. However, the quantitative distribution of phylotypes between bacteriocin groups differed significantly for the overwhelming majority of genes (88.8% of gene/bacteriocin group combinations; P < 0.0018; pairwise χ2 test with Holm–Bonferroni correction, excluding four tests with less than five observed cases; fig. 4). This was especially noticeable for group D–E, which had markedly less diversity across all genes (average Shannon diversity of phylotypes within genes for group D–E = 0.130; 0.733–1.28 for other bacteriocin groups). Variation within genes was concentrated in genes blpN, pncG, blpL, and pncW, which all showed a larger amount of phylotypic diversity across groups (Bray–Curtis dissimilarity > 0.82; average across genes = 0.65; standard deviation = 0.19).
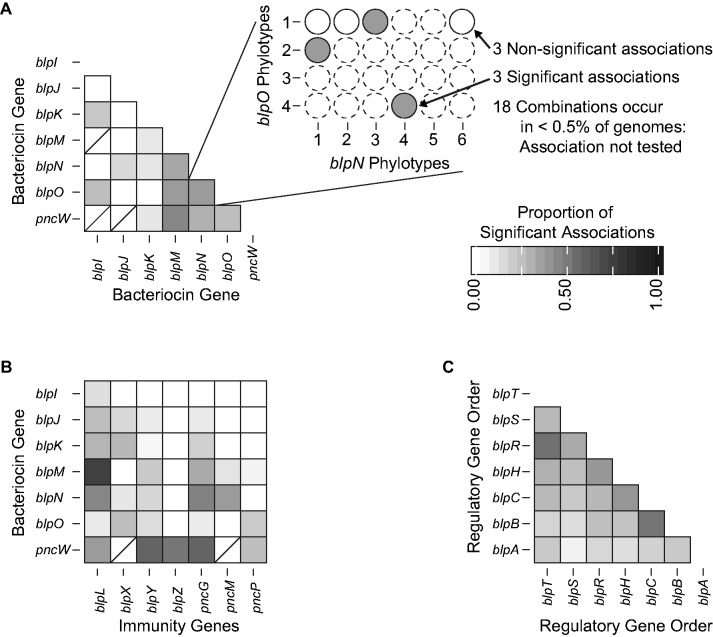
Fig. 4.—
Distribution of phylotypes across bacteriocin groups. For each blp operon gene, the number of genomes with each phylotype is shown, with the phylotypes colored by their overall ranked abundance within each gene across all bacteriocin groups. Phylotypes found in less than 1% of randomly sampled genomes are shown in white. Genes are ordered by gene class and Bray–Curtis dissimilarity between bacteriocin groups.
Allelic Co-occurrence
A potential constraint on realized blp diversity is the requirement for functionally interdependent proteins to co-occur within genomes, thereby giving rise to patterns of genic/allelic association that occur more frequently than would be predicted by chance and after correcting for phylogeny. Within the blp operon, we predict correlated changes between three blp gene categories. First, bacteriocin genes are predicted to have correlated changes with their partner bacteriocin, with which it creates a functional type IIb (two-peptide) bacteriocin outside the cell. Second, bacteriocin genes should have correlated changes with proteins that provide self-immunity to the functional bacteriocin. Third, the BlpC signal molecule is expected to change in concert with its receptor, BlpH. Because few of these associations have been verified empirically, our aim was to identify pairs of proteins whose functional association can be tested in subsequent work, and also to ask whether genes that are predicted to interact do so with high levels of allelic fidelity.
In figure 5, we show patterns of association between three categories of blp operon genes, which indicate that locus and phylotypic levels of coassociation were highly nonrandom. Importantly, many combinations of phylotypes or genes were extremely rare or absent; for example, 904 out of 1,034 pairs of putative bacteriocin phylotypes in figure 5 co-occurred in less than 0.5% of genomes. However, among the 129 tested pairwise combinations of bacteriocin phylotypes, 26 pairs showed significant associations (fig. 5A). The presence of associations is consistent with the hypothesis that these are type IIb bacteriocins, which require two partner peptides to form functional bacteriocins; however, it is also important to note that these patterns may be partially driven by physical linkage between genes. Finally, this analysis highlights that the fidelity between partner peptides was not absolute. As an example, blpN had significant associations with specific phylotypes of blpJ, blpK, blpM, blpO, and pncW; while experimental evidence has shown that one phylotype of blpN formed a functional bacteriocin with one phylotype of blpM (Dawid et al. 2007), these additional associations raise the possibility of multiple partner peptides with various blpN phylotypes.
Fig. 5.—
Significant associations between blp operon gene phylotypes. Within each pair of genes, the proportion of pairwise phylotypes that had significant associations is shown: (A) Between putative bacteriocin genes; (B) between putative bacteriocins and immunity genes; and (C) between regulatory blp operon genes, shown in consensus gene order. Comparisons between phylotypes co-occurring in less than 0.5% of genomes were not tested. Pairs of genes with fewer than three tested phylotype combinations are shown with a slash. Genes with fewer than three phylotypes each occurring in at least 0.5% of genomes are not shown.
In 88 out of 491 tested pairwise combinations of putative bacteriocin and immunity phylotypes, we found significant positive associations that are candidates for immunity against specific bacteriocins (fig. 5B). This includes established and putative immunity genes that have associations with many bacteriocin genes (blpL, blpY, blpZ, and pncP, all associated with 1–7 putative bacteriocin genes), as well as newly discovered pncG (Bogaardt et al. 2015), which is associated with six putative bacteriocins. As with pairs of blp bacteriocin phylotypes, there was not strict fidelity of one-bacteriocin:one-immunity gene, suggesting the possibility that genes for immunity may provide cross-protection against several bacteriocins.
In testing for blpC–blpH association, we found strong patterns of nonrandom co-occurrence (fig. 5C), as expected. Also, consistent with the effects of physical linkage, we observed that associations between genes within the entire regulatory region scale with gene proximity. Specifically, regulatory blp operon genes physically next to each other had a higher proportion of significant associations than other regulatory blp operon genes (average proportion significant association for neighboring genes = 0.465; nonneighboring genes = 0.294; P = 0.024; Mann–Whitney test).
Phylogenetic Constraints
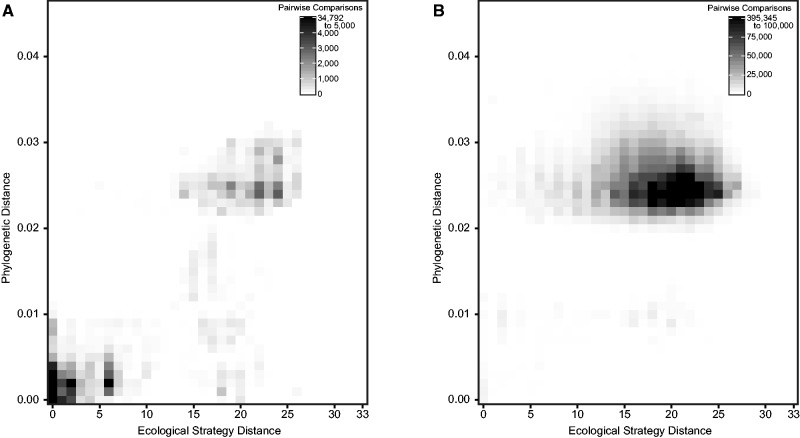
As the D–E group comprised a single clade (fig. 1), we next examined if phylogeny constrained the number of realized ecological strategies. We first classified genomes into clonal complexes based on MLST, and calculated the effective distance between ecological strategies by quantifying the number of differences between pairs of strains in phylotypes for blpC, blpH, bacteriocin genes, and immunity genes. The results in figure 6 show that there is a strong influence of phylogenetic distance on ecological distance, and that strains within clonal complexes were more similar to one another with respect to ecological strategies than they were to strains in different clonal complexes, whereas strains in the same clonal complex had an average distance of 6.74 between potential ecological strategies (fig. 6A), and strains in different clonal complexes had an average ecological strategy distance of 18.76 (fig. 6B; P < 10−99, Mann–Whitney test). Interestingly, a sizable minority of strain pairwise comparisons (28.5%) within the same clonal complex differed as much or more in ecological strategies than the upper 95% of all comparisons across clonal complexes (ecological strategy distance ≥ 11), as seen in figure 6A. Of these, 87.1% pairwise strain comparisons were quite phylogenetically diverged (to the same degree as across clonal complex comparisons with a phylogenetic distance greater than 0.01), perhaps reflecting the fact that clonal complexes are themselves imperfect representations of diversity across the genome (Turner et al. 2007). However, the remaining 12.9% were highly related, and in 77.5% of these comparisons, the ecological divergence resulted from switches in bacteriocin groups, which are likely due to recombination of the entire operon. Further studies are needed to analyze if these recombinants were positively selected, or if this variation was a result of neutral drift.
Fig. 6.—
Distribution of pairwise strain comparisons across phylogenetic and ecological strategy distance. For strains that were placed into clonal complexes, we calculated the pairwise distance along the phylogenetic tree and the pairwise distance between ecological strategies (e.g., the number of different gene-phylotypes for blpH, blpC, putative bacteriocins, and immunity genes) of (A) strains belonging to the same clonal complex and (B) strains belonging to different clonal complexes. The axes divisions are identical for each graph, although the scales differ. Figure 6A has 14 divisions with over 5,000 pairwise comparisons, while figure 6B has 24 divisions with over 100,000 pairwise comparisons.
Discussion
Comprehensive Sampling Revealed Extensive Genic and Allelic Diversity
Bacteriocins may be crucial regulators of bacterial community dynamics, yet there is little understanding of the blp operon that mediates these interactions in S. pneumoniae. Here we used a comprehensive bioinformatics approach to understand diversity in this operon. At every level of classification, we found an unexpectedly large amount of variation: In the number of putative bacteriocin and immunity genes (supplementary fig. S3, Supplementary Material online), the presence or absence of specific blp operon genes (fig. 3), gene order (fig. 3), and the tremendous breadth of phylotypic variants (fig. 4)—all of which resulted in an exceptionally large diversity of blp operon arrangements. Yet despite the fact that thousands of S. pneumoniae genomes were required to reveal both the genic composition as well as the full allelic diversity of the genes in this operon, rarefaction analysis indicated that although we identified most of the diversity, more remains to be discovered (fig. 2B and supplementary fig. S2, Supplementary Material online). Nearly 50% of the combinations of all blp ecological strategies were unique (fig. 2B), suggesting that new arrangements emerge frequently; moreover, it was clear that the only way to fully understand the diversity of this locus was by taking advantage of the vast sequencing data sets of this species. In so doing, we identified the novel putative bacteriocin gene blpI2 and five potential genes blpU1–blpU5. Additionally, we found nine common (0.5% or greater) variants of the BlpC signal molecule, compared with the previously reported four to six common BlpC mature peptides (Bogaardt et al. 2015; Pinchas et al. 2015). These analyses extend results from another recent examination of the blp operon (Bogaardt et al. 2015), which also reported on the astounding diversity in this operon as well as organized the previously chaotic blp operon gene nomenclature (Bogaardt et al. 2015).
Possible Constraints on the Number of Realized Ecological Strategies
We began with the naive null hypothesis that assumed phylotypes are functionally distinct (which they are not designed to indicate) and that all phylotypes can freely associate with one another. As described in figure 2, this straw-man hypothesis then produced an estimate of approximately 1015 unique ecological strategies. Because each arrangement could potentially specify a distinct mode of signaling, killing, and susceptibility, each represents a potential strategy by which strains interact and compete with one another. Yet instead of 1015 possible strategies, we found significantly fewer, with rarefaction analysis (excluding singletons) saturating at around 250 combinations (fig. 2). Our analyses provide several explanations for why the data do not conform to these extreme, simplifying assumptions. First, we showed that genomes could be classified into distinct groups that contain unique sets of genes (fig. 3). These “variable unique genes” had a conserved orientation in the blp operon; however, at present little is known of either the specific functional relevance of these genes or the factors that lead to their patterns of association. Interestingly, each group, with the exception of the D–E group, was broadly distributed across the strain phylogeny (fig. 1). This is consistent with the idea that horizontal gene transfer had mobilized genes within this region to unrelated strains. However, although this could explain the phylogenetic dispersion of bacteriocin groups, it would not explain the associations between the genes themselves. Nor does it explain other group-specific differences in putative bacteriocin and immunity gene number and the differential presence/absence of specific genes in the blp operon (fig. 3). Identifying the functional reasons, if any, for these associations remains an important area for further investigation.
A second type of constraint that would limit the number of realized blp strategies is physical or functional linkage interactions among genes and phylotypes within the blp locus: BlpC binds to its cognate receptor, BlpH; type IIb bacteriocins are believed to act co-ordinately in pairs; and cells expressing specific bacteriocins must be protected from suicide by immunity. As expected, given these presumed associations, we found strong correlations between specific phylotypes of blp operon genes (fig. 5). Equally informative was the fact that many phylotypes were never found together, and interestingly this differed across various regions of the operon. For example, there were extensive associations among genes regulating blp activation, in addition to nearly complete conservation of gene order in this region; however, there was markedly less coassociation between putative bacteriocins or between bacteriocins and immunity. Each of these associations, particularly for cases not influenced by physical linkage, is suggestive of functional relationships that can be examined experimentally.
Although these patterns of coassociation can be interpreted as a mechanism constraining overall diversity in the blp operon, it is also important to realize that the coassociations can also reveal a potential cause of functional diversification. This is most easily seen for the bacteriocins that are believed to function as two-peptide pairs. One possibility is that these pairs are highly precise, showing complete fidelity at both the genic and phylotypic levels. Alternatively, if different phylotypic variants retain activity with distinct partners, as recently described in a two-component lantibiotic system (Zhao and van der Donk 2016), then this could potentially expand the target range of each “class” of type IIb bacteriocins. For example, BlpM and BlpN have been shown experimentally to interact to generate lethal activity to the target cell (Dawid et al. 2007); yet in the D–E group, the frequency of strains containing blpM was only 12.2% compared with 94.3% containing blpN (fig. 3), suggesting either a very slow decay rate for ineffective bacteriocin peptides or the possibility that these genes combine with other peptides to become active. Additionally, within each of these genes there was marked phylotypic diversity. blpM had 5 common phylotypes and blpN had 6, leading to 30 potential combinations; of these, only 9 were realized, consistent with the ideas of functional constraints or genetic linkage. At the same time, many phylotypes occurred with multiple partners, suggesting that either strict fidelity is not required to retain function or alternatively that tight associations have not been selected for; such infidelity between bacteriocin peptide partners has not been described previously. Regulatory genes showed a similar lack of strict phylotypic association (fig. 5C); however, it is important to note that there was evidence that these associations could be partially caused by physical gene linkage, a possibility that also holds true for bacteriocin–bacteriocin associations (fig. 5A).
A potential epistatic interaction could exist between the blpAB ABC transporter and both bacteriocins and the quorum sensing signal BlpC, as nonfunctional BlpAB would not export these peptides. Although only 23.5% of strains had intact blpAB, the competence-related ABC transporter ComAB has been shown to export BlpC (Kjos et al. 2016). The export of bacteriocins by ComAB has not been examined, but strains with interrupted blpAB had on average only 0.14 fewer putative blp bacteriocins than strains with intact blpAB (Kjos et al. 2016). This suggests that another ABC transporter can export blp bacteriocins, as there were few signs of evolutionary decay within putative blp bacteriocin genes.
Phylogenetic conservation also provides an explanation for the limited number of observed ecological strategies in our sequenced genomes, as reflected in the significant similarity among the ecological strategies within clonal complexes (fig. 6). It is interesting, however, that variation within clonal complexes is still present, indicating that even closely related strains may frequently switch bacteriocin groups or diversify via recombination at this locus; further investigation will be needed to examine if these changes among related ST are driven by neutral or selective factors.
Mechanisms of Diversification
Although various constraints undoubtedly limit the number of realized blp ecological strategies, 255 moderately common strategies still bear explaining; what creates and maintains these strategies? From an ecological and evolutionary standpoint, we hypothesize that antagonistic relationships between competing strains in the nasopharynx select for bacteriocin diversity. Interference competition, in which individuals expend resources to actively inhibit others’ growth, leads to a winner-takes-all system of low diversity in a well-mixed environment. However, interference competition with spatial structure has the potential to increase global diversity by creating several locally optimal ecological strategies but with no single, globally best ecological strategy (Riley and Gordon 1999; Riley and Wertz 2002; Abrudan et al. 2012; Hawlena et al. 2012). Assuming that bacteriocin production and immunity come with fitness costs, trade-offs between killing, immunity, and faster growth can lead to nontransitive dynamics that promote and preserve diversity across local sites in silico (Czárán et al. 2002), in vitro (Kerr et al. 2002; Majeed et al. 2011), and in vivo (Kirkup and Riley 2004; Bakkal et al. 2010). Novel ecological strategies are constantly selected for, as the frequency of specific, local strategies is ever-changing. This would explain the variation seen at three specific levels in the blp operon: In the rapid molecular evolution of bacteriocins (table 3); in the diversity of phylotypes within genes (fig. 4); and in the changing gene composition of the blp locus (fig. 3). The lower variation within and between genes in the D–E group compared with other bacteriocin groups (figs. 3 and 4) may suggest that a different evolutionary force was acting on this bacteriocin group; while the D–E group was tightly clustered (fig. 1), more investigation is needed to determine if this is evidence of an expanding or a declining lineage.
At a more mechanistic level, different factors could explain the diversification of the blp operon, in turn helping to explain the vast (and possibly limitless) number of unique ecological strategies. First, the mutation rate of individual genes could be high, although there is no evidence to support this possibility. Second, and more likely, a key driver of allelic and genic diversity at the blp locus may be recombination, as S. pneumoniae is naturally transformable with high rates of horizontal gene transfer occurring in both liquid and surface associated communities (Johnston et al. 2014). For example, the first 12 residues of 2 blpN variants have recombined with blpM and blpK (table 2). Recombination can explain the presence of two distinct presumed leader sequences within blpN and blpK (supplementary table S5, Supplementary Material online) as well the blpM variant found in 1.1% of genomes (table 2), which encodes for two bacteriocin peptides within one gene. As noted above, the presence of similar, transposase-derived sequences following blpA and before blpK is a potential region of homologous recombination that can underlie the transfer of variable unique genes between strains, as well as the intragenomic mobility of blpK itself. In contrast to other genes in the operon, blpK is located in two positions, either within the canonical operon or alternatively adjacent to comAB. The recent discovery of a circular bacteriocin next to comAB (Bogaardt et al. 2015) and experimental evidence demonstrating the ability of ComAB to export BlpC (Kjos et al. 2016) suggests a strong relationship between comAB and bacteriocin production.
Our analyses have uncovered unprecedented diversity at the blp operon and highlighted numerous questions that can be examined empirically. This is important for understanding the role of these toxins in interference competition and pneumococcal strain prevalence, but also as a way to potentially identify new bacteriocins and bacteriocin targets that may be of clinical value. This holds true for classical type IIb bacteriocins but also for new bacteriocins that fall into different functional types. Toward that end, it is notable that the number of putative bacteriocin genes per genomes did not peak at even numbers (supplementary fig. S3, Supplementary Material online), which would be expected if blp bacteriocin genes were always found with their dedicated partner when forming a functional type IIb bacteriocin. Additionally, there were significantly higher numbers of immunity genes than putative bacteriocin genes, especially after considering that bacteriocin genes are predicted to function in pairs; this suggests that there is not a one-to-one relationship between bacteriocin and immunity genes. Finally, the relationship between the CAAX amino protease immunity genes (e.g., pncP, blpG, and blpY; Pei and Grishin 2001; Kjos et al. 2010, 2011; Bogaardt et al. 2015) and the shorter, membrane protein immunity genes (e.g., blpL, blpX, pncG, pncM, and blpZ) is unclear and needs to be elucidated to fully understand how immunity protects against specific bacteriocins.
In summary, thousands of S. pneumoniae genomes were required to saturate the diversity at the blp locus. Although trillions of ecological strategies are possible within this diversity, hundreds are instead realized. The reason for this discrepancy is that bacteriocins are found in discrete groups, that correlations between functionally or physically linked genes limit combinations among genes and phylotypes, and that clonally related genotypes share highly similar potential blp ecological strategies. Diversity lies mainly in the presence or absence of blp operon genes that then form extremely rare ecological strategies found in less than 5.2 × 10−4 of genomes (i.e., singleton strategies in our genome set). The blp system in S. pneumoniae provides an opportunity to further study interference competition in which hundreds of millions of strategies can potentially exist.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Bioinformatic work was carried out on the Dutch national e-infrastructure with the support of the SURF Foundation. This work made use of the facilities of N8 HPC provided and funded by the N8 consortium and EPSRC (grant no. EP/K000225/1). The Center is co-ordinated by the Universities of Leeds and Manchester. We acknowledge the helpful comments on an earlier draft of this article by Morten Kjos as well as three anonymous referees who provided extremely constructive suggestions to improve the clarity of the manuscript. The work was supported by the Biotechnology and Biological Sciences Research Council (BB/J006009/1) to D.E.R. and I.S.R.
Literature Cited
- Abrudan MI, Brown S, Rozen DE. 2012. Killing as means of promoting biodiversity. Biochem Soc Trans. 40:1512–1516. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkal S, Robinson SM, Ordonez CL, Waltz DA, Riley MA. 2010. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 156:2058–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr Infect Dis J. 19:187–195. [DOI] [PubMed] [Google Scholar]
- Bogaardt C, van Tonder AJ, Brueggemann AB. 2015. Genomic analyses of pneumococci reveal a wide diversity of bacteriocins—including pneumocyclicin, a novel circular bacteriocin. BMC Genomics 16:554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert D, et al. 2001. Pneumococcal carriage in children in the Netherlands: a molecular epidemiological study. J Clin Microbiol. 39:3316–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger SD, Frey P, Aebi S, Hinds J, Muhlemann K. 2010. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS One 5:e11638.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancey ST, et al. 2015. Composite mobile genetic elements disseminating macrolide resistance in Streptococcus pneumoniae. Front Microbiol. 6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C, et al. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 46:305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Finkelstein JA, et al. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 45:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Mitchell AM, et al. 2013. Dominant role of nucleotide substitution in the diversification of serotype 3 pneumococci over decades and during a single infection. PLoS Genet. 9:e1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts FT, et al. 2005. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet 365:1139–1146. [DOI] [PubMed] [Google Scholar]
- Czárán TL, Hoekstra RF, Pagie L. 2002. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci U S A. 99:786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SM, Deloria-Knoll M, Kassa HT, O’Brien KL. 2013. Impact of pneumococcal conjugate vaccines on nasopharyngeal carriage and invasive disease among unvaccinated people: review of evidence on indirect effects. Vaccine 32:133–145. [DOI] [PubMed] [Google Scholar]
- Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 75:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Saizieu A, et al. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 182:4696–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 186:1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez JA, Fresnadillo Martínez MJ. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother. 50(Suppl. S):59–73. [DOI] [PubMed] [Google Scholar]
- Gladman S, Seemann T. 2008. VelvetOptimiser. http://www.vicbioinformatics.com/software.velvetoptimiser.shtml [last accessed November 20, 2013].
- Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 102:8710–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 16:229–240. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Bashey F, Lively CM. 2012. Bacteriocin‐mediated interactions within and between coexisting species. Ecol Evol. 2:2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat. 6:65–70. [Google Scholar]
- Hoover SE, et al. 2015. A new quorum‐sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol Microbiol. 97:229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Campo N, Bergé MJ, Polard P, Claverys JP. 2014. Streptococcus pneumoniae, le transformiste. Trends Microbiol. 22:113–119. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174. [DOI] [PubMed] [Google Scholar]
- Kirkup BC, Riley MA. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412–414. [DOI] [PubMed] [Google Scholar]
- Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol. 192:2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos M, et al. 2011. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology 157:3256–3267. [DOI] [PubMed] [Google Scholar]
- Kjos M, et al. 2016. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 12:e1005422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugman KP, et al. 2003. A trial of a 9-valent pneumococcal conjugate vaccine in children with and those without HIV infection. N Engl J Med. 349:1341–1348. [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Aberer AJ, Stamatakis A. 2015. ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics 31:2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux T, Nuhn M, Hakenbeck R, Reichmann P. 2007. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 189:7741–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed H, Gillor O, Kerr B, Riley MA. 2011. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 5:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee L, et al. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 39:2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen-Meyer J, Oppegård C, Rogne P, Haugen HS, Kristiansen PE. 2010. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob Proteins. 2:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KL, et al. 2003. Efficacy and safety of seven-valent conjugate pneumococcal vaccine in American Indian children: group randomised trial. Lancet 362:355–361. [DOI] [PubMed] [Google Scholar]
- O’Brien KL, et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 53:673–684. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 26:275–277. [DOI] [PubMed] [Google Scholar]
- Pinchas MD, LaCross NC, Dawid S. 2015. An electrostatic interaction between BlpC and BlpH dictates pheromone specificity in the control of bacteriocin production and immunity in Streptococcus pneumoniae. J Bacteriol. 197:1236–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: http://www.R–project.org/.
- Regev-Yochay G, et al. 2004. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis. 38:632–639. [DOI] [PubMed] [Google Scholar]
- Reichmann P, Hakenbeck R. 2000. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol Lett. 190:231–236. [DOI] [PubMed] [Google Scholar]
- Riley MA, Gordon DM. 1999. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7:129–133. [DOI] [PubMed] [Google Scholar]
- Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 56:117–137. [DOI] [PubMed] [Google Scholar]
- RStudio Team. 2015. RStudio: integrated development environment for R. Boston (MA): RStudio, Inc. Available from: http://www.rstudio.com/.
- Sauver JS, et al. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg Infect Dis. 6:622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spijkerman J, et al. 2012. Long-term effects of pneumococcal conjugate vaccine on nasopharyngeal carriage of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis. PLoS One 7:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol Lett. 241:129–134. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Tocheva AS, et al. 2011. Declining serotype coverage of new pneumococcal conjugate vaccines relating to the carriage of Streptococcus pneumoniae in young children. Vaccine 29:4400–4404. [DOI] [PubMed] [Google Scholar]
- Turner KME, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie AL, et al. 2014. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS One 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, van der Donk WA. 2016. Structural characterization and bioactivity analysis of the two-component lantibiotic Flv system from a ruminant bacterium. Cell Chem Biol. 23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]