Abstract
In several species genetic differentiation across environmental gradients or between geographically separate populations has been reported to center at “genomic islands of divergence,” resulting in heterogeneous differentiation patterns across genomes. Here, genomic regions of elevated divergence were observed on three chromosomes of the highly mobile fish Atlantic cod (Gadus morhua) within geographically fine-scaled coastal areas. The “genomic islands” extended at least 5, 9.5, and 13 megabases on linkage groups 2, 7, and 12, respectively, and coincided with large blocks of linkage disequilibrium. For each of these three chromosomes, pairs of segregating, highly divergent alleles were identified, with little or no gene exchange between them. These patterns of recombination and divergence mirror genomic signatures previously described for large polymorphic inversions, which have been shown to repress recombination across extensive chromosomal segments. The lack of genetic exchange permits divergence between noninverted and inverted chromosomes in spite of gene flow. For the rearrangements on linkage groups 2 and 12, allelic frequency shifts between coastal and oceanic environments suggest a role in ecological adaptation, in agreement with recently reported associations between molecular variation within these genomic regions and temperature, oxygen, and salinity levels. Elevated genetic differentiation in these genomic regions has previously been described on both sides of the Atlantic Ocean, and we therefore suggest that these polymorphisms are involved in adaptive divergence across the species distributional range.
Keywords: chromosomal rearrangements, gene flow, marine organisms, population genomics, ecological adaptation, structural polymorphisms
Introduction
Elevated genetic differentiation across environmental gradients or between geographically spaced populations is in some species observed to center to a few genomic regions, or “genomic islands of divergence,” contrasting the overall genomic differentiation (Wu 2001; Turner et al. 2005; Nosil et al. 2009). The occurrence of these patterns may be explained by spatially varying selection in combination with heterogeneity in recombination rates, as may be caused by structural features such as chromosomal rearrangements or centromeres, will affect the patterns of observed differentiation across genomes (Cutter and Payseur 2013). Consequently, heterogeneous differentiation across chromosomes might be a reflection of variable recombination rate rather than suppression of gene flow due to selection (Cruickshank and Hahn 2014), which has recently been documented in a number of species and clades (Renaut et al. 2013; Barb et al. 2014; Burri et al. 2015; Feulner et al. 2015).
Often described as “recombination repressors,” large chromosomal inversions have repeatedly been identified as drivers of genomic differentiation by repressing recombination between “noninverted” and “inverted” alleles (Sturtevant and Beadle 1936; Dobzhansky and Epling 1948), allowing for sympatric divergent evolution (White et al. 2007; Fang et al. 2012; Guerrero et al. 2012; Pyhäjärvi et al. 2013). Chromosomal inversions have the potential to span a large number of molecular polymorphisms, and may thereby attain a selective advantage from the combination of nucleotides they hold (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). In addition to the alleles initially captured by the polymorphism, both structural problems during meiosis and disruption of genes by inversion breakpoints have also been proposed as potential targets of selection.
Inversions has been identified as drivers of adaptation in a wide number of taxa (Hoffmann and Rieseberg 2008; Nosil et al. 2009; Kirkpatrick 2010). For example, in three-spined sticklebacks (Gasterosteus aculeatus) several inversions, ranging up to 1.7 Mb, have been shown to facilitate recurrent transitions between marine and freshwater forms worldwide (Jones et al. 2012). A large inversion in the common fruit fly (Drosophila melanogaster), first discovered over 20 years ago, is known to contribute to climatic adaptation (Krimbas and Powell 1992; Rane et al. 2015). In monkeyflowers (Mimulus guttatus), a large inversion spanning over 30 cM has been shown to distinguish between annual and perennial forms on the American west coast (Lowry and Willis 2010; Twyford and Friedman 2015).
While selection acting chromosomes of the same arrangement will create an excess of linkage disequilibrium (LD) in a narrow region extending on both sides of a beneficial genetic variant, but a lack of LD across the two sides (Fay and Wu 2000; Kim and Stephan 2002; Sabeti et al. 2002; Kim and Nilsen 2004; McVean 2007), a large inversion will cause reduced recombination between noninverted and inverted alleles, and excess of LD may be observable also between loci at opposite margins of the rearrangement (Navarro and Ruiz 1997; Navarro et al. 1997; Munte et al. 2005; Bansal et al. 2007; Fang et al. 2012). As the genomic region showing reduced recombination will be physically determined by the two inversion breakpoints, it should reach the same physical extent within the distribution of the polymorphism.
In marine organisms the potential for gene flow is high. Recent studies, however, have revealed the presence of genetic structuring, even in mobile species (reviewed in Hauser and Carvalho 2008; Salmenkova 2011).
The Atlantic cod (Gadus morhua), a highly mobile fish distributed throughout the continental shelf on both sides of the North Atlantic Ocean, exploiting a wide range of marine environments through a variety of life-history strategies and behaviors. On both sides of the Atlantic Ocean, genomic regions of elevated differentiation on Atlantic cod linkage groups 2, 7, and 12 have been observed across latitudinal and climatic gradients (Bradbury et al. 2010), and elevated LD have been described within the same genomic regions (Bradbury et al. 2014).
In this study, we show that for these genomic regions of elevated differentiation on Atlantic cod, the observed differentiation reflects vast molecular divergence between pairs of morphs, or alleles, extending 5, 9.5, and 13 Mb on linkage groups 2, 7, and 12, respectively. These alleles segregate within both coastal and oceanic sampling locations in a North-sea—Skagerrak study area. We further show that these three genomic regions mirror the divergence and recombination signatures of large polymorphic inversions, and assign alternate inversion alleles within individual samples. Based on the allelic shifts observed herein between coastal and oceanic environments, and previously observed patterns of genome differentiation across the species distributional range, we finally relate these extensive polymorphisms to adaptive evolution on both sides of the Atlantic Ocean.
Materials and Methods
Species, Study Area, and Sampling
Atlantic cod are seasonal batch spawners with pelagic egg and larval stages (Kjesbu 1989). Its distribution spans the North Atlantic from Novaya Zemlya (Russia), Spitsbergen (Norway and Russia) and Greenland in the north, to Bay of Biscay (France and Spain) and Cape Hatteras (USA) in the south. In the Skagerrak, which separate southern Norway from Denmark, and in the North Sea, spawning generally takes place during January to April and juveniles settle a few weeks later. Coastal cod typically become sexually mature at ages 2–4 years (Olsen et al. 2004; Yoneda and Wright 2004), and average generation length in the Skagerrak area is around 4 years (Knutsen et al. 2011). In all parts of its distribution, cod in oceanic areas typically move more extensively than cod in sheltered areas (Fox et al. 2008; Neuenfeldt et al. 2013), which holds also for the North Sea—Skagerrak system (Danielssen 1969; Espeland et al. 2008, Rogers et al. 2013), and genetic divergence between cod inhabiting oceanic waters and cod from coastal locations has been reported on both sides of the Atlantic ocean (Ruzzante et al. 1999; Knutsen et al. 2004, 2011; Westgaard and Fevolden 2007).
Here altogether six coastal sites were chosen, representing the three coastal study systems Kristiansand (K), Lillesand (L), and Risør (R). From each of these three study systems, samples (n = 47–48) were taken from an inner, supposed sheltered, fjord (abbreviated KI, LI, and RI) and from a nearby, more exposed outer-coast location (KO, LO, and RO). For all six coastal localities, juveniles (young-of-the-year) were collected in the autumn by beach seine, as part of an on-going long-term survey of the Norwegian Skagerrak coast (Olsen et al. 2009; Rogers et al. 2011). Fish were selected from three different years (i.e., cohorts) when available, to obtain representative samples from each locality. In addition, two oceanic North Sea samples was taken of adult fish (several age classes) off the Danish coast (NSS; n = 43) and off the Norwegian coast (NSN; n = 48). Previous mapping of North Sea—Skagerrak spawning grounds (Fox et al. 2008), as well as studies of the effects of oceanic—coastal currents on drift of egg and larvae (Stenseth et al. 2006; Knutsen et al. 2007; Ciannelli et al. 2010), have shown that these oceanic spawning grounds have the potential to genetically influence the coastal study areas represented herein. All sampled fish (n = 378: table 1) were frozen in the field and stored whole until DNA extraction.
Table 1.
Genotyped Samples Used in the Study
| Name | Location | N | Years | Called | HWE | MAF | Het |
|---|---|---|---|---|---|---|---|
| KI | Inner | 48 | 2000, 2005, 2008 | 99.3 | 0.06 | 0.77 | 0.358 |
| KO | Outera | 48 | 2000, 2005, 2008 | 99.1 | 0.06 | 0.53 | 0.361 |
| LI | Inner | 47 | 2004, 2005, 2010 | 99.3 | 0.09 | 0.69 | 0.358 |
| LO | Outer | 48 | 2004, 2005, 2010 | 99.3 | 0.09 | 0.76 | 0.355 |
| RI | Inner | 48 | 1997, 1998, 2004, 2005 | 99.3 | 0.05 | 0.67 | 0.356 |
| RO | Outer | 48 | 1997, 1998, 2004, 2005 | 99.6 | 0.03 | 0.59 | 0.355 |
| NSN | Oceanic | 48 | 2012 | 98.7 | 0.05 | 0.72 | 0.362 |
| NSS | Oceanic | 43 | 2002 | 99.7 | 0.01 | 0.70 | 0.360 |
NOTE.—Name, location, number of individuals (N), years of sampling, percentage of called genotypes (out of 9,187 SNPs), percentage of SNPs out of Hardy–Weinberg (HWE; False-discovery rate <0.1), percentage of SNPs with minor-allele frequency (MAF) below 0.01, and average observed heterozygocities (Het) are given for each sample.
The KO sample proved more similar to the inner-coast samples when comparing overall population differentiation (table 2), and was in further analyses not included as an outer-coast sample.
Single Nucleotide Polymorphism Array Development
A custom single nucleotide polymorphism (SNP) array, manufactured by Illumina (San Diego, CA), was constructed as part of the Norwegian Cod SNP Consortium (CSC) composed of four Norwegian research organizations (Norwegian University of Life Sciences, University of Oslo, Institute of Marine Research, and NOFIMA). Genomes from seven individual cod samples collected from a wide geographic range across the North East Atlantic (including cod from the Risør inner-coast) were shotgun sequenced as paired-end read libraries using Illumina GAii instrumentation. For each sample an average of 79% of reads were aligned to the reference genome (gadMor1; Star et al. 2011) using the Burrows–Wheeler Aligner (Li and Durbin 2009). SNPs were detected using SAMtools (Li et al. 2009). A list of 2,877,794 putative SNPs was reduced using a variety of filters including their physical distribution, functional associations, and a need to display a minor-allele frequency greater than 0.1 in the sequenced samples.
From a total of 10,605 assayed SNPs, quality and validity assessment performed at Center for Integrative Genetics, Norwegian University of Life Sciences, left 9,420 working assays (Kent M, unpublished data). Of these 9,420 SNPs, 260 were previously published (Moen et al. 2008; Hubert et al. 2010), 672 were in proximity to selected candidate genes, and 1,595 were nonsynonymous coding SNPs. There was an average of 409 SNPs per chromosome, with a maximum of 554 (linkage group 7) and a minimum of 279 (linkage group 19).
Genotyping and Initial Data Exploration
Genotypes were clustered using the Illumina GenomeStudio software 2011.1, and 9,187 SNPs were found to be actual polymorphic loci called for at least 95% of individuals across the 378 samples. All 9,187 SNPs are identified in supplementary table S1, Supplementary Material online, by their accession number in the dbSNP database (Sherry et al. 2001).
The R software 3.1.3 (R Core Team 2014) was used for subsequent analyses. The GenABEL R package (Aulchenko et al. 2007) was applied to test for deviations from Hardy–Weinberg equilibrium across, while minor-allele frequency and average observed heterozygocity was estimated with the hierfstat R package (Goudet 2005). LD was estimated as composite LD (Schaid 2004), a method suitable for unphased genotype data, for all pairs of SNPs with the R software utilizing the algorithm proposed by Gao et al. (2009). SNP pairs with LD >0.5 were classified as being in high LD, and physical extent of LD blocks was estimated by summing the lengths of the scaffolds in the Atlantic cod genome assembly (Star et al. 2011) anchoring SNPs spanned by the blocks.
Population Differentiation and Genome Divergence
Population differentiation were characterized by FST (Weir and Cockerham 1984) and estimated between pairs of sampling locations as well as for groups of sampling locations with the wc function in the hierfstat R package (Goudet 2005). For examining patterns of divergence across the genome, SNPs were mapped to the 23 Atlantic cod linkage groups (supplementary table S1, Supplementary Material online; nomenclature of Hubert et al. [2010]), and approximately positioned within linkage groups 2, 7, and 12 based on preliminary linkage data (supplementary table S1, Supplementary Material online; Lien S, unpublished data).
Chromosomal phases for linkage groups 2, 7, and 12 were predicted employing an algorithm for unrelated individuals with the Beagle software (Browning and Browning 2007). Genealogies of phased chromosomes were explored by unrooted neighbor-joining (BioNJ; Gascuel 1997) of euclidean distances by a method allowing for incomplete distance matrices (Criscuolo and Gascuel 2008), implemented in the dist and bionjs functions in the R package ape (Paradis et al. 2004). Genealogies were found separately for SNPs within the blocks of LD on linkage groups 2, 7, and 12, and for the remainder of these linkage groups. Random balanced subsets of chromosomes (n = 100) were used for the final neighbor-joining trees.
A probabilistic sliding-window analysis for detection of inversions (Sindi and Raphael 2010) was conducted with codeHaplo and scanInv functions in the R package inveRsion (Càceres et al. 2012) using a window size of 4 cM and block size 3. Individuals were assigned to inversion genotypes by principle component analysis (PCA) based on SNPs within the LD blocks on linkage groups 2, 7, and 12 using the snpgdsPCA function in the R package SNPRelate (Zheng et al. 2012), following the approach suggested by Sindi and Raphael (2010). Differentiation characterized by FST (Weir and Cockerham 1984) for the inversion alleles were estimated with the basic.stats function in the hierfstat R package (Goudet 2005). Sliding window analyses (window size 5 cM, step size one SNP) of net divergence (DXY; Nei and Li 1979), as well as across-loci analyses of nucleotide diversity, were calculated with the DNAsp software version 5.0 based on phased chromosomes (Librado and Rozas 2009).
Results
Patterns of genome differentiation (FST; Weir and Cockerham 1984) in Atlantic cod were observed on a fine geographic scale in its southern range of distribution (i.e., North Sea—Skagerrak area) based on 9,187 genome-wide distributed SNPs. Samples included inner-coast (KI, LI, and RI), outer-coast (KO, LO, and RO), and oceanic locations (NSS and NSN), and a total of 378 individual fish (table 1). The average amount of differentiation over loci among sample pairs indicated a pattern of spatial genetic structure with inner-coast samples being very similar between themselves, yet divergent from the outer-coast and oceanic samples, which were generally more similar to each other (table 2). This inner–outer dichotomy was more pronounced in the full (9,187 loci) SNP data set than in a LD filtered data set (LD < 0.5; 8,107 loci). The KO sample, initially classified as an outer-coast sample, was observed to be more similar to the inner-coast samples than the remaining outer-coast samples (table 2), and hence excluded from further analyses of between-locations genome divergence.
Table 2.
Overall Genetic Differentiation between Samples
|
Inner Fjord |
Outer Coast |
Ocean |
||||||
|---|---|---|---|---|---|---|---|---|
| Sample | KI | LI | RI | KO | LO | RO | NSS | NSN |
| KI | — | 0.0001 | 0.0003 | 0.0006 | 0.0093 | 0.0071 | 0.0110 | 0.0118 |
| LI | −0.0001 | — | 0.0001 | 0.0002 | 0.0078 | 0.0060 | 0.0090 | 0.0096 |
| RI | 0.0002 | 0.0001 | — | 0.0003 | 0.0064 | 0.0046 | 0.0076 | 0.0085 |
| KO | 0.0005 | 0.0003 | 0.0003 | — | 0.0052 | 0.0036 | 0.0063 | 0.0071 |
| LO | 0.0116 | 0.0107 | 0.0077 | 0.0068 | — | 0.0004 | 0.0005 | 0.0010 |
| RO | 0.0091 | 0.0085 | 0.0063 | 0.0046 | 0.0012 | — | 0.0003 | 0.0012 |
| NSS | 0.0177 | 0.0167 | 0.0143 | 0.0113 | 0.0060 | 0.0020 | — | 0.0006 |
| NSN | 0.0189 | 0.0172 | 0.0164 | 0.0131 | 0.0109 | 0.0058 | 0.0025 | — |
Note.—Population differentiation (FST) between all pairs of sampling locations. Lower triangle contain estimates based on all 9,187 loci remaining after initial quality filtering, upper triangle contain estimates based on a subset of 8,107 loci where LD filtering (<0.5) within sympatric SNP pairs is applied.
Genomic Differentiation and LD
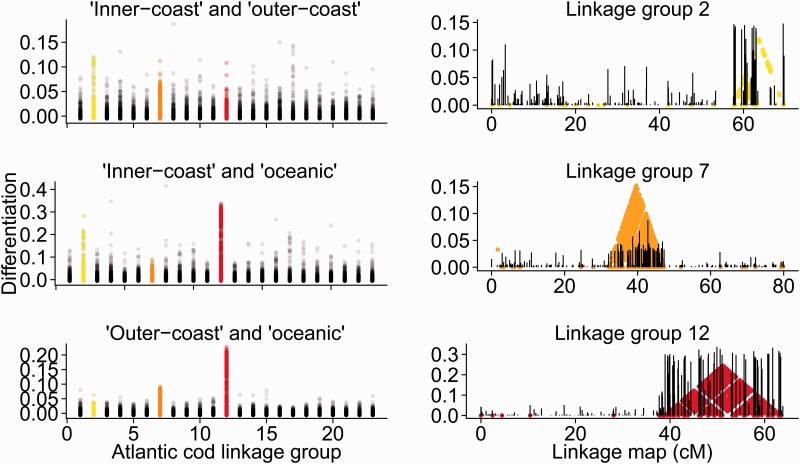
A genomic region of elevated differentiation, including 205 SNPs in a region of 13 megabases (Mb), was observed for linkage group 12 (Lg12) between inner-coast and oceanic samples and between outer-coast and oceanic samples (fig. 1). Genomic regions of elevated differentiation were also observed for Lg2 and Lg7, represented by 85 and 193 SNP, embraced within 5 and 9.5 Mb, respectively. Extensive blocks of elevated LD on Lg2, Lg7, and Lg12 were evident both across samples within the study area (fig. 1) and within individual sampling locations (supplementary figs S1–S3, Supplementary Material online), spanning the same chromosomal segments within all locations. For each of the three genomic regions, a single, LD block was identified, with high LD also between loci at opposite margins of these regions of reduced observed recombination (fig. 1), separated by several megabases. As the current genome assembly is fragmented (Star et al. 2011), the estimates of the extent of these genomic regions are likely to be conservative. For the remaining 20 Atlantic cod chromosomes, less than 0.01% of syntenic SNP pairs were in high LD (LD > 0.5).
Fig. 1.—
Genomic differentiation and LD. Left; allelic differentiation (FST) between “inner-coast” (pooled samples KI, LI, and RI) and “outer-coast” (pooled LO and RO) locations, between “inner-coast” and “oceanic” (pooled NSs and NSN) locations, and finally, between “outer-coast” and “oceanic” locations, for SNPs mapped to the 23 Atlantic cod linkage groups. Linkage groups 2, 7, and 12 are indicated in yellow, orange and red, respectively. Right; allelic differentiation (FST; black bars) across linkage group 2 (between inner-coast and outer coast locations), 7 (between outer-coast and oceanic locations), and 12 (inner-coast and oceanic locations) is shown for SNPs according to preliminary linkage maps (cM), with SNP pairs showing high LD (LD > 0.5), color coded as in the left panel.
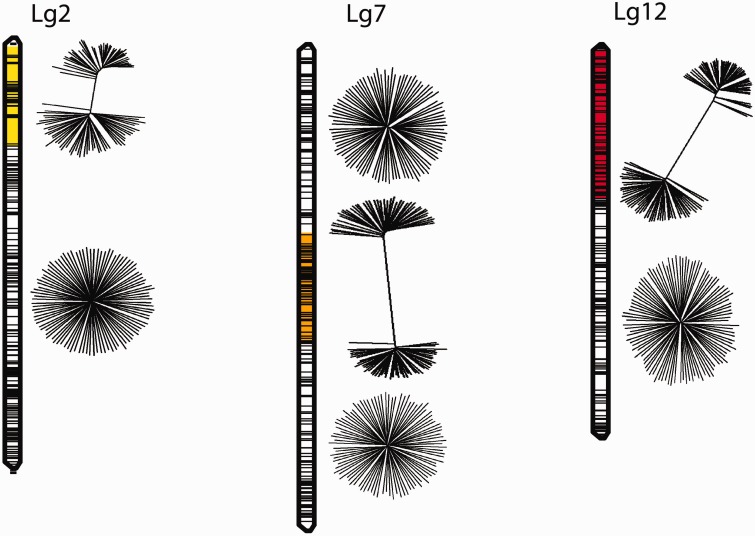
Chromosomal Rearrangements
Divergent genealogies with two clearly separable morphs, or alleles, were revealed for phased chromosomes within the genomic regions of high divergence on Lg2, Lg7, and Lg12, contrasting genealogies based on SNPs positions for the remainder of these three linkage groups (fig. 2). The observed patterns of LD and divergence were in agreement with genomic signatures previously described for large inversions (White et al. 2007; Fang et al. 2012; Guerrero et al. 2012; Pyhäjärvi et al. 2013). This observation was supported by a probabilistic scan by the method of Sindi and Raphael (2010) (supplementary fig. S4, Supplementary Material online), which models a population as a mixture of noninverted and inverted haplotypes and identifies putative inversion breakpoints by characteristic differences in haplotype frequencies.
Fig. 2.—
Genealogies within genomic regions of high divergence. Chromosomal regions spanned by the detected inversions are indicated for linkage group 2 (Lg2; yellow), 7 (Lg7; orange), and 12 (Lg12; red), and SNPs are positioned according to preliminary genetic maps (horizontal black bars). Genealogies of phased chromosomes are illustrated as neighbor-joining trees to the right of each linkage group, separately for the observed genomic regions of high divergence and for the remainder of the same linkage groups.
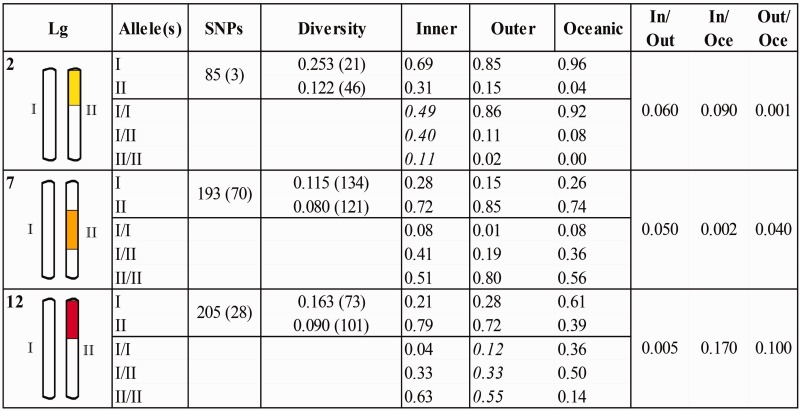
PCA based on all SNP genotypes within the genomic regions of elevated differentiation on linkage groups 2, 7, and 12 were used to assign inversion alleles in individual samples (fig. 3; supplementary fig. S5, Supplementary Material online), following the approach suggested by Ma and Amos (2012) for identification of alternate inversion alleles. The PCA assignments were in agreement with the genealogies observed for phased chromosomes within each of the three genomic regions (fig. 2). For all the three chromosomal rearrangements one of the two alleles showed reduced nucleotide diversity (typically about half) relative to the other for SNPs positioned within the rearrangements. This might indicate relative age of an ancestral, higher-diversity allele (I), and a derived, lower-diversity allele (II). A larger proportion of SNPs were found to be fixated within the lower-diversity alleles than within the higher-diversity alleles (fig. 3).
Fig. 3.—
Alleles of the chromosomal rearrangements. Observed alleles (I and II) of the chromosomal rearrangements on linkage groups (Lg) 2, 7, and 12 are characterized by the number of SNPs they span and the number of fixed SNPs (in parenthesis) between inversion alleles and nucleotide diversity and the number of fixed SNPs (in parenthesis) within each inversion allele. Genotypic frequencies of the alleles (I/I, I/II, and II/II) within inner-coast (Inner), outer-coast (Outer), and oceanic samples, as well as allelic differentiation (FST) of inversion alleles between inner-coast and outer-coast samples (In/Out), inner-coast and oceanic samples (In/Oce), and outer-coast and oceanic samples (Out/Oce) is also presented.
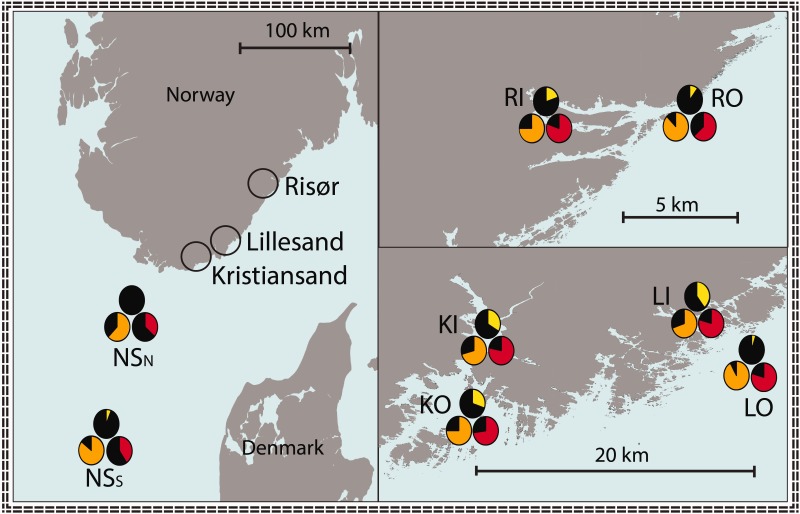
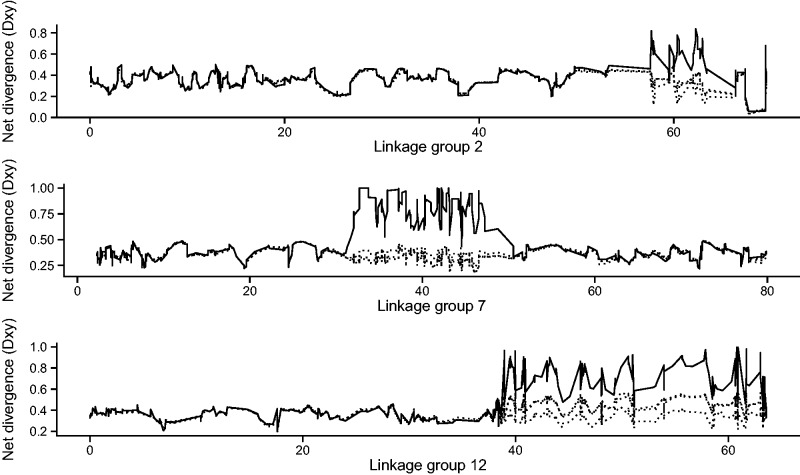
For Lg2 and Lg12, shifts in frequency of rearranged chromosomes were observed from oceanic to coastal sampling sites, where outer-coast samples were intermediate to the inner-coast and oceanic samples (figs. 3 and 4). For the two alleles on Lg2, elevated differentiation (FST) was observed between inner-coast and oceanic samples and between inner-coast and outer-coast samples, whereas for the alleles on Lg12 elevated FST was observed between inner-coast and oceanic samples and between outer-coast and oceanic samples (fig. 3). Sliding window analyses of net divergence (DXY) between phased chromosomes showed high divergence between alleles I and II relative to divergence between sampling locations (fig. 5), and each pair of inversion alleles were differentiated by a set of fixed SNPs (fig. 3; supplementary table S1, Supplementary Material online).
Fig. 4.—
Distribution of rearranged chromosomes in the North sea—Skagerrak study area. Frequencies of rearranged chromosomes (allele II) are presented as pie charts for each sampling location, where linkage group 2, 7, and 12 are indicated in yellow, orange, and red, respectively. Left; North sea North (NSN) and North sea South (NSS). Right; the three coastal locations Kristiansand, Lillesand, and Risør, each containing an inner-coast and an associated outer-coast sampling site.
Fig. 5.—
Divergence between chromosomes of alternate arrangements. Sliding window analyses of net divergence (DXY) between chromosomes of alternate arrangements (solid line) and between individuals of different geographic origins (dotted lines) are shown across linkage groups 2 (top), 7 (middle), and 12 (bottom) (cM).
Discussion
Utilizing genome scans for understanding evolutionary and ecological processes, described as reverse ecology (Li et al. 2008), represent an unbiased approach for identifying the mechanisms behind adaptive evolution in natural populations (Crawford and Nielsen 2013; Ellegren 2014; Storz et al. 2015). Here, within a geographically fine-scaled marine study area, a newly developed panel of over 9,000 genome-wide distributed SNPs was utilized to identify and accurately position three large genomic regions of elevated genetic differentiation, extending 5, 9.5, and 13 Mb on Atlantic cod linkage groups 2, 7, and 12. Both the level of differentiation and the size of these genomic regions were unanticipated, as divergent selection should not be observable beyond the kilobase-scale in the presence of gene flow (Kim and Stephan 2002). Instead, reduced genetic exchange between pairs of divergent alleles spanning these three genomic regions, likely to have emerged through chromosomal inversions, was found to cause the observed genetic differentiation.
Evidence is accumulating regarding the importance of recombination heterogeneity in adaptive radiation and speciation in a number of species and clades (Hoffman and Rieseberg 2008; Nosil et al. 2009), and the novel view of the Atlantic cod genome presented herein represents a striking example of how repressed recombination may allow adaptive divergence in the face of gene flow.
Chromosomal Rearrangements as Promoters of Genome Divergence
Large chromosomal inversions may lead to the segregation of noninverted and inverted chromosomes, where meiotic recombination may be almost completely repressed between the alternate inversion alleles (Sturtevant and Beadle 1936; Dobzhansky and Epling 1948; Navarro and Ruiz 1997; Navarro et al. 1997). In time, repressed recombination will create a block of LD spanning the rearranged chromosomal segment, physically bound by the inversion breakpoints (Munte et al. 2005; Bansal et al. 2007; Fang et al. 2012).
Here, elevated genomic differentiation coinciding with extensive LD blocks, spanning 5, 9.5, and 13 Mb, were observed on Atlantic cod linkage groups 2, 7, and 12 within a geographically fine-scaled study area. High LD was also observed between SNPs separated by several megabases, positioned at opposite margins of each block. LD blocks of the same extent were observed within all sampling locations, supporting the presence of two inversion breakpoints as the physical determinants of the genomic regions of reduced recombination. In contrast, divergent selection on chromosomes of the same arrangement is expected to result in separate, smaller, LD blocks extending in both directions from loci targeted by selection, but not create excess of LD extending beyond the kilobase-scale, or to neutral loci at opposite margins of the region of observed reduced recombination (Fay and Wu 2000; Kim and Stephan 2002; Sabeti et al. 2002; Kim and Nilsen 2004; McVean 2007). Besides inversions, both translocations of large chromosomal segments, as well as the presence of centromeres, are known to reduce recombination rates. However, these alternative recombination repressors are observed to reduce recombination proximal to a single translocation breakpoint (Liu et al. 1994; Livingston et al. 1999, 2000) or centromeric region (Smith et al. 2005; Turner et al. 2005), and LD should therefore be more pronounced at the center of the regions of reduced recombination than between distant loci at opposite margins of these regions. While large translocations could theoretically create excess of LD in a similar manner to selection acting on noninverted chromosomes (Liu et al. 1994; Livingston et al. 1999, 2000), their genomic signatures remain poorly characterized.
Here, highly divergent genealogies was observed between pairs of morphs, or alleles, within previously reported genomic regions of high differentiation on linkage groups 2, 7, and 12 (fig. 2; supplementary fig. S5, Supplementary Material online), segregating within both coastal and off-shore sampling locations (fig. 3). These genealogies demonstrate that repressed genetic exchange or repression of recombination, between these alternate alleles is responsible for the observed genomic differentiation and excess of LD. For all the three genomic regions, high net divergence (DXY) was demonstrated to be driven by divergent evolution between chromosomes of alternate arrangements, mirroring previously observed divergence patterns for large polymorphic inversions (White et al. 2007; Fang et al. 2012; Guerrero et al. 2012; Pyhäjärvi et al. 2013; Twyford and Friedman 2015). For Lg2, less pronounced inversion signatures were observed, as assessed by LD (fig. 1; supplementary fig. S1, Supplementary Material online) and DXY (fig. 5), compared with Lg7 and Lg12, which might be due to more gene exchange, reduced selective pressure or SNP ascertainment bias, as recently discussed by Twyford and Friedman (2015). Alternatively, recurrent chromosomal rearrangements within the same genomic region, as observed in Drosophila subobscura (Puerma et al. 2014), might explain the less pronounced signature within this linkage group. Finally, the presence of chromosomal rearrangements within all three linkage groups was supported by a probabilistic scan for inverted haplotypes (supplementary fig. S4, Supplementary Material online). This method models a population as a mixture of noninverted and inverted haplotypes, identifying putative inversion breakpoints by characteristic differences in haplotype frequencies. The approach has been shown to accurately identify inversions in both real and simulated data (Sindi and Raphael 2010).
Distribution and Divergence of Inversion Alleles
Within each of the three rearranged genomic regions, one morph or allele showed reduced genetic variability relative to the other. Comparative analyses of samples across larger spatial scales, as well as full sequencing data to avoid ascertainment bias, should shed more light on the evolutionary relationships between the alleles of the herein described polymorphisms.
Elevated genomic differentiation within the same three genomic regions described herein have been observed on both sides of the Atlantic Ocean (Bradbury et al. 2010), indicating that the inversions predate the split between eastern and western Atlantic cod, about 100 kyr ago (Bigg et al. 2008). Although emergence at such an early stage in the evolutionary history of cod was supported by the relatively high diversities found within both allele I and II on all three linkage groups (fig. 3), the possibility of subsequent dispersal cannot be excluded at this stage. Allelic clines for SNPs within these genomic regions on Lg2, Lg7, and Lg12 have been shown to correlate with latitudinal and climatic gradients on both sides of the Atlantic Ocean (Bradbury et al. 2010, 2014), which is here interpreted as allelic clines of the inversions described herein. Molecular variation on Lg12 has been linked to both environmental gradients (Bradbury et al. 2010) and behavior (Hemmer-Hansen et al. 2013; Karlsen et al. 2013), and more recently, to sea temperature and oxygen levels (Berg et al. 2015). Coastal cod are observed to show differences in behavior from oceanic cod (Nordeide et al. 2011), and are likely to experience different environmental conditions. These previous findings are thus consistent with the allelic shifts between coastal and oceanic locations observed for the rearrangements on Lg12, where intermediate frequencies were observed in outer-coast locations compared with inner-coast and oceanic locations (fig. 4). Similarly, recently reported associations between molecular variation on Lg2 and salinity gradients (Berg et al. 2015) may explain the observed allelic shifts from coastal to oceanic locations for the rearrangement on this linkage group (fig. 4). It is thus plausible that fitness effects relating to environmental heterogeneity across the distributional range of this species are maintaining the alternate inversion alleles described here. Such fitness effects should be identifiable by mapping allelic distributions to environmental conditions over a larger geographical scale, as well as through experimental studies relating to functionality of inversion alleles.
Implications for Demography
In natural populations genome divergence is opposed by gene flow and rarely observed without the presence of spatial barriers or very strong fitness effects of segregating genetic variation (reviewed in Nosil et al. 2009; Abbott et al. 2013). Most of the observed genomic differentiation previously described between geographic regions or behavioral types of cod centers to a few distinct genomic regions (Bradbury et al. 2010; Hemmer-Hansen et al. 2013; Karlsen et al. 2013; Berg et al. 2015), three of which we here suggest to be caused by polymorphic chromosomal rearrangements.
These rearrangements span large numbers of molecular polymorphisms, many of which show high divergence between inverted and noninverted chromosomes. In addition to demographic processes, we thus predict that the genetic structuring observed within this species may be influenced by selective processes acting on the herein described inversions.
Supplementary Material
Supplementary table S1 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Research Council of Norway (project “CODFLICT”), the county of Aust-Agder Fylkeskommune, the European Regional Development Fund (Interreg IVa, “MarGen” project), and The Norwegian Ministry of Fishery and Coastal Affairs. The authors are grateful to Svein E. Enersen, Alf Ring Kleiven, Øystein Paulsen, Knut Hansen, Petter Baardsen, Kate Enersen, and Hanne Sannæs for technical assistance and sampling. SNP-genotypes for the study were provided by the Norwegian Cod SNP Consortium (CSC).
Literature Cited
- Abbott R, et al. 2013. Hybridization and speciation. J Evol Biol. 26:229–246. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23:1294–1296. [DOI] [PubMed] [Google Scholar]
- Bansal V, Bashir A, Bafna V. 2007. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 17:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb JG, et al. 2014. Chromosomal evolution and patterns of introgression in Helianthus. Genetics 197:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg PR, et al. 2015. Adaptation to low salinity promotes genomic divergence in Atlantic cod (Gadus morhua L.). Genome Biol Evol. 7:1644–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigg GR, et al. 2008. Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proc Biol Sci. 275:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury IR, et al. 2010. Parallel adaptive evolution of Atlantic cod on both sides of the Atlantic Ocean in response to temperature. Proc Biol Sci. 277:3725–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury IR, et al. 2014. Long distance linkage disequilibrium and limited hybridization suggest cryptic speciation in Atlantic cod. PLoS One 9:e106380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25:1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Càceres A, Sindi SS, Raphael BJ, Cáceres M, González JR. 2012. Identification of polymorphic inversions from genotypes. BMC Bioinformatics 13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciannelli L, et al. 2010. Small-scale genetic structure in a marine population in relation to water circulation and egg characteristics. Ecology 91:2918–2930. [DOI] [PubMed] [Google Scholar]
- Crawford JE, Nielsen R. 2013. Detecting adaptive trait loci in non-model systems: divergence or admixture mapping? Mol Ecol. 22:6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gascuel O. 2008. Fast NJ-like algorithms to deal with incomplete distance matrices. BMC Bioinformatics 9:166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23:3133–3157. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielssen DS. 1969. On the migrations of the cod in the Skagerak shown by tagging experiments in the period 1954-1965. 15(3):331–338. [Google Scholar]
- Ellegren H. 2014. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29:51–63. [DOI] [PubMed] [Google Scholar]
- Espeland SH, et al. 2008. New perspectives on fish movement: kernel and GAM smoothers applied to a century of tagging data on coastal Atlantic cod. Mar Ecol Prog Ser. 372:231–241. [Google Scholar]
- Fang Z, et al. 2012. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. 2000. Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feulner PGD, et al. 2015. Genomics of divergence along a continuum of parapatric population differentiation. PLoS Genet. 11:e1004966.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, et al. 2008. Mapping the spawning grounds of North Sea cod (Gadus morhua) by direct and indirect means. Proc Biol Sci. 275:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starmer J, Martin ER. 2008. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 32:361–369. [DOI] [PubMed] [Google Scholar]
- Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 14:685–695. [DOI] [PubMed] [Google Scholar]
- Goudet J. 2005. Hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes. 5:184–186. [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M. 2012. Coalescent patterns for chromosomal inversions in divergent populations. Philos Trans R Soc B Biol Sci. 367:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser L, Carvalho GR. 2008. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish. 9:333–362. [Google Scholar]
- Hemmer-Hansen J, et al. 2013. A genomic island linked to ecotype divergence in Atlantic cod. Mol Ecol. 22:2653–2667. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Ann Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert S, Higgins B, Borza T, Bowman S. 2010. Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua). BMC Genomics 11:191.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen BO, et al. 2013. Genomic divergence between the migratory and stationary ecotypes of Atlantic cod. Mol Ecol 22:5098–5111. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167:1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8:e1000501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjesbu OS. 1989. The spawning activity of cod, Gadus morhua L. J Fish Biol. 34:195–206. [Google Scholar]
- Knutsen H, et al. 2004. Transport of North Sea cod larvae into the Skagerrak coastal populations. Proc R Soc Lond B Biol Sci. 271:1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen H, et al. 2007. Egg distribution, bottom topography and small-scale cod population structure in a coastal marine system. Mar Ecol Prog Ser. 333:249–255. [Google Scholar]
- Knutsen H, et al. 2011. Are low but statistically significant levels of genetic differentiation in marine fishes “biologically meaningful”? A case study of coastal Atlantic cod. Mol Ecol. 20:768–783. [DOI] [PubMed] [Google Scholar]
- Krimbas CB, Powell JR. 1992. Drosophila inversion polymorphism. Boca Raton (FL): CRC Press. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Costello JC, Holloway AK, Hahn MW. 2008. “Reverse Ecology” and the power of population genomics. Evolution 62:2984–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu CJ, et al. 1994. An RFLP-based genetic map of pearl millet (Pennisetum glaucum). Theor Appl Genet. 89:481–487. [DOI] [PubMed] [Google Scholar]
- Livingstone KD, Churchill G, Jahn MK. 2000. Linkage mapping in populations with karyotypic rearrangements. J Hered. 91:423–428. [DOI] [PubMed] [Google Scholar]
- Livingstone KD, Lackney VK, Blauth JR, van Wijk R, Jahn MK. 1999. Genome mapping in capsicum and the evolution of genome structure in the solanaceae. Genetics 152:1183–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Amos CI. 2012. Investigation of Inversion Polymorphisms in the Human Genome Using Principal Components Analysis. PLoS ONE. 7:e40224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. 2007. The Structure of Linkage Disequilibrium Around a Selective Sweep. Genetics 175:1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T, et al. 2008. Identification and characterisation of novel SNP markers in Atlantic cod: evidence for directional selection. BMC Genet. 9:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munte A, Rozas J, Aguadé M, Segarra C. 2005. Chromosomal Inversion Polymorphism Leads to Extensive Genetic Structure A Multilocus Survey in Drosophila subobscura. Genetics 169:1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. 2003. Chromosomal speciation and molecular divergence–accelerated evolution in rearranged chromosomes. Science 300:321–324. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A. 1997. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146:695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Ruiz A. 1997. On the fertility effects of pericentric inversions. Genetics 147:931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76:5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenfeldt S, et al. 2013. Analysing migrations of Atlantic cod Gadus morhua in the north-east Atlantic Ocean: then, now and the future. J Fish Biol. 82:741–763. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 98:12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeide JT, Johansen SD, Jorgensen TE, Karlsen BO, Moum T. 2011. Population connectivity among migratory and stationary cod Gadus morhua in the Northeast Atlantic-A review of 80 years of study. Mar Ecol Prog Ser. 435:269–283. [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Olsen EM, Carlson SM, Gjøsæter J, Stenseth NC. 2009. Nine decades of decreasing phenotypic variability in Atlantic cod. Ecol Lett. 12:622–631. [DOI] [PubMed] [Google Scholar]
- Olsen EM, et al. 2004. Life-history variation among local populations of Atlantic cod from the Norwegian Skagerrak coast. J Fish Biol. 64:1725–1730. [Google Scholar]
- Puerma E. et al. 2014. Characterization of the breakpoints of a polymorphic inversion complex detects strict and broad breakpoint reuse at the molecular level. Mol Biol Evol. 31:2331–2341. [DOI] [PubMed] [Google Scholar]
- Pyhäjärvi T, Hufford MB, Mezmouk S, Ross-Ibarra J. 2013. Complex patterns of local adaptation in teosinte. Genome Biol Evol. 5:1594–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA. 2015. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Molecular Ecology. 24:2423–2432. [DOI] [PubMed] [Google Scholar]
- Renaut S, et al. 2013. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 4:1827.. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:351–358. [DOI] [PubMed] [Google Scholar]
- Rogers LA, et al. 2011. Climate and population density drive changes in cod body size throughout a century on the Norwegian coast. Proc Natl Acad Sci U S A. 108:1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LA, et al. 2013. Centennial-scale fluctuations and regional complexity characterize Pacific salmon population dynamics over the past five centuries. Proc Natl Acad Sci U S A. 110:1750–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzante DE, Taggart CT, Cook D. 1999. A review of the evidence for genetic structure of cod (Gadus morhua) populations in the NW Atlantic and population affinities of larval cod off Newfoundland and the Gulf of St. Lawrence. Fish Res. 43:79–97. [Google Scholar]
- Sabeti PC, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837. [DOI] [PubMed] [Google Scholar]
- Salmenkova EA. 2011. New view on the population genetic structure of marine fish. Russian J Genet. 47:1279–1287. [PubMed] [Google Scholar]
- Schaid DJ. 2004. Linkage disequilibrium testing when linkage phase is unknown. Genetics 166:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindi SS, Raphael BJ. 2010. Identification and frequency estimation of inversion polymorphisms from haplotype data. J Comput Biol J Comput Mol Cell Biol. 17:517–531. [DOI] [PubMed] [Google Scholar]
- Smith AV, Thomas DJ, Munro HM, Abecasis GR. 2005. Sequence features in regions of weak and strong linkage disequilibrium. Genome Res. 15:1519–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth NC, Jorde PE, Chan KS, et al. 2006. Ecological and genetic impact of Atlantic cod larval drift in the Skagerrak. Proc R Soc Lond B Biol Sci. 273:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Bridgham JT, Kelly SA, Garland T. 2015. Genetic approaches in comparative and evolutionary physiology. Am J Physiol Regul Integr Comp Physiol. 309:R197–R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, Beadle GW. 1936. The relations of inversions in the X chromosome of Drosophila Melanogaster to crossing over and disjunction. Genetics 21:554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3:e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford AD, Friedman J. 2015. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution 69:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-Statistics for the analysis of population structure. Evolution 38:1358.. [DOI] [PubMed] [Google Scholar]
- Westgaard J-I, Fevolden S-E. 2007. Atlantic cod (Gadus morhua L.) in inner and outer coastal zones of northern Norway display divergent genetic signature at non-neutral loci. Fish Res. 85:306–315. [Google Scholar]
- White BJ, et al. 2007. Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae. PLoS Genet. 3:e217.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. 2001. The genic view of the process of speciation. J Evol Biol. 14:851–865. [Google Scholar]
- Yoneda M, Wright PJ. 2004. Temporal and spatial variation in reproductive investment of Atlantic cod Gadus morhua in the northern North Sea and Scottish west coast. Mar Ecol Prog Ser. 276:237–248. [Google Scholar]
- Zheng X et al.. 2004. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.